Bil1 binds to the regulatory sequence of Bud6, unmasking its ability to stimulate the activity of the formin Bni1 and promote actin nucleation.
Abstract
Formins associate with other nucleators and nucleation-promoting factors (NPFs) to stimulate collaborative actin assembly, but the mechanisms regulating these interactions have been unclear. Yeast Bud6 has an established role as an NPF for the formin Bni1, but whether it also directly regulates the formin Bnr1 has remained enigmatic. In this paper, we analyzed NPF-impaired alleles of bud6 in a bni1Δ background and found that Bud6 stimulated Bnr1 activity in vivo. Furthermore, Bud6 bound directly to Bnr1, but its NPF effects were masked by a short regulatory sequence, suggesting that additional factors may be required for activation. We isolated a novel in vivo binding partner of Bud6, Yor304c-a/Bil1, which colocalized with Bud6 and functioned in the Bnr1 pathway for actin assembly. Purified Bil1 bound to the regulatory sequence in Bud6 and triggered NPF effects on Bnr1. These observations define a new mode of formin regulation, which has important implications for understanding NPF-nucleator pairs in diverse systems.
Introduction
The assembly of filamentous actin arrays is a tightly regulated process in cells and requires the convergence of multiple signaling pathways to activate machinery that directly stimulates actin filament nucleation and elongation (Heasman and Ridley, 2008; Pollard and Cooper, 2009; Padrick and Rosen, 2010). Several actin nucleators and their cofactors or nucleation-promoting factors (NPFs) have been identified (Pollard, 2007; Chesarone and Goode, 2009; Dominguez, 2009; Firat-Karalar and Welch, 2011). These nucleators and NPFs often work together in pairs to overcome cellular barriers to actin assembly (Blanchoin and Michelot, 2012; Breitsprecher et al., 2012). Perhaps the best understood nucleator-NPF pair is the Arp2/3 complex and its actin monomer-binding cofactor Wiskott–Aldrich syndrome protein, which is tightly regulated on multiple levels (Dominguez, 2010; Rottner et al., 2010). Formins have also recently been shown to work with other nucleators and NPFs (Quinlan et al., 2007; Okada et al., 2010; Graziano et al., 2011; Blanchoin and Michelot, 2012; Block et al., 2012; Breitsprecher et al., 2012), but far less is known about the regulation and molecular basis of these interactions.
Formins are large signal-responsive proteins that promote actin assembly using their C-terminal halves. The FH2 domain is required and sufficient for nucleation (Pruyne et al., 2002; Sagot et al., 2002b) and forms a flexibly tethered homodimer that binds with high affinity to the barbed ends of filaments (Pruyne et al., 2002; Zigmond et al., 2003; Moseley et al., 2004; Xu et al., 2004; Otomo et al., 2005). Nucleation by the FH2 domain can be strongly enhanced by the adjacent actin monomer-binding C-terminal tail regions, which often contain diaphanous autoregulatory domain (DAD) and/or WH2 domains (Gould et al., 2011; Vizcarra et al., 2011; Heimsath and Higgs, 2012). After nucleation, the dimeric FH2 domain processively tracks the growing barbed end of the filament, protecting it from capping proteins (Zigmond et al., 2003; Kovar and Pollard, 2004; Moseley et al., 2004; Breitsprecher et al., 2012). On the N-terminal side of the FH2 domain is the FH1 domain, which delivers profilin-bound actin monomers to the growing barbed end to accelerate elongation (Romero et al., 2004; Kovar et al., 2006; Vavylonis et al., 2006; Courtemanche and Pollard, 2012).
To date, formins have been shown to physically associate with three other nucleators or NPFs: Spire and adenomatous polyposis coli (APC) in metazoans and Bud6 in fungi. APC and Dia (Diaphanous) colocalize and interact in developing Drosophila melanogaster embryos, in which they are both required for actin-based ingression of pseudocleavage furrows (Webb et al., 2009). Corroborating this genetic relationship, vertebrate APC and mDia1 directly interact in vitro and together stimulate actin assembly by a “rocket launcher” mechanism (Wen et al., 2004; Okada et al., 2010; Breitsprecher et al., 2012). Spire colocalizes and interacts with the formin Fmn2/Capu in both Drosophila and mouse oocytes and is required for Fmn2/Capu-dependent assembly of actin meshworks (Pfender et al., 2011; Schuh, 2011). However, the mechanism underlying their collaboration is still poorly understood, as purified Spire inhibits rather than enhances the activity of Fmn2/Capu in vitro (Quinlan et al., 2007; Vizcarra et al., 2011). In yeast, Bud6 and the formin Bni1 colocalize and interact at polarity sites, where they collaborate to promote actin cable assembly (Amberg et al., 1997; Segal et al., 2000), and purified Bud6 and Bni1 interact in vitro to stimulate actin nucleation (Moseley et al., 2004; Moseley and Goode, 2005; Graziano et al., 2011). Thus, although these formin-binding nucleators or NPFs exhibit no obvious sequence homology, they share key properties: each binds to the tail region of a formin, each binds with high affinity to multiple actin monomers, and each promotes actin nucleation without significantly altering the rate of elongation (Bosch et al., 2007; Quinlan et al., 2007; Pechlivanis et al., 2009; Okada et al., 2010; Graziano et al., 2011; Sitar et al., 2011; Vizcarra et al., 2011; Zeth et al., 2011; Breitsprecher et al., 2012; Tu et al., 2012).
In addition to Bni1, budding yeast contains a second formin, Bnr1, which localizes to the bud neck (Kamei et al., 1998). Genetic evidence of bud6Δ defects in a bni1Δ background has suggested that Bud6 either directly or indirectly promotes Bnr1-mediated actin cable assembly, just as it does for Bni1, and indeed, Bud6 localizes to the bud neck in addition to sites of polarized growth (Tong et al., 2001; Delgehyr et al., 2008). However, the same purified fragment of Bud6 (residues 489–788) that stimulated Bni1 activity instead partially inhibited Bnr1 activity (Moseley and Goode, 2005). Thus, the interactions of Bud6–Bnr1, like Spire-Fmn2/Capu, have remained enigmatic. Together, these observations point to a deficit in our understanding of how formin collaborations are regulated and suggest that additional factors (absent in these in vitro assays) may be required for nucleation by such pairs.
Here, we addressed this issue by investigating the genetic and biochemical relationship between Bud6 and Bnr1. We uncovered a crucial difference in how Bud6 interacts with Bnr1 versus Bni1 and identified a regulatory sequence in Bud6 that specifically masks its NPF effects on Bnr1. Using proteomic approaches, we isolated a novel in vivo binding partner of Bud6, Bil1, encoded by a previously uncharacterized gene. Bil1 not only colocalized and cofunctioned with Bud6 to promote Bnr1-mediated actin cable assembly in vivo but bound directly to the regulatory region of Bud6 and unmasked Bud6 NPF effects on Bnr1 in vitro. Together, our results resolve the enigma of Bud6–Bnr1 activity and define a new ligand-activated mechanism for controlling collaborative actin assembly.
Results
Bud6 functions in vivo as an NPF for Bnr1
Bni1 and Bnr1 are the only two formins in Saccharomyces cerevisiae. Single deletions of either BNI1 or BNR1 cause minimal defects in cell growth, but loss of both genes is lethal (Vallen et al., 2000; Ozaki-Kuroda et al., 2001). Bni1 and Bnr1 localize to the bud tip and bud neck, respectively, and from these positions promote the assembly of two dynamic sets of actin cables that serve as tracks for myosin-dependent transport of secretory vesicles and other cargoes to the bud tip (Sagot et al., 2002a; Pruyne et al., 2004; Buttery et al., 2007).
To ask whether Bud6 functions in Bnr1-mediated cable assembly, we first generated an isogenic set of mutant strains (bni1Δ, bnr1Δ, bud6Δ, bud6Δbni1Δ, and bud6Δbnr1Δ) that were compared for cell growth and actin organization (Fig. 1). This analysis showed that bni1Δbud6Δ double mutants are more severely impaired for growth at 37°C compared with bni1Δ or bud6Δ single mutants (Fig. 1 A) in agreement with previous studies (Tong et al., 2001; Delgehyr et al., 2008). In addition, bni1Δbud6Δ double mutants showed increased numbers of cells with diminished cable staining, highly disorganized cables, and/or depolarized actin patches at 25°C (Fig. 1 C, quantification in D), confirming the results of Delgehyr et al. (2008) showing that Bud6 is required for proper Bnr1-dependent actin cable assembly and organization.
Figure 1.
Bud6 NPF activity is required in vivo for proper actin cable assembly by Bnr1. (A and B) 10-fold serial dilutions of yeast strains grown at 25 and 37°C on YEPD plates. (C) F-actin staining of the indicated strains. Cells were grown to log phase at 25°C in YEPD, fixed, and stained with Alexa Fluor 488–phalloidin. The image containing the bni1Δbud6-35 strain is a composite of three individual images. Bar, 5 µm. (D and E) Quantification of actin cable phenotypes for strains in C. For each strain, >200 budded cells were scored (approximately equal numbers of cells pooled from two independent experiments) and placed into categories: (a) Numerous and robust cables in mother; cables sometimes visible in bud. (b) Fewer or thinner cables in mother; cables visible only occasionally in the bud. (c) Highly depolarized patches and/or highly disorganized cables in the mother (e.g., cable meshworks or cables running perpendicular rather than parallel to the mother–bud axis).
Next, we used specific NPF-impaired alleles of bud6 to test whether the requirement for Bud6 in stimulating Bnr1 in vivo stems from its NPF activities. Mutants separately impaired in binding to the formin tail (bud6-35) and G-actin (bud6-8) were integrated at the BUD6 locus in a bni1Δ background (Graziano et al., 2011). The resulting double mutants, bni1Δbud6-35 and bni1Δbud6-8, were compared with single mutants for cell growth and actin organization. bud6-35 and bud6-8 each caused strong defects in growth and actin organization in the bni1Δ background. Double mutants failed to grow at 37°C (Fig. 1 B), and >90% of double mutant cells at 25°C lacked visible cables (Fig. 1, C and E, quantification). These results demonstrate that Bud6’s NPF activities are required in vivo for Bnr1-mediated actin cable assembly. In addition, we note that the bud6-35 and bud6-8 alleles showed slightly stronger defects in cell growth and actin organization than bud6Δ, which we previously determined is caused by recessive gain-of-function effects (Graziano et al., 2011).
The C terminus of Bud6 binds to Bni1 and Bnr1 but only stimulates Bni1 in vitro
To test whether Bud6 directly binds Bnr1, we used a construct, Bud6(L) (residues 489–788), which is the longest soluble fragment of Bud6 that we have been able to purify. Coomassie-stained gels of purified Bud6(L) and other key polypeptides used in this study are shown in Fig. S1. Bud6(L) was previously shown to bind C-Bni1 (FH1-FH2-C) and G-actin and to directly enhance C-Bni1 activity in a manner that depends on both interactions (Moseley et al., 2004; Moseley and Goode, 2005; Graziano et al., 2011). Recently, the crystal structure of a major portion of Bud6(L) was solved (residues 552–688), revealing a barrel-shaped antiparallel dimeric “core” domain that binds to the Bni1 tail region (Protein Data Bank accession nos. 3OKQ and 3ONX; Fig. 2 A, yellow bar; Tu et al., 2012). Furthermore, the flanking C-terminal sequence in Bud6 (residues 699–788) was shown to bind G-actin (Fig. 2 A, blue bar), whereas no function has been assigned to the remaining N-terminal sequences in Bud6(L) (residues 489–549; Fig. 2 A, red bar).
Figure 2.
Biochemical activities of purified Bud6(L) on yeast formins. (A) Domain layout of Bud6. Bud6(L) (residues 489–788) and Bud6(S) (residues 550–788) each contain the dimeric “core” domain (yellow; residues 550–688) that binds Bni1 and the “flank” domain that binds G-actin (blue; residues 689–788). Bud6(L) additionally contains a short N-terminal sequence of unknown function (red). (B and C) Quantitative binding assays using supernatant depletion analysis. The indicated concentrations of GST-Bud6(L) immobilized on glutathione beads were incubated with 1.0 µM C-Bni1 (B) or C-Bnr1 (C). For each data point, an equivalent reaction was performed using GST alone immobilized on an equal volume of glutathione beads to correct for nonspecific binding. Coomassie-stained gels below each plot show representative reactions for each of the datasets collected. Each band represents formin remaining in the supernatant after incubation with immobilized protein and glutathione agarose precipitation. The data in each panel were pooled from two independent experiments. In the top gel of C, intervening lanes were removed for presentation purposes (white line), but all bands are from the same gel. The lines in B and C are fits to the equation B = a × c/(c + K), in which B is the fraction of C-Bni1 (B) or C-Bnr1 (C) bound, a is the amplitude of the binding interaction, c is the molar concentration of GST-Bud6(L), and K is the binding affinity (i.e., Kd). (D and E) 2 µM monomeric actin was polymerized in the presence of 10 nM C-Bni1 (D) or 2 nM Bnr1 (E), and the indicated concentrations of Bud6(L) in the absence (left) or presence (right) of 5 µM profilin. Data shown in each panel are from one representative example of four independent experiments. (F and G) Concentration-dependent effects of Bud6(L) on C-Bni1 or C-Bnr1 in the absence (F) or presence (G) of 5 µM profilin. The data in each panel were pooled from four independent experiments. The green lines in F and G are fits to the equation B = a × c/(c + K), in which B is the fold change in formin activity, c is the molar concentration of Bud6(L), a is the fold change in formin activity at saturating concentrations of Bud6(L), and K is the concentration of Bud6(L) at which B = 0.5 × a (i.e., the Kapp). The orange lines in F and G are drawn to guide the eyes. See Materials and methods for calculation of rates. AU, arbitrary unit.
To test binding of Bud6 to Bnr1, we used supernatant depletion assays with variable concentrations of GST-Bud6(L) immobilized on glutathione agarose and a fixed concentration of soluble C-Bnr1. As a positive control for binding, we tested GST-Bud6(L) interactions with C-Bni1, and as a negative control, we tested binding to GST alone on beads. Our analysis revealed that GST-Bud6(L) binds specifically to C-Bni1 and C-Bnr1 with similar affinities (Kd ∼4–8 µM; Fig. 2, B and C). In a previous study, we did not detect Bud6–Bnr1 interactions when monitoring bound material after multiple bead washes (Moseley and Goode, 2005), but the approach used here is a more reliable and quantitative one for detecting protein–protein interactions (Pollard, 2010). From these data, we conclude that Bud6 directly interacts with both Bnr1 and Bni1, which agrees with previous two-hybrid studies (Evangelista et al., 1997; Kikyo et al., 1999) and with our aforementioned genetic observations showing that Bud6 NPF activity contributes to Bnr1 function in vivo.
We next asked how Bud6(L) affects C-Bnr1 actin assembly activity, both in the presence and absence of profilin. As a positive control, we measured Bud6(L) effects on C-Bni1. As expected, Bud6(L) potently enhanced C-Bni1 activity in the presence or absence of profilin (Fig. 2 D), with a maximal sixfold increase in the rate of actin assembly (Fig. 2, F and G). In contrast, Bud6(L) failed to enhance C-Bnr1 activity in the presence of profilin (Fig. 2 E, quantification in F and G). Furthermore, in the absence of profilin, Bud6(L) inhibited C-Bnr1 in a concentration-dependent manner. These effects are likely caused by Bud6(L) binding C-Bnr1 rather than sequestering actin monomers, given that inhibition was observed at concentrations of actin monomers >10-fold higher than the concentration of Bud6(L).
Regulatory sequences in Bud6 “mask” its NPF effects on Bnr1
The discrepancy between our genetic data showing that Bud6 is an NPF for Bnr1 in vivo and our biochemical data showing that Bud6(L) inhibits rather than activates C-Bnr1 was puzzling and led us to consider whether Bud6 NPF effects on Bnr1 might be masked. The aforementioned structural study on Bud6 recently defined the boundaries of a dimeric “core” domain (550–688) and demonstrated that a truncated Bud6 construct, referred to here as Bud6(S) (residues 550–788), is sufficient to activate Bni1 in vitro (Fig. 2 A; Tu et al., 2012). Therefore, we tested the effects of Bud6(S) on Bni1- and Bnr1-mediated actin assembly. Bud6(S) potently enhanced C-Bni1 activity in the absence of profilin (Fig. 3 A, left), similar to Bud6(L) (Fig. 2). Furthermore, the presence of profilin did not significantly alter the magnitude of the stimulatory effects at higher concentrations of Bud6(S) (Fig. 3 A, right), though it did decrease the potency of Bud6(S), presumably through competition for actin monomers (Fig. 3 B, Kapp ∼11 nM without profilin and Kapp ∼90 nM with profilin). Thus, Bud6(L) and Bud6(S) both increased by about sixfold the maximal rate of C-Bni1–mediated actin assembly in the presence or absence of profilin (compare Fig. 2 [F and G] with Fig. 3 B).
Figure 3.
Bud6(S) stimulates C-Bni1 and C-Bnr1 in actin assembly assays. (A and C) 2 µM monomeric actin was polymerized in the presence of 10 nM C-Bni1 (A) or 2 nM C-Bnr1 (C), and the indicated concentrations of Bud6(S) in the absence (left) or presence (right) of 5 µM profilin. Data shown in each panel are from one representative example of four independent experiments. (B and D) Concentration-dependent effects of Bud6(S) on C-Bni1 (B) or C-Bnr1 (D) actin assembly activity, performed as in A and C. Filled triangles indicate reactions performed in the presence of 5 µM yeast profilin; hollow triangles indicate identical reactions lacking yeast profilin. Data shown are from one representative example of four independent experiments. See Materials and methods for calculation of rates. Both lines in B and the solid line in D are fits to the equation B = a × c/(c + K), in which B is the percent increase in formin activity, c is the molar concentration of Bud6(S), a is the percent increase in formin activity at saturating concentrations of Bud6(S), and K is the concentration of Bud6(S) at which B = 0.5 × a (i.e., the Kapp). The dashed line in D is drawn to guide the eyes. (E and F) Quantitative GST pull-down assays performed as in Fig. 2 (B and C) using indicated concentrations of immobilized GST-Bud6(S) and 1.0 µM of soluble C-Bni1 (E) or C-Bnr1 (F). The data in each panel were pooled from two independent experiments. The lines in E and F are fits to the equation B = a × c/(c + K), in which B is the fraction of C-Bni1 (E) or C-Bnr1 (F) bound, a is the amplitude of the binding interaction, c is the molar concentration of GST-Bud6(S), and K is the binding affinity (i.e., Kd). (G) Quantitative binding assays using 150 nM pyrene-labeled actin (100% labeling efficiency) and indicated amounts of either Bud6(L) or Bud6(S). The lines in G are fits to the equation B = ([c + a + k] − [(c + a + k)2 − 4 × c × a]0.5)/(2 × a), in which B is the fraction of G-actin bound, c is the molar concentration of Bud6, a is the molar concentration of G-actin (170 nM in all cases), and k is the binding affinity (i.e., Kd). AU, arbitrary unit.
Next, we tested the effects of Bud6(S) on C-Bnr1. In the presence of profilin, Bud6(S) potently stimulated C-Bnr1 (Fig. 3 C, right) with a Kapp of ∼30 nM (Fig. 3 D), whereas Bud6(L) failed to stimulate C-Bnr1 (Fig. 2 E–G). Interestingly, the ability of Bud6(S) to stimulate C-Bnr1 depended on the presence of profilin (Fig. 3 C, left), which differs from the profilin-independent stimulatory effects of Bud6(S) on C-Bni1 (see Discussion).
To better understand how Bud6(S) stimulates C-Bnr1, we compared binding of Bud6(S) to C-Bni1 (Kd ∼5.2 µM) and C-Bnr1 (Kd ∼2.6 µM; Fig. 3, E and F). Although profilin was required for the aforementioned stimulatory effects of Bud6(S) on C-Bnr1, the strength of the association between C-Bnr1 with Bud6(S) did not change appreciably when profilin, G-actin, or profilin and G-actin were present in the reactions (Fig. S2). Together with the data in Fig. 2 (B and C), these results show that although Bud6(S) and Bud6(L) bind to both formins with similar affinities, the presence of the additional N-terminal sequence in Bud6(L) (residues 489–549; from here on referred to as the regulatory region) “masks” the NPF effects on Bnr1 and does so without altering Bud6–Bnr1 affinity. Furthermore, we found that Bud6(S) and Bud6(L) each bind G-actin with affinities in the low nanomolar range (Fig. 3 G), suggesting that differences in actin monomer binding affinity do not explain differences in their NPF effects on C-Bnr1.
Bud6 stimulates actin nucleation rather than elongation by Bnr1
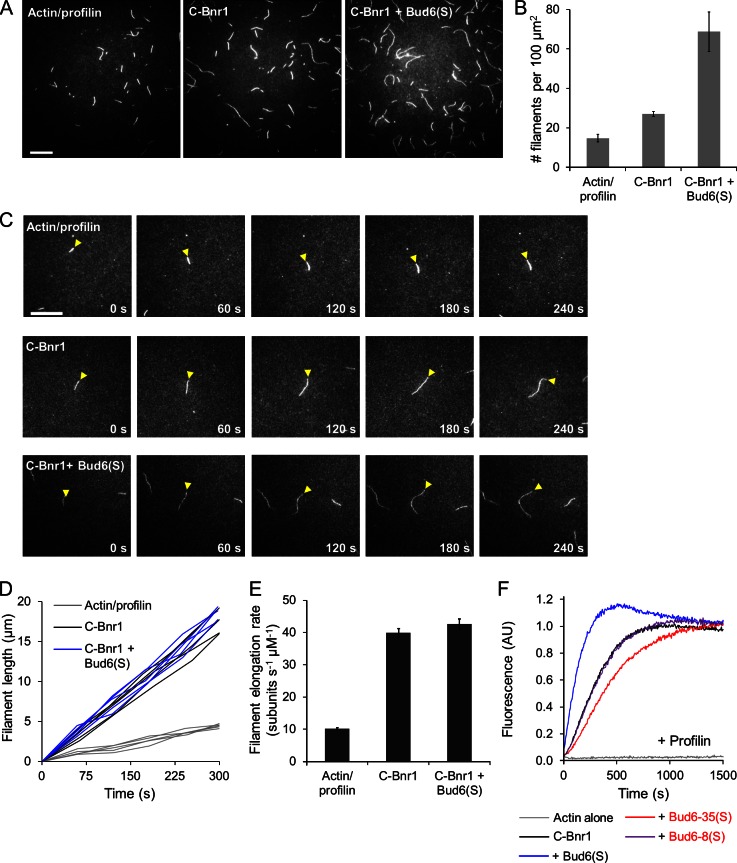
We previously used total internal reflection fluorescence (TIRF) microscopy (TIRF-M) to show that Bud6 enhances the actin nucleation rather than elongation activity of Bni1 (Graziano et al., 2011). The aforementioned observations prompted us to use TIRF-M to independently assess the effects of Bud6 on Bnr1-dependent actin filament nucleation versus elongation. First, we measured nucleation rates by quantifying filament density in reactions that had proceeded for a short time. Control reactions containing actin monomers and profilin produced a mean filament density of 14.8 ± 2.0 per 100 µm2 (Fig. 4 A, quantification in B). Addition of 0.2 nM C-Bnr1 increased filament density to 27.0 ± 1.2 per 100 µm2, whereas the combination of C-Bnr1 and Bud6(S) produced a filament density of 68.8 ± 10.1 per 100 µm2. Thus, Bud6(S) dramatically increases the rate of Bnr1-dependent filament nucleation.
Figure 4.
Bud6(S) functions as an NPF for Bnr1 but does not affect the elongation rate of Bnr1-capped filaments. (A) Actin filament densities observed by TIRF-M (450 s after the start of the reaction) for reactions containing 0.5 µM actin + 5.0 µM yeast profilin with no further proteins (left), 0.2 nM C-Bnr1 (middle), or 0.2 nM C-Bnr1 + 100 nM Bud6(S) (right). The data shown are one representative example of three independent experiments. (B) Quantification of filament densities for reactions in A. Each bar represents a mean of nine fields of view from three independent experiments; error bars show SEM. (C) Time-lapse TIRF-M performed as in A. Yellow arrowheads indicate observed filaments. The data shown are one representative example of three independent experiments. (D) Plot of filament length versus time for individual filaments observed in C. Filaments were measured every 60 s and were normalized to a length of 0 µm at 0 s. The data shown are one representative example of three independent experiments. (E) Mean elongation rates for filaments observed as in C. Rates were determined by measuring the mean slopes of filament growth as in D. Data shown are from three independent experiments, in which for each condition in each experiment, >15 filaments were measured; error bars show SEM. (F) Bulk actin assembly assays performed as in Fig. 3 C using 2 µM actin, 5 µM yeast profilin, 2 nM C-Bnr1, and where indicated, 200 nM Bud6(S), Bud6-35(S), or Bud6-8(S). The data shown are one representative example of two independent experiments. AU, arbitrary unit. Bars, 20 µm.
We next measured filament elongation rates by quantifying change in filament length over time. In control reactions containing actin monomers and profilin, filaments elongated at a mean rate of 10.2 ± 0.3 subunits/s (Fig. 4 C, quantification in E). Addition of C-Bnr1 increased the elongation rate to 39.9 ± 1.2 subunits/s, and the combination of C-Bnr1 and Bud6(S) yielded filaments that elongated at a mean rate of 42.7 ± 1.5 subunits/s. Collectively, these results indicate that Bud6(S) strongly enhances Bnr1’s nucleation but not elongation activity, demonstrating the Bud6 is by definition an NPF for both formins.
Other NPFs, including Wiskott–Aldrich syndrome protein, APC, and Bud6 itself (when acting on Bni1), function by recruiting actin monomers to the nucleator. We therefore tested the actin assembly enhancing activities of mutant Bud6(S) polypeptides with disrupted Bud6–actin and Bud6–formin interactions, Bud6-8(S) and Bud6-35(S), respectively. Bud6-35(S) and Bud6-8(S) each failed to enhance C-Bnr1 activity (Fig. 4 F). Note that the concentration of Bud6-35(S) used in these reactions (200 nM) caused a slight inhibition of actin assembly because of actin monomer sequestration. These effects were not observed for Bud6-8(S), which does not bind actin monomers. These data from bulk assays support our TIRF-M data and demonstrate that Bud6 stimulates nucleation rather than elongation by Bni1 and Bnr1.
The location of the Bud6 binding site (BBS) is different in Bnr1 and Bni1
Distinct effects of Bud6(L) on Bni1 versus Bnr1 (Fig. 2, D–G) prompted us to ask whether Bud6 binds to the same or distinct regions of each formin. Our previous work showed that the tail region of Bni1 contains the BBS, which is marked by tandem serines and a cluster of positively charged residues located on the C-terminal side of the DAD domain (Moseley and Goode, 2005). We asked whether the tail region of Bnr1 similarly mediates Bud6 binding.
In supernatant depletion assays, C-Bni1Δtail failed to bind either GST-Bud6(L) or GST-Bud6(S) (Fig. 5 A, left), consistent with the position of the BBS in the Bni1 tail region. In contrast, C-Bnr1Δtail bound to both GST-Bud6(L) and GST-Bud6(S) and with only slightly reduced affinity to C-Bnr1 (Fig. 5 A, right; compare with Fig. 2 C and Fig. 3 F). Thus, the tail region of Bnr1 is not essential for Bud6 binding. Consistent with this view, Bud6(L) inhibited C-Bnr1Δtail activity (Fig. 5 B, left). Furthermore, we found that Bnr1 FH2 alone (lacking FH1 or tail but including the long αT helix leading up to the DAD; Xu et al., 2004) bound to GST-Bud6(S) with a similar affinity to C-Bnr1Δtail (Fig. 5 C). Together, these results indicate that the primary BBS in Bnr1 is located outside of the tail region (see Discussion).
Figure 5.
The C-terminal region of Bnr1 is not required for Bud6 binding. (A) Quantitative GST pull-down assays using immobilized GST-Bud6(L) or GST-Bud6(S) and soluble C-Bni1Δtail (left) or C-Bnr1Δtail (right). (B) 2 µM monomeric actin was assembled alone, with 2 nM C-Bnr1Δtail, or with 2 nM C-Bnr1Δtail + 200 nM Bud6(L). Reactions were performed in the absence (left) or presence (right) of 5 µM yeast profilin. (C) Quantitative GST pull-down assays as in A using immobilized GST-Bud6(S) and soluble Bnr1 FH2. For each panel of this figure, the data shown are single representative examples from two independent experiments. The lines in A and C are fits to the equation B = a × c/(c + K), in which B is the fraction of C-Bnr1Δtail (A) or Bnr1 FH2 (C) bound, a is the amplitude of the binding interaction, c is the molar concentration of GST-Bud6, and K is the binding affinity (i.e., Kd). AU, arbitrary unit.
Interestingly, we also observed that although Bud6(S) binds to C-Bnr1Δtail, it fails to enhance C-Bnr1Δtail activity (Fig. 5 B). Thus, formin binding by Bud6 is not sufficient to confer its NPF effects, and the tail region also plays a pivotal role in this process.
Isolation of a novel Bud6-binding protein that activates NPF effects on Bnr1
Given the aforementioned observations, we hypothesized that additional binding partners of Bud6 may be required to unmask its NPF effects on Bnr1 in vivo. To address this, we integrated a C-terminal GFP tag on BUD6, immunoprecipitated Bud6-GFP from cell extracts, and identified Bud6-associated proteins by tandem mass spectrometry analysis (see Materials and methods). One protein, encoded by the gene YOR304C-A, was selected for further analysis because it had previously been identified in proteomic studies as localizing to polarity sites and interacting with Bud6 (Huh et al., 2003; Krogan et al., 2006; Yu et al., 2008; Narayanaswamy et al., 2009). YOR304C-A encodes a protein that is only 76 amino acids long, and we refer to it as Bil1 (Bud6-interacting ligand).
Bil1 was expressed and purified in Escherichia coli as a GST fusion protein and tested for its ability to bind Bud6(S) and Bud6(L). GST-Bil1 bound to Bud6(L) but not Bud6(S) (Fig. 6 A), indicating that the regulatory region is essential for Bil1 binding. Next, we tested Bil1 (GST tag removed) in the presence of Bud6(L) or Bud6(S) for effects on Bnr1-mediated actin assembly. Bil1 showed no effects on actin assembly in the absence of other factors (Fig. S3 A) and no effects on Bnr1-mediated actin assembly in the absence of Bud6 (Fig. S3 B). However, in the presence of Bud6(L), Bil1 caused a concentration-dependent increase in the rate of C-Bnr1–mediated actin assembly (Fig. 6 B, quantification in C). In contrast, Bil1 showed no effects on C-Bnr1 in the presence of Bud6(S) (Fig. S3 C and quantification in Fig. 6 C). TIRF-M analysis confirmed that Bil1 increased the number of filaments nucleated by C-Bnr1 in the presence of Bud6(L) (Fig. 6 D, quantification in E), while showing minimal effects on rate of filament elongation (Fig. 6 F).
Figure 6.
Bil1 unmasks Bud6(L) stimulatory effects on Bnr1. (A) Quantitative GST pull-down assays using indicated concentrations of purified immobilized GST-Bil1 and soluble Bud6(L) or soluble Bud6(S). Data shown were pooled from three independent experiments. The line in A is a fit to the equation B = a × c/(c + K), in which B is the fraction of Bud6 bound, a is the amplitude of the binding interaction, c is the molar concentration of GST-Bil1, and K is the binding affinity (i.e., Kd). (B) Bulk actin assembly assays. 2 µM monomeric actin was assembled in the presence of 5 µM yeast profilin and 2 nM C-Bnr1. Where indicated, 100 nM Bud6(L) and/or 500 nM Bil1 was included. The data shown are one representative example of four independent experiments. AU, arbitrary unit. (C) Concentration-dependent effects of Bil1 on Bnr1-mediated actin assembly in the presence of either Bud6(L) or Bud6(S). Data shown were pooled from four independent experiments. The line in C is a fit to the equation B = a × c/(c + K), in which B is the percent increase in actin assembly, c is the molar concentration of Bil1, a is the percent increase in actin assembly at saturating concentrations of Bil1, and K is the concentration of Bil1 at which B = 0.5 × a (i.e., the Kapp). (D) Actin filament densities observed using TIRF-M for reactions containing 0.5 µM monomeric actin and 0.2 nM C-Bnr1. Where indicated, 100 nM Bud6(L) and/or 500 nM Bil1 was included. The data shown are one representative example of two independent experiments. Bar, 20 µm. (E) Quantification of filament densities observed by TIRF-M under conditions described in D. Each bar represents a mean of nine fields of view from three independent experiments; error bars show SEM. (F) Mean elongation rates of filaments observed in D, calculated as in Fig. 4 F. Data shown are from two independent experiments, where for each condition in each experiment >15 filaments were measured; error bars show SEM. (G and H) Working model for Bud6 regulation of actin assembly by Bni1 (G) and Bnr1 (H). Bud6 dimers (yellow), which are bound to actin monomers (gray), interact with the Bud6 binding site (BBS) on the formin (green). Because of differences in the position of the BBS in Bni1 versus Bnr1, the interaction of Bud6 leads to distinct functional effects. In the case of Bni1, Bud6 binding results in enhanced actin nucleation, whereas in the case of Bnr1, Bud6 binding obstructs nucleation as a result of the proximity of the regulatory (R) region of Bud6 to the FH2 domain. Binding of Bil1 (blue) to the R region of Bud6 relieves the obstruction, triggering enhanced nucleation. Profilin (orange) is also required for the Bud6 stimulatory effects on Bnr1, although the mechanistic basis for this requirement is still unclear.
Using all of the aforementioned observations, we constructed a working model for the differential regulation of Bni1 and Bnr1 by Bud6 (Fig. 6, G and H). A key feature of this model is that the location of the BBS (Fig. 6, G and H, red boxes) is different in Bni1 and Bnr1, such that it is farther from the FH2 domain in Bni1. The BBS interacts with a recently defined formin binding site (Fig. 6, G and H, dotted red circles) on the dimeric Bud6core domain (Tu et al., 2012). The R (regulatory) region of Bud6(L) is much closer to the FH2 domain in Bnr1 versus Bni1. We propose that the proximity of the R region to the FH2 domain obstructs nucleation, explaining why Bud6(L) stimulates Bni1 in the absence of additional factors (Fig. 6 G) but can stimulate Bnr1 only after addition of Bil1, which binds to and neutralizes the R region (Fig. 6 H).
Bil1 colocalizes with Bud6 and functions in the Bnr1 pathway for actin assembly
To investigate the in vivo functions of Bil1, we first generated strains in which we separately integrated C-terminal GFP tag on BIL1 and BUD6 and compared localization patterns at different stages of bud growth (Fig. 7 A and Fig. S4). Bud6-GFP primarily localized to the bud tip of small-budded cells and then progressively shifted to the bud neck as the daughter cell increased in size (Fig. S4, top), consistent with previous studies (Jin and Amberg, 2000; Segal et al., 2000). Bil1-GFP showed a similar pattern of localization to Bud6-GFP at all stages of bud development (Fig. S4, bottom), although Bil1-GFP fluorescence intensity was noticeably lower than Bud6-GFP, which agrees with previous quantitation of Bud6-GFP and Bil1-GFP levels in cells (Huh et al., 2003). In addition, we examined Bil1-GFP localization in bud6Δ cells, and Bud6-GFP localization in bil1Δ cells. This revealed that Bil1-GFP shifts to the cytosol in bud6Δ cells (Fig. 7 A, bottom), whereas Bud6-GFP localization is unaffected by bil1Δ (Fig. 7 A, top). Thus, Bil1 colocalizes with Bud6 and depends on Bud6 for localization, whereas Bud6 localization is not dependent on Bil1. We also generated a strain with integrated Bil1-GFP and Bud6-RFP to compare localization in the same cells (Fig. 7 B). Bil1-GFP and Bud6-RFP again were enriched at the bud neck and bud cortex and displayed colocalization at the neck (Fig. 7 B, arrows). Each protein was also visible as less bright spots in the mother and bud. However, we were unable to determine the degree to which these secondary spots colocalized because they were highly dynamic as previously reported for Bud6-RFP (Buttery et al., 2007) and many of them moved faster than the two-color acquisition rate.
Figure 7.
Analysis of Bil1 localization and function in vivo. (A) Comparisons of Bud6-GFP localization in wild-type and bil1Δ cells (top), and Bil1-GFP localization in wild-type and bud6Δ cells (bottom). Bar, 10 µm. (B) Representative images of Bil1-GFP and Bud6-RFP localization in the same cells. Strong overlapping signals were observed at the bud neck (cyan arrowheads). Bar, 10 µm. (C) 10-fold serial dilutions of the indicated strains grown on YEPD plates at either 25 or 37°C. (D) Representative images of F-actin staining for the indicated strains. Bar, 5 µm. (E) Quantification of actin cable phenotypes (same categories as in Fig. 1, D and E). Note that data for some of the strains that appear in Fig. 1 D again appear here to facilitate comparison of phenotypes among relevant strains. The data for all of these strains were collected in the same sets of experiments. For each strain, >200 cells were scored (approximately equal numbers of cells pooled from two independent experiments). (F) Tracking of GFP-Sec4 particles in cells. Moving particles were followed over a 60-s observation window and placed into one of two categories: (1) normal anterograde movements (particles moving directionally toward the bud) or (2) random/retrograde movements. For each different yeast strain, two to three individual particles were tracked in 50 cells (n > 140 particles total; from approximately equal numbers of cells imaged in two independent experiments).
Finally, to investigate the importance of Bil1 in the Bni1- and Bnr1-dependent pathways for actin cable assembly, we generated a bil1Δ mutation and compared its effects on cell growth in wild-type, bni1Δ, and bnr1Δ backgrounds (Fig. 7 C). bil1Δ caused no obvious defects in growth in the wild-type or bnr1Δ backgrounds but strongly impaired cell growth at 37°C in the bni1Δ background, similar to bud6Δ (Fig. 7 C). Furthermore, bil1Δ exacerbated actin organization defects in the bni1Δ but not the bnr1Δ background, increasing the numbers of cells with diminished cable staining, highly disorganized cables, and/or depolarized actin patches (Fig. 7, D and E, quantification). We also generated a triple mutant bni1Δbud6Δbil1Δ strain, which displayed defects in cell growth and actin organization similar to the bni1Δbud6Δ strain (Fig. S4). These results suggest that BIL1 and BUD6 function in the same pathway regulating BNR1.
To additionally test cable function in mutant strains, we tracked the movements of secretory vesicles (marked with GFP-Sec4), which are trafficked by the type V myosin Myo2 on actin cables. For each strain, we scored the percentage of anterograde movements (toward the bud tip) versus random and/or retrograde movements (Fig. 7 F and Videos 1–6). Consistent with our actin staining results, bni1Δbud6Δ and bni1Δbil1Δ mutants exhibited a higher percentage of random and/or retrograde vesicle movements (84 and 87%, respectively) compared with wild-type cells (31%) or compared with single mutant bni1Δ, bud6Δ, and bil1Δ cells (64, 61, and 65%, respectively). Although bil1Δ cells displayed no clear defects in cable organization by actin staining, they did show defects in secretory vesicle movement, suggesting that bil1Δ may alter the architecture of actin cables in a manner that is not readily detected by light microscopy. Collectively with our biochemical data, these in vivo results provide strong support for the model that Bil1 promotes Bnr1-dependent actin cable assembly.
Discussion
In this work, we set out to answer the long-standing question of whether Bud6 directly regulates Bnr1 activity. Early genetic studies and two-hybrid experiments pointed to a role for Bud6 in promoting Bnr1-dependent actin cable assembly either directly or indirectly (Amberg et al., 1997; Kikyo et al., 1999; Delgehyr et al., 2008). However, a subsequent biochemical study using purified Bud6(L) (residues 489–788) showed that Bud6(L) stimulated Bni1- but not Bnr1-dependent actin assembly (Moseley and Goode, 2005). These results left it unclear whether Bud6 directly regulates Bnr1 function. We investigated this issue by taking a combined genetic and biochemical approach. Integrated bud6 alleles that are specifically defective in formin binding (bud6-35) and G-actin binding (bud6-8) caused growth defects and further reduced levels of actin cable staining in a bni1Δ background, where Bnr1 is the only formin expressed. These data demonstrate that Bud6 directly promotes Bnr1-mediated actin cable assembly because all cables in these cells are generated by Bnr1. Furthermore, we found that purified Bud6(L) binds to both Bnr1 and Bni1 with similar affinity. However, although Bud6(L) enhanced nucleation by Bni1 in vitro, it partially inhibited Bnr1, making it unclear how Bud6 could stimulate Bnr1 in vivo. Analysis with a shorter construct, Bud6(S), resolved this issue, as we observed that Bud6(S) stimulated both Bni1 and Bnr1 activity. These observations suggested that Bud6 stimulation of Bnr1 might require additional cellular factors. Using affinity-tagged endogenously expressed Bud6, we isolated from cell extracts a novel Bud6-binding partner, Bil1, which we showed binds to a regulatory sequence in Bud6 (residues 489–549) and unmasks Bud6(L) NPF effects on Bnr1. Bil1 also colocalized with Bud6 at the bud neck, where Bnr1 resides, and depended on Bud6 for its localization, consistent with their direct interaction. Finally, genetic analysis demonstrated that BIL1 functions in the Bnr1-dependent pathway of cable assembly and, consequently, that bil1Δ causes defects in the movements of secretory vesicles.
These findings have several broad implications. First, they resolve earlier discrepancies between genetic and biochemical data on Bud6–Bnr1 interactions, demonstrating that Bud6 directly binds and stimulates Bnr1 and that Bud6-Bnr1 interactions are important for actin cable assembly and function in vivo. Second, they reveal an unanticipated “ligand-gated” mechanism for activating this NPF–formin collaboration in actin nucleation, which may serve as a paradigm for understanding the regulation of other formin collaborations. Third, they show that two different formins expressed in the same cell type can be stimulated by a single NPF through distinct regulatory mechanisms and that this may be achieved simply by shifting the location of the NPF binding site on the formin.
Mechanism of ligand-induced activation of Bud6–Bnr1
We found that Bud6(L) and Bud6(S) each bind to Bni1 and Bnr1 with similar affinities. However, Bud6(L) only stimulated Bni1 activity, whereas Bud6(S) stimulated both Bni1 and Bnr1. These observations suggested that the sequence by which Bud6(L) and Bud6(S) differ (489–549) masks the NPF effects of Bud6 on Bnr1 but not Bni1.
How is differential regulation of two formins by the same NPF achieved? An important clue is that the same point mutations (bud6-35), located in the dimeric core domain of Bud6, abolish interactions with both Bni1 (Graziano et al., 2011) and Bnr1 (Fig. 4 F). This suggests that Bud6 binds to each formin through interactions of a similar molecular nature. A second clue is that the position of the BBS is different in Bnr1 compared with Bni1. Our data show that Bud6 binds to C-Bni1 but not C-Bni1Δtail, indicating that the BBS is contained in the tail region of Bni1, consistent with previous mapping (Moseley and Goode, 2005). In contrast, Bud6 bound with similar affinities to C-Bnr1, C-Bnr1Δtail, and Bnr1 FH2, demonstrating that the BBS in Bnr1 is contained in the FH2 region. This construct consists of the donut-shaped dimeric FH2 domain plus two long αT helices that extend away from its surface and connect to the C-terminal DAD-containing tail region (Fig. 6, G and H; Xu et al., 2004). Although the exact position of the BBS in Bnr1 has yet to be determined, it is notable that the tandem serine residues found in Bni1 that are just C terminal to its DAD domain and critical for Bud6 binding are not conserved in the tail region of Bnr1 (Moseley and Goode, 2005). However, tandem serines followed by a cluster of positively charged residues instead appear along the αT helix of Bnr1, i.e., substantially closer to the main body of the FH2 domain. Binding of Bud6 to that site would put the Bud6 regulatory region (489–549) in a better position to obstruct nucleation by the FH2 domain (in the absence of Bil1), perhaps by inhibiting FH2 dimer capture of actin seeds. Thus, a shift in the position of the BBS could explain how two formins can be differentially regulated by one NPF and explain why ligand-gated activation is required for one Bud6–formin pair but not the other (Fig. 6, G and H, model). Consistent with this model, we have previously shown that fusing the BBS-containing tail region of Bni1 onto the FH1-FH2-αT of Bnr1 enables Bud6(L) to stimulate Bnr1 (Moseley and Goode, 2005). This chimera should have two BBS elements, the distal one being far enough from the FH2 domain to allow Bud6(L) to provide the observed NPF effects on the chimera.
One other difference we noted between the NPF effects of Bud6 on Bni1 versus Bnr1 was that profilin was required specifically for Bud6 NPF effects on Bnr1 and not Bni1. We do not yet understand the basis for this mechanistic difference between Bni1 and Bnr1. However, previous studies have shown that the FH1 domains in some formins contribute to actin nucleation specifically in the presence of profilin (Li and Higgs, 2003; Paul and Pollard, 2008; Gould et al., 2011). Thus, it is possible that FH1–profilin–actin complexes could deliver actin monomers to Bud6 or work in parallel with Bud6 to recruit monomers for Bnr1-dependent nucleation.
Bil1 in vivo functions
What in vivo function is served by Bil1 regulation of Bud6 effects on Bnr1? One possibility is that Bil1 functions as a molecular “thermostat,” which can define the fraction of Bud6 (from the cellular pool) available for productive nucleation interactions with Bnr1. In support of this view, Bil1 levels are significantly lower (approximately fourfold) than Bud6 throughout bud development (Huh et al., 2003). Thus, Bil1 may restrict levels of cable assembly by Bud6–Bnr1 (a powerful nucleation unit), scaling them to Bil1 levels, while not affecting the pool of Bud6 that is available to stimulate Bni1. Such a thermostat mechanism would permit cells to alter Bud6 levels for the purpose of controlling Bni1-dependent cable assembly without overstimulating Bnr1, and indeed, Bud6 levels fluctuate in a cell cycle–regulated manner (Moseley and Goode, 2005).
A second possibility is that Bil1 links Bud6–Bnr1 activation to upstream regulatory pathways, enabling a cable assembly response to signals controlling membrane trafficking, organelle inheritance, cell cycle stage, bud size, or response to environmental stress. Bni1 and Bnr1 nucleate actin assembly from two distinct sites and produce separate sets of cables with different elongation speeds (Yu et al., 2011). Small proteins, such as Bil1 (8.6 kD), can be translated rapidly in response to cues, and thus, Bil1 production/degradation may provide a rapid way to tune Bnr1 function (Bnr1 itself is 156.8 kD, and therefore, its up-regulation would be much slower). Thus, a mechanism involving Bil1 could allow the two formins to more readily adapt to environmental stresses, such as cell wall damage, which was recently shown to trigger recruitment of Bnr1 to the cortical wound site (Kono et al., 2012).
Relationship between Bud6 and other formin NPFs
Our results lend new insights into how collaborative nucleator pairs in other systems may be regulated. Although the three known formin-interacting nucleators/NPFs (Bud6, Spire, and APC) share little if any sequence homology, they appear to have related mechanisms and functions. Each binds to the C-terminal region of the formin and to multiple actin monomers and is thought to catalyze the formation of an actin seed, which is then captured at its barbed end by the FH2 domain. In addition, Bud6, Spire, and APC each are reported to bind microtubules and regulate microtubule-dependent processes in vivo, and Bud6 and APC also interact with the microtubule plus end–tracking protein EB1 (Su et al., 1995; Kita et al., 2006; Rosales-Nieves et al., 2006; Delgehyr et al., 2008; Ten Hoopen et al., 2012). Thus, actin- and microtubule-based roles for these NPFs and the formins they bind are rapidly being defined. However, by comparison, relatively little is known about how NPF–formin interactions are regulated in vivo. Our results suggest that these collaborative associations may remain idle until activated by specific ligands. This paradigm may also be relevant to understanding Spire–formin interactions. Genetic data indicate that Spire cooperates with the formin Capu/Fmn2 to generate cytoplasmic actin networks in both Drosophila and mouse oocytes (Pfender et al., 2011; Schuh, 2011). However, purified Spire partially inhibits rather than enhances Capu/Fmn2 activity in vitro (Quinlan et al., 2007; Vizcarra et al., 2011). This paradox resembles the one that surrounded Bud6 and Bnr1 at the onset of this study, raising the possibility that even though Spire binds to the formin, it contains regulatory sequences obstructing its NPF effects, which must be released by binding of a specific ligand and/or posttranslational modification.
Materials and methods
Plasmids and strains
Vectors previously described (Moseley et al., 2004; Moseley and Goode, 2005; Tu et al., 2012) were used for galactose-inducible expression in S. cerevisiae of N-terminal His6 fusions of C-Bni1 (residues 1,227–1,953), C-Bni1Δtail (residues 1,227–1,797), C-Bnr1 (residues 757–1,375), and C-Bnr1Δtail (residues 757–1,292) and for E. coli expression of GST-tobacco etch virus (TEV)-Bud6(L) (residues 478–788) and GST-TEV-Bud6(S) (residues 550–788). The E. coli expression vector for Bnr1 FH2 domain was a gift from M. Rosen (Howard Hughes Medical Institute, University of Texas Southwestern Medical Center at Dallas, Dallas, TX). To construct vectors for E. coli expression of GST-TEV-Bud6-35(S) and GST-TEV-Bud6-8(S), we PCR amplified sequences encoding residues 550–788 from vectors expressing GST-TEV-Bud6-35(L) and GST-TEV-Bud6-8(L) (pBG1363 and pBG1364, respectively; Graziano et al., 2011) and subcloned these inserts into the BamHI and NotI sites of pET-GST-TEV (Moseley et al., 2004) to produce pBG1365 and pBG1366. A plasmid for expressing Bil1 in E. coli (pBG1367) was generated by PCR amplifying the BIL1 ORF from the wild-type yeast strain BGY12 and subcloning this fragment into the BamHI and NotI sites of pET-GST-TEV.
All yeast strains used in this study were isogenic to BGY12 (MATα; his3-11,15; ura3-53; leu2-3,112; ade2-1; trp1-1; psi+; ssd-; GAL+) or BGY10 (MATa; his3-11,15; ura3-53; leu2-3,112; ade2-1; trp1-1; psi+; ssd-; GAL+) unless otherwise noted. Strains bni1Δ (BGY1247), bnr1Δ (BGY1248), bud6Δ (BGY1413), bud6-35 (BGY1411), and bud6-8 (BGY1412) were generated in previous studies (Chesarone et al., 2009; Graziano et al., 2011). We generated bni1Δbud6-35 (BGY1476) and bni1Δbud6-8 (BGY1477) by crossing BGY1411 and BGY1412 to BGY1247 followed by sporulation and tetrad dissection. Similarly, bni1Δbud6Δ (BGY1478) and bnr1Δbud6Δ (BGY1479) were generated by crossing BGY1413 to BGY1247 and BGY1248, respectively. The bil1Δ strain (BGY1480) was generated by homologous recombination using a HIS3MX6 cassette (Longtine et al., 1998). To generate bud6Δbil1Δ (BGY1481), bni1Δbil1Δ (BGY1482), and bnr1Δbil1Δ (BGY1483) strains, BGY1480 was crossed to BGY1413, BGY1247, and BGY1248, respectively. To generate bud6Δbil1Δbni1Δ (BGY1489) BIL1 was deleted (as described for BGY1480) in BGY1478. Bud6-GFP (BGY1484) or Bil1-GFP (BGY1485) strains were generated by recombination using C-terminal GFP::HIS3MX6 cassettes (Longtine et al., 1998) in BGY1257 (MATα, ura3Δ, leu2Δ, his3Δ, met15Δ). Bud6-GFP bil1Δ (BGY1486) and Bil1-GFP bud6Δ (BGY1487) strains were generated by replacing the BIL1 ORF in BGY1484 and the BUD6 ORF in BGY1485 with KANMX6 cassettes. The strain expressing Bil1-GFP and Bud6-RFP (BGY1488) was generated by introducing a C-terminal GFP::HIS3MX6 cassette at BIL1 via homologous recombination (Longtine et al., 1998) in strain PY5434 (a gift from D. Pellman, Dana-Farber Cancer Institute, Boston, MA; Buttery et al., 2007).
Protein purification
Rabbit skeletal muscle actin (RMA) was purified as previously described (Spudich and Watt, 1971). In brief, RMA was purified first by generating an acetone powder from ground muscle tissue, which was stored in aliquots at −80°C. Aliquots of acetone powder were then pulverized using a coffee grinder, resuspended in G buffer, and cleared by low speed centrifugation. The actin was polymerized overnight and then pelleted. The pellet was disrupted by douncing, dialyzed against G buffer for 2–3 d, and then aliquoted and stored at −80°C until use. Every 2–3 wk, fresh RMA was prepared by thawing an aliquot of actin, dialyzing against G buffer for 1–2 d, clearing by ultracentrifugation, and gel filtering the supernatant on a 16/60 S200 column (GE Healthcare). Column fractions were stored at 4°C.
For bulk actin assembly assays, RMA was fluorescently labeled with pyrenyl-iodoacetamide on cysteine 374 (Pollard and Cooper, 1984). RMA prepared as in the previous paragraph, excluding the gel filtration step, was dialyzed against pyrene buffer (25 mM Tris-HCl, pH 7.5, 100 mM KCl, 0.02% NaN3, 0.3 mM ATP, and 2 mM MgSO4) for 3–4 h and then diluted with pyrene buffer to 1 mg/ml (23.8 µM). A sevenfold molar excess of pyrenyl-iodoacetamide was added, the actin solution was incubated overnight at 4°C, and then, aggregates were cleared by low-speed centrifugation. The supernatant (containing F-actin) was centrifuged for 3 h at 4°C at 45,000 rpm in a rotor (Ti70; Beckman Coulter) to pellet F-actin. The actin pellets were disrupted by douncing, dialyzed against G buffer for 1–2 d, and gel filtered on a 16/60 S200 column as in the previous paragraph. Peak fractions were pooled, aliquoted, snap frozen, and stored at −80°C until use.
For TIRF-M, RMA was labeled with Oregon green (Kuhn and Pollard, 2005). The labeling protocol was identical to that described for pyrene labeling (see preceding paragraph), except that a 10-fold molar excess of Oregon green 488 iodoacetamide (Molecular Probes) was used instead of the sevenfold molar excess of pyrenyl-iodoacetamide.
S. cerevisiae profilin were expressed and purified from BL21(DE3) E. coli as previously described (Moseley et al., 2004). Bacterial cells were grown in Luria broth to log phase and induced with 0.4 mM IPTG for 3–4 h at 37°C. Cells were pelleted and stored at −80°C. Frozen pellets were thawed, resuspended in 20 mM Tris-HCl, pH 8.0, with standard protease inhibitors, and lysed with lysozyme treatment and sonication. Cell lysates were cleared by centrifugation at 80,000 rpm at 4°C for 20 min using a rotor (TLA-100.3; Beckman Coulter). The supernatant was loaded onto a 5-ml column (HiTrap Q; GE Healthcare) and eluted with a 75-ml salt gradient (0–400 mM NaCl) in 20 mM Tris-HCl, pH 8.0. Peak fractions were pooled, concentrated to 5 ml, and loaded onto a gel filtration column (26/60 Superdex 75; GE Healthcare). Again, peak fractions were pooled, snap frozen in liquid N2, and stored at −80°C.
Bud6 polypeptides were expressed in BL21(DE3) E. coli and purified as previously described (Graziano et al., 2011). Bacterial cells were grown in terrific broth to late log phase and induced using 0.4 mM IPTG for 3–4 h at 37°C. Cells were pelleted and frozen at −80°C. Frozen pellets were thawed, resuspended in lysis buffer (50 mM Tris, pH 8.5, 150 mM NaCl, 5 mM EDTA, 1.5% sarkosyl, 5 mM DTT, and standard protease inhibitors), and lysed with lysozyme treatment and sonication. Cell lysates were cleared by centrifugation at 12,000 rpm for 10 min in a rotor (Sorvall S600; Thermo Fisher Scientific). Triton X-100 (final concentration 3.3% [vol/vol]) was added to the supernatant, and the mixture was then mixed with 1 ml preswollen glutathione agarose in PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.47 mM KH2PO4, pH 7.4). After incubation at 4°C for 3–4 h, beads were washed four times with PBS and then washed twice with HEKD (20 mM Hepes, pH 7.5, 1 mM EDTA, 50 mM KCl, and 1 mM DTT). The resulting GST-Bud6–bound beads were used in quantitative supernatant depletion pull-down assays (e.g., Fig. 2 B). Alternatively, Bud6 polypeptides were cleaved from GST and released from beads by digestion with TEV protease for 2 h at room temperature and snap frozen. GST-TEV-Bil1 was expressed in E. coli and purified as described for Bud6 polypeptides except that the cell pellet was resuspended and lysed in a different buffer: 20 mM Tris-HCl, pH 8.0, 50 mM NaCl, 1% (vol/vol) Triton X-100, 1 mM DTT, and standard protease inhibitors.
C-Bni1 (residues 1,227–1,953), C-Bnr1 (residues 757–1,375), C-Bni1Δtail (residues 1,227–1,797), and C-Bnr1Δtail (residues 757–1,292) were expressed in S. cerevisiae strain BJ2168 on 2-µm plasmids as 6His fusion proteins under galactose-inducible promoters as previously described (Moseley et al., 2006). For each formin polypeptide, 2–4 liters of yeast cells were grown in synthetic medium lacking uracil and 2% raffinose to OD600 = 0.6–0.9, and then, protein expression was induced by the addition of galactose (final 2%), and cells were grown for another 8–9 h at 30°C. Cells were then washed in H2O, frozen under liquid N2, and lysed by mechanical disruption using a coffee grinder and liquid N2. The resulting frozen yeast lysate powder was stored at −80°C until use. Purification of C-Bni1 and C-Bni1Δtail was performed as previously described (Gould et al., 2011; Graziano et al., 2011). In brief, frozen lysed yeast powder was resuspended in buffer A (20 mM NaPO4, pH 7.4, 150 mM NaCl, 0.5 mM DTT, 1% NP-40, and standard protease inhibitors) and then cleared by ultracentrifugation at 80,000 rpm for 20 min in a rotor (TLA-100.3). The supernatant was incubated with Ni–nitrilotriacetic acid (NTA) beads for 1.5 h at 4°C, and then, the beads were washed with buffer A (no protease inhibitors or NP-40), and proteins were eluted with buffer A + 300 nM imidazole. Eluted proteins were purified further on a gel filtration column (Superose 12; GE Healthcare) equilibrated in buffer HEKG10D (20 mM Hepes, pH 7.5, 1 mM EDTA, 50 mM KCl, 10% [vol/vol] glycerol, and 1 mM DTT), and peak fractions were pooled, aliquoted, snap frozen, and stored at −80°C. C-Bnr1 and C-Bnr1Δtail were purified similarly, except that the cleared yeast lysates were filtered through a 0.45-µm syringe-driven filter unit (Millex; EMD Millipore), and the 6His-fusion proteins were isolated on a purification system (Profinia; Bio-Rad Laboratories) using the native IMAC (immobilized metal ion affinity chromatography) + desalting program (1-ml IMAC column + 5-ml desalting column). The desalting buffer used was HEKG10D. The eluted C-Bnr1 and C-Bnr1Δtail polypeptides (4 ml) were concentrated using centrifugal filter units (molecular mass cutoff of 30 kD; Amicon Ultra; EMD Millipore) to a final volume of ∼0.7 ml.
Bnr1 FH2 (residues 868–1,291) was expressed as a dual end–tagged His10-Smt3-FH2 fusion protein in E. coli induced with 0.4 mM IPTG overnight at 18°C. Cells were harvested and resuspended in lysis buffer (30 mM imidazole, 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, pH 7.4, 0.5 mM DTT, 1% [vol/vol] NP-40, and standard protease inhibitors), treated with lysozyme, and sonicated, and then, the clarified supernatant was incubated with 0.5 ml Ni-NTA agarose (QIAGEN) for 1.5 h at 4°C. The Ni-NTA agarose was washed four times with wash buffer (350 mM NaCl, 50 mM imidazole, 20 mM Tris-HCl, pH 8.0, 10% [vol/vol] glycerol, and 1 mM β-mercaptoethanol), and proteins were eluted with 2 ml elution buffer (350 mM NaCl, 400 mM imidazole, 20 mM Tris-HCl, pH 8.0, 10% [vol/vol] glycerol, and 1 mM β-mercaptoethanol). Bnr1 FH2 was cleaved from the His10-Smt3 tag by incubation with UlpI protease overnight at 4°C while being dialyzed against the HEK350G10 buffer (20 mM Hepes, pH 7.5, 1 mM EDTA, 350 mM KCl, and 10% [vol/vol] glycerol). The dialyzed proteins were separated by gel filtration on a column (Superose 12) using a fast protein liquid chromatography (ÄKTA; GE Healthcare). Peak FH2-containing fractions were pooled, aliquoted, snap frozen in liquid N2, and stored at −80°C.
Actin assembly assays
Before each experiment, gel-filtered monomeric actin in G buffer was cleared by ultracentrifugation for 1 h at 4°C at 90,000 rpm in a rotor (TLA-100), and the top ∼75% of the supernatant was carefully recovered. Assembly reactions (60 µl) contained 2 µM G-actin (5% pyrene labeled) and 5 µM yeast profilin where indicated. G-actin was converted to Mg2+-ATP-actin 2 min before use, and then, 42 µl G-actin was mixed rapidly with 15 µl proteins/control buffer followed by 3 µl of 20× initiation mix (40 mM MgCl2, 10 mM ATP, and 1 M KCl) before the start of reactions. Pyrene-actin fluorescence was monitored in a plate reader (Infinite M200; Tecan) at excitation and emission wavelengths of 365 and 407 nm, respectively. Kapp corresponds to the concentration of Bud6 or Bil1 (in the presence of Bud6) required for half-maximal stimulation of formin-mediated actin assembly and was determined by first measuring the slopes of the polymerization curves for the initial 200 s of each reaction and then dividing the slopes of the reactions containing the variable component (e.g., Bud6) by the slopes of the reactions lacking the component. The resulting “normalized” rates were plotted as a function of the concentration of the variable component. For all assembly assays, at least two independent experiments were performed using different stocks of purified proteins. The data shown in Fig. 3 G are representative results from one of the individual experiments.
Quantitative supernatant depletion pull-down assays
During purification of GST-Bud6 constructs from E. coli (Graziano et al., 2011), the amount of GST-Bud6 bound to beads was quantified by SDS-PAGE and Coomassie staining. Then, variable concentrations of immobilized GST-Bud6 were incubated for 10–15 min at room temperature with a fixed concentration of soluble formin (1–2 µM). All reactions were performed in HEND (20 mM Hepes, pH 7.5, 1 mM EDTA, 100 mM NaCl, and 1 mM DTT). Beads were pelleted, and supernatants were recovered. The amount of formin remaining in the supernatant of each reaction was determined by SDS-PAGE and Coomassie staining and compared with the amount of formin remaining in the supernatants of control reactions containing beads with GST alone. This value was then used to calculate the amount of formin that had been bound by GST-Bud6. Quantification of Coomassie-stained bands was performed using an infrared imaging system (Odyssey; LI-COR Biosciences). The Kd for each Bud6–formin pair was calculated plotting the fraction of formin bound as a function of the concentration of Bud6 and fitting the data to a hyperbola (Pollard, 2010). All binding experiments were performed on a separate 2–3 d using different stocks of purified proteins.
Cell imaging
Fixation, staining, imaging, and analysis of yeast strains were performed as previously described (Graziano et al., 2011). In brief, yeast strains were grown to early/mid–log phase (OD600 0.3–0.5) in YEPD (yeast extract, peptone, and 2% glucose) supplemented with adenine, fixed in 4.5% formaldehyde for 45–60 min, and stained with Alexa Fluor 488–phalloidin (Molecular Probes). Cells were imaged in mounting media (10 mM NaPO4, pH 7.4, 75 mM NaCl, 4.3 mM p-phenylenediamine, 0.01 mg/ml DAPI, and 45% glycerol [vol/vol]) at room temperature (25°C) using a microscope (Axioskop 2 mot plus; Carl Zeiss) equipped with a 100× Plan Apochromat objective, NA 1.40 (Carl Zeiss), and a digital charge-coupled device camera (ORCA-ER; Hamamatsu Photonics). All images of fixed cells were 1,500-ms exposures of a single focal plane in which multiple cells were present within the field of view. Images were acquired using OpenLab software (PerkinElmer) and analyzed using ImageJ (National Institutes of Health). For live-cell imaging of Bil1-GFP and Bud6-GFP strains, cells were grown in synthetic complete media + 2% glucose to early/mid–log phase and imaged in the same medium at room temperature. For single-color imaging (Fig. 7 A and Fig. S4), 600-ms exposures were acquired with 2 × 2 binning; but otherwise, the same microscope configuration and analysis software was used as described for the imaging of fixed cells.
For two-color imaging of Bil1-GFP and Bud6-RFP (Fig. 7 B), cells were grown in synthetic complete media + 2% glucose to early/mid–log phase and imaged in the same medium at room temperature. Images were acquired using Elements AR software (Nikon) with laser excitation at 488 and 561 nm and a 100×, NA 1.45 Plan Apochromat objective on an upright microscope (Ni-E; Nikon) equipped with a spinning-disk head (CSU-W1; Yokogawa Corporation of America), 525/40- and 617/73-nm emission filters, and an electron multiplying charge-coupled device camera (iXon 897U; Andor Technology). Image analysis was performed as described in the previous paragraph.
For live-cell imaging of GFP-Sec4 particle movements, yeast strains carrying the GFP-Sec4 plasmid (URA3 marked) were grown in synthetic media lacking uracil and 2% glucose to early/mid–log phase. Cells were then imaged in the same medium at room temperature on a spinning-disk confocal system (Marianas; 3I, Inc.), consisting of a microscope (Observer.Z1; Carl Zeiss) equipped with a 100× Plan Apochromat objective, NA 1.4, a spinning-disk confocal head (CSU-X1), and an electron multiplying charge-coupled device camera (QuantEM 512SC; Photometrics). Exposures of 150 ms were acquired every 200 ms over a time course of 60 s. Image acquisition and analysis were performed using SlideBook 5.0 (3I, Inc.) and ImageJ.
TIRF-M
Before all TIRF-M experiments, coverslips were coated with PEG-5000 and assembled into flow cells. Immediately before each experiment, flow cells were incubated for ∼3 min in TBSA (Tris-HCl, pH 8.0, and 1% BSA) and then washed with TIRF buffer (10 mM imidazole, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.2 mM ATP, 10 mM DTT, 15 mM glucose, 20 µg/ml glucose oxidase, and 0.5% methylcellulose [4,000 cP], pH 7.4). Proteins in TIRF buffer were then mixed with 0.5 µM G-actin (10% Oregon green labeled) and added to the flow cell. Images for each reaction were acquired at 5-s intervals for a total of 600 s using an inverted microscope (Ti200; Nikon) with a 150-mW argon laser (Mellot Griot), a 60× TIRF objective, NA 1.49 (Nikon), and an electron multiplying charge-coupled device camera (iXon; Andor Technology). Focus was maintained using the Perfect Focus System (Nikon). Temperature was maintained at 20–25°C. Elongation rates of filaments were determined by measuring the length of a single filament at 60-s intervals over a period of 300 s. Bnr1-elongated filaments were distinguished by their approximately fourfold faster elongation rate versus filaments elongating in the absence of formin. For each condition, three independent experiments were performed. To determine the number of filaments nucleated, each field of view was examined 450 s after G-actin was added to the reaction. For each condition, filament densities were calculated in a total of nine fields of view spanning three independent reactions. Analysis was performed using NIS Elements (Nikon) and ImageJ. All TIRF experiments were performed on a separate 2–3 d using different stocks of purified proteins. In Fig. 4 (B and E) and Fig. 6 (E and F), the graphs show data averaged from two to three experiments. In Fig. 4 D, each line represents an individual actin filament taken from the same experiment.
Actin binding assays
Binding reactions were performed using 150 nM RMA (100% pyrene labeled), 600 nM latrunculin B, and varying concentrations of either Bud6(S) or Bud6(L). All reactions contained a final volume of 60 µl comprising 5 µl RMA in G buffer and 55 µl proteins in HK buffer (20 mM Hepes and 50 mM KCl). Upon reaching equilibrium, pyrene fluorescence was monitored as previously described (see subsection Actin assembly assays). Binding curves were generated by plotting Fluorescence/Fluorescencemax (after subtracting the baseline pyrene fluorescence obtained from reactions containing no Bud6) as a function of Bud6 concentration. To determine the Kd for each reaction, data points from the plots were fitted using a quadratic equation (Pollard, 2010). Experiments were performed on two separate occasions using different stocks of purified proteins. The data shown in Fig. 3 G are representative results from one of the individual experiments.
Bud6-GFP pull-down and mass spectrometry analysis
Rabbit polyclonal α-GFP antibody was generated by established protocols using GFP expressed and purified from E. coli (Cristea et al., 2005). A plasmid expressing 6His-GFP was transformed into E. coli strain BL21, cells were grown to log phase, and cells were induced at 25°C overnight using 1 mM IPTG. Cells were pelleted and stored at −80°C. Pellets were thawed, resuspended in lysis buffer (PBS, pH 7.4, and standard protease inhibitors) and lysed by lysozyme treatment and sonication. Lysates were cleared by centrifugation at 12,000 rpm for 15 min at 4°C in a rotor (Sorvall S600), and then, ammonium sulfate was added to a final concentration of 50%, incubated on ice for 30 min, and centrifuged again (12,000 rpm for 15 min at 4°C). The soluble fraction of the ammonium sulfate cut was passed over a 5-ml Ni-NTA column three times. The Ni-NTA column was washed with lysis buffer (no protease inhibitors), and then, 6His-GFP was eluted using PBS + 350 mM imidazole, and the eluted protein was further purified on a column equilibrated in PBS (26/60 Superdex 75). Peak fractions were pooled and concentrated. The purified 6His-GFP was injected into New Zealand white rabbits (Covance), and the bleeds were tested by Western blotting for specificity. Antibodies were then affinity purified for use in proteomic analyses (see following paragraph).
Large cultures of a yeast strain carrying an integrated GFP tag at the C terminus of Bud6 (BGY1484) and a control strain with no tag (BGY1257) were grown to late log phase, frozen, and lysed by mechanical perturbation under liquid nitrogen. Cell lysates from the two strains were clarified by ultracentrifugation and then incubated for 1 h with protein A beads coated with affinity purified α-GFP antibodies, which had been cross-linked to the beads using dimethyl pimelimidate (Sigma-Aldrich). Beads were then pelleted and washed, and the bound proteins were eluted with 0.5 M ammonium hydroxide and 0.5 mM EDTA. Samples were TCA precipitated, digested trypsin, and analyzed by tandem mass spectrometry and MudPIT (Multidimensional Protein Identification Technology). Among the proteins that associated specifically with Bud6-GFP, we focused on Yor304c-a (Bil1) because it was uncharacterized and previously had been identified as a potential Bud6-binding partner in a proteome-wide tandem affinity purification–tagging study (Krogan et al., 2006).
Online supplemental material
Fig. S1 shows Coomassie-stained SDS-PAGE gels of the key purified proteins used in this study. Fig. S2 shows quantitative GST pull-down assays performed as in Fig. 3 (E and F) using the indicated concentrations of immobilized GST-Bud6(S) and 1.5 µM soluble C-Bnr1. Fig. S3 shows biochemical effects of Bil1 on actin assembly. Fig. S4 shows localization of endogenously expressed Bud6-GFP or Bil1-GFP throughout the cell cycle. Fig. S5 shows genetic analysis of BIL1 function in a bni1Δbud6Δ strain. Video 1 shows GFP-Sec4 vesicle movements in wild-type yeast cells. Video 2 shows GFP-Sec4 vesicle movements in bni1Δ cells. Video 3 shows GFP-Sec4 vesicle movements in bud6Δ cells. Video 4 shows GFP-Sec4 vesicle movements in bil1Δ cells. Video 5 shows GFP-Sec4 vesicle movements in bni1Δbud6Δ cells. Video 6 shows GFP-Sec4 vesicle movements in bni1Δbil1Δ cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201212059/DC1.
Supplementary Material
Acknowledgments
We thank Tony Bretscher, David Pellman, and Mike Rosen for strains and plasmids. We thank John Yates and the Yeast Resource Center for performing the mass spectrometry analysis and Ed Dougherty for assistance at the Brandeis Confocal Imaging Laboratory. We are also grateful to David Pruyne, Avital Rodal, Satoshi Yoshida, Dennis Breitsprecher, Julian Eskin, and Casey Ydenberg for helpful discussions and critical reading of the manuscript.
This work was supported by a grant from National Institutes of Health (GM083187) to B.L. Goode.
Footnotes
Abbreviations used in this paper:
- APC
- adenomatous polyposis coli
- BBS
- Bud6 binding site
- DAD
- diaphanous autoregulatory domain
- NPF
- nucleation-promoting factor
- NTA
- nitrilotriacetic acid
- RMA
- rabbit skeletal muscle actin
- TEV
- tobacco etch virus
- TIRF
- total internal reflection fluorescence
- TIRF-M
- TIRF microscopy
References
- Amberg D.C., Zahner J.E., Mulholland J.W., Pringle J.R., Botstein D. 1997. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol. Biol. Cell. 8:729–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L., Michelot A. 2012. Actin cytoskeleton: a team effort during actin assembly. Curr. Biol. 22:R643–R645 10.1016/j.cub.2012.07.026 [DOI] [PubMed] [Google Scholar]
- Block J., Breitsprecher D., Kühn S., Winterhoff M., Kage F., Geffers R., Duwe P., Rohn J.L., Baum B., Brakebusch C., et al. 2012. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr. Biol. 22:1005–1012 10.1016/j.cub.2012.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M., Le K.H., Bugyi B., Correia J.J., Renault L., Carlier M.F. 2007. Analysis of the function of Spire in actin assembly and its synergy with formin and profilin. Mol. Cell. 28:555–568 10.1016/j.molcel.2007.09.018 [DOI] [PubMed] [Google Scholar]
- Breitsprecher D., Jaiswal R., Bombardier J.P., Gould C.J., Gelles J., Goode B.L. 2012. Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science. 336:1164–1168 10.1126/science.1218062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttery S.M., Yoshida S., Pellman D. 2007. Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol. Biol. Cell. 18:1826–1838 10.1091/mbc.E06-09-0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone M.A., Goode B.L. 2009. Actin nucleation and elongation factors: mechanisms and interplay. Curr. Opin. Cell Biol. 21:28–37 10.1016/j.ceb.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone M., Gould C.J., Moseley J.B., Goode B.L. 2009. Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Dev. Cell. 16:292–302 10.1016/j.devcel.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche N., Pollard T.D. 2012. Determinants of Formin Homology 1 (FH1) domain function in actin filament elongation by formins. J. Biol. Chem. 287:7812–7820 10.1074/jbc.M111.322958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea I.M., Williams R., Chait B.T., Rout M.P. 2005. Fluorescent proteins as proteomic probes. Mol. Cell. Proteomics. 4:1933–1941 10.1074/mcp.M500227-MCP200 [DOI] [PubMed] [Google Scholar]
- Delgehyr N., Lopes C.S., Moir C.A., Huisman S.M., Segal M. 2008. Dissecting the involvement of formins in Bud6p-mediated cortical capture of microtubules in S. cerevisiae. J. Cell Sci. 121:3803–3814 10.1242/jcs.036269 [DOI] [PubMed] [Google Scholar]
- Dominguez R. 2009. Actin filament nucleation and elongation factors—structure-function relationships. Crit. Rev. Biochem. Mol. Biol. 44:351–366 10.3109/10409230903277340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R. 2010. Structural insights into de novo actin polymerization. Curr. Opin. Struct. Biol. 20:217–225 10.1016/j.sbi.2009.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M., Blundell K., Longtine M.S., Chow C.J., Adames N., Pringle J.R., Peter M., Boone C. 1997. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 276:118–122 10.1126/science.276.5309.118 [DOI] [PubMed] [Google Scholar]
- Firat-Karalar E.N., Welch M.D. 2011. New mechanisms and functions of actin nucleation. Curr. Opin. Cell Biol. 23:4–13 10.1016/j.ceb.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould C.J., Maiti S., Michelot A., Graziano B.R., Blanchoin L., Goode B.L. 2011. The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr. Biol. 21:384–390 10.1016/j.cub.2011.01.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano B.R., DuPage A.G., Michelot A., Breitsprecher D., Moseley J.B., Sagot I., Blanchoin L., Goode B.L. 2011. Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor. Mol. Biol. Cell. 22:4016–4028 10.1091/mbc.E11-05-0404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman S.J., Ridley A.J. 2008. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9:690–701 10.1038/nrm2476 [DOI] [PubMed] [Google Scholar]
- Heimsath E.G., Jr, Higgs H.N. 2012. The C terminus of formin FMNL3 accelerates actin polymerization and contains a WH2 domain-like sequence that binds both monomers and filament barbed ends. J. Biol. Chem. 287:3087–3098 10.1074/jbc.M111.312207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Jin H., Amberg D.C. 2000. The secretory pathway mediates localization of the cell polarity regulator Aip3p/Bud6p. Mol. Biol. Cell. 11:647–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei T., Tanaka K., Hihara T., Umikawa M., Imamura H., Kikyo M., Ozaki K., Takai Y. 1998. Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae. J. Biol. Chem. 273:28341–28345 10.1074/jbc.273.43.28341 [DOI] [PubMed] [Google Scholar]
- Kikyo M., Tanaka K., Kamei T., Ozaki K., Fujiwara T., Inoue E., Takita Y., Ohya Y., Takai Y. 1999. An FH domain-containing Bnr1p is a multifunctional protein interacting with a variety of cytoskeletal proteins in Saccharomyces cerevisiae. Oncogene. 18:7046–7054 10.1038/sj.onc.1203184 [DOI] [PubMed] [Google Scholar]
- Kita K., Wittmann T., Näthke I.S., Waterman-Storer C.M. 2006. Adenomatous polyposis coli on microtubule plus ends in cell extensions can promote microtubule net growth with or without EB1. Mol. Biol. Cell. 17:2331–2345 10.1091/mbc.E05-06-0498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono K., Saeki Y., Yoshida S., Tanaka K., Pellman D. 2012. Proteasomal degradation resolves competition between cell polarization and cellular wound healing. Cell. 150:151–164 10.1016/j.cell.2012.05.030 [DOI] [PubMed] [Google Scholar]
- Kovar D.R., Pollard T.D. 2004. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl. Acad. Sci. USA. 101:14725–14730 10.1073/pnas.0405902101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D.R., Harris E.S., Mahaffy R., Higgs H.N., Pollard T.D. 2006. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 124:423–435 10.1016/j.cell.2005.11.038 [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., et al. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 440:637–643 10.1038/nature04670 [DOI] [PubMed] [Google Scholar]
- Kuhn J.R., Pollard T.D. 2005. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 88:1387–1402 10.1529/biophysj.104.047399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Higgs H.N. 2003. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13:1335–1340 10.1016/S0960-9822(03)00540-2 [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Moseley J.B., Goode B.L. 2005. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J. Biol. Chem. 280:28023–28033 10.1074/jbc.M503094200 [DOI] [PubMed] [Google Scholar]
- Moseley J.B., Sagot I., Manning A.L., Xu Y., Eck M.J., Pellman D., Goode B.L. 2004. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell. 15:896–907 10.1091/mbc.E03-08-0621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.B., Maiti S., Goode B.L. 2006. Formin proteins: purification and measurement of effects on actin assembly. Methods Enzymol. 406:215–234 10.1016/S0076-6879(06)06016-2 [DOI] [PubMed] [Google Scholar]
- Narayanaswamy R., Moradi E.K., Niu W., Hart G.T., Davis M., McGary K.L., Ellington A.D., Marcotte E.M. 2009. Systematic definition of protein constituents along the major polarization axis reveals an adaptive reuse of the polarization machinery in pheromone-treated budding yeast. J. Proteome Res. 8:6–19 10.1021/pr800524g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Bartolini F., Deaconescu A.M., Moseley J.B., Dogic Z., Grigorieff N., Gundersen G.G., Goode B.L. 2010. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J. Cell Biol. 189:1087–1096 10.1083/jcb.201001016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T., Tomchick D.R., Otomo C., Panchal S.C., Machius M., Rosen M.K. 2005. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 433:488–494 10.1038/nature03251 [DOI] [PubMed] [Google Scholar]
- Ozaki-Kuroda K., Yamamoto Y., Nohara H., Kinoshita M., Fujiwara T., Irie K., Takai Y. 2001. Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:827–839 10.1128/MCB.21.3.827-839.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick S.B., Rosen M.K. 2010. Physical mechanisms of signal integration by WASP family proteins. Annu. Rev. Biochem. 79:707–735 10.1146/annurev.biochem.77.060407.135452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A.S., Pollard T.D. 2008. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol. 18:9–19 10.1016/j.cub.2007.11.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechlivanis M., Samol A., Kerkhoff E. 2009. Identification of a short Spir interaction sequence at the C-terminal end of formin subgroup proteins. J. Biol. Chem. 284:25324–25333 10.1074/jbc.M109.030320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfender S., Kuznetsov V., Pleiser S., Kerkhoff E., Schuh M. 2011. Spire-type actin nucleators cooperate with Formin-2 to drive asymmetric oocyte division. Curr. Biol. 21:955–960 10.1016/j.cub.2011.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D. 2007. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 36:451–477 10.1146/annurev.biophys.35.040405.101936 [DOI] [PubMed] [Google Scholar]
- Pollard T.D. 2010. A guide to simple and informative binding assays. Mol. Biol. Cell. 21:4061–4067 10.1091/mbc.E10-08-0683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Cooper J.A. 1984. Quantitative analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry. 23:6631–6641 10.1021/bi00321a054 [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Cooper J.A. 2009. Actin, a central player in cell shape and movement. Science. 326:1208–1212 10.1126/science.1175862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A., Boone C. 2002. Role of formins in actin assembly: nucleation and barbed-end association. Science. 297:612–615 10.1126/science.1072309 [DOI] [PubMed] [Google Scholar]
- Pruyne D., Gao L., Bi E., Bretscher A. 2004. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell. 15:4971–4989 10.1091/mbc.E04-04-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan M.E., Hilgert S., Bedrossian A., Mullins R.D., Kerkhoff E. 2007. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J. Cell Biol. 179:117–128 10.1083/jcb.200706196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero S., Le Clainche C., Didry D., Egile C., Pantaloni D., Carlier M.F. 2004. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 119:419–429 10.1016/j.cell.2004.09.039 [DOI] [PubMed] [Google Scholar]
- Rosales-Nieves A.E., Johndrow J.E., Keller L.C., Magie C.R., Pinto-Santini D.M., Parkhurst S.M. 2006. Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat. Cell Biol. 8:367–376 10.1038/ncb1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K., Hänisch J., Campellone K.G. 2010. WASH, WHAMM and JMY: regulation of Arp2/3 complex and beyond. Trends Cell Biol. 20:650–661 10.1016/j.tcb.2010.08.014 [DOI] [PubMed] [Google Scholar]
- Sagot I., Klee S.K., Pellman D. 2002a. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4:42–50 [DOI] [PubMed] [Google Scholar]
- Sagot I., Rodal A.A., Moseley J., Goode B.L., Pellman D. 2002b. An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4:626–631 [DOI] [PubMed] [Google Scholar]
- Schuh M. 2011. An actin-dependent mechanism for long-range vesicle transport. Nat. Cell Biol. 13:1431–1436 10.1038/ncb2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M., Bloom K., Reed S.I. 2000. Bud6 directs sequential microtubule interactions with the bud tip and bud neck during spindle morphogenesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 11:3689–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitar T., Gallinger J., Ducka A.M., Ikonen T.P., Wohlhoefler M., Schmoller K.M., Bausch A.R., Joel P., Trybus K.M., Noegel A.A., et al. 2011. Molecular architecture of the Spire-actin nucleus and its implication for actin filament assembly. Proc. Natl. Acad. Sci. USA. 108:19575–19580 10.1073/pnas.1115465108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J.A., Watt S. 1971. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246:4866–4871 [PubMed] [Google Scholar]
- Su L.K., Burrell M., Hill D.E., Gyuris J., Brent R., Wiltshire R., Trent J., Vogelstein B., Kinzler K.W. 1995. APC binds to the novel protein EB1. Cancer Res. 55:2972–2977 [PubMed] [Google Scholar]
- Ten Hoopen R., Cepeda-García C., Fernández-Arruti R., Juanes M.A., Delgehyr N., Segal M. 2012. Mechanism for astral microtubule capture by cortical Bud6p priming spindle polarity in S. cerevisiae. Curr. Biol. 22:1075–1083 10.1016/j.cub.2012.04.059 [DOI] [PubMed] [Google Scholar]
- Tong A.H., Evangelista M., Parsons A.B., Xu H., Bader G.D., Pagé N., Robinson M., Raghibizadeh S., Hogue C.W., Bussey H., et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 294:2364–2368 10.1126/science.1065810 [DOI] [PubMed] [Google Scholar]
- Tu D., Graziano B.R., Park E., Zheng W., Li Y., Goode B.L., Eck M.J. 2012. Structure of the formin-interaction domain of the actin nucleation-promoting factor Bud6. Proc. Natl. Acad. Sci. USA. 109:E3424–E3433 10.1073/pnas.1203035109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen E.A., Caviston J., Bi E. 2000. Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 11:593–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D., Kovar D.R., O’Shaughnessy B., Pollard T.D. 2006. Model of formin-associated actin filament elongation. Mol. Cell. 21:455–466 10.1016/j.molcel.2006.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcarra C.L., Kreutz B., Rodal A.A., Toms A.V., Lu J., Zheng W., Quinlan M.E., Eck M.J. 2011. Structure and function of the interacting domains of Spire and Fmn-family formins. Proc. Natl. Acad. Sci. USA. 108:11884–11889 10.1073/pnas.1105703108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb R.L., Zhou M.N., McCartney B.M. 2009. A novel role for an APC2-Diaphanous complex in regulating actin organization in Drosophila. Development. 136:1283–1293 10.1242/dev.026963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Eng C.H., Schmoranzer J., Cabrera-Poch N., Morris E.J., Chen M., Wallar B.J., Alberts A.S., Gundersen G.G. 2004. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 6:820–830 10.1038/ncb1160 [DOI] [PubMed] [Google Scholar]
- Xu Y., Moseley J.B., Sagot I., Poy F., Pellman D., Goode B.L., Eck M.J. 2004. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 116:711–723 10.1016/S0092-8674(04)00210-7 [DOI] [PubMed] [Google Scholar]
- Yu H., Braun P., Yildirim M.A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., et al. 2008. High-quality binary protein interaction map of the yeast interactome network. Science. 322:104–110 10.1126/science.1158684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.H., Crevenna A.H., Bettenbühl M., Freisinger T., Wedlich-Söldner R. 2011. Cortical actin dynamics driven by formins and myosin V. J. Cell Sci. 124:1533–1541 10.1242/jcs.079038 [DOI] [PubMed] [Google Scholar]
- Zeth K., Pechlivanis M., Samol A., Pleiser S., Vonrhein C., Kerkhoff E. 2011. Molecular basis of actin nucleation factor cooperativity: crystal structure of the Spir-1 kinase non-catalytic C-lobe domain (KIND)−formin-2 formin SPIR interaction motif (FSI) complex. J. Biol. Chem. 286:30732–30739 10.1074/jbc.M111.257782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond S.H., Evangelista M., Boone C., Yang C., Dar A.C., Sicheri F., Forkey J., Pring M. 2003. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr. Biol. 13:1820–1823 10.1016/j.cub.2003.09.057 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.