Abstract
Alcohol is the most abused substance worldwide and a significant source of liver injury; the mechanisms of alcohol-induced liver disease are not fully understood. Significant cellular toxicity and impairment of protein synthesis and degradation occur in alcohol-exposed liver cells, along with changes in energy balance and modified responses to pathogens. Autophagy is the process of cellular catabolism through the lysosomal-dependent machinery, which maintains a balance among protein synthesis, degradation, and recycling of self. Autophagy is part of normal homeostasis and it can be triggered by multiple factors that threaten cell integrity including starvation, toxins, or pathogens. Multiple factors regulate autophagy; survival and preservation of cellular integrity at the expense of inadequately-folded proteins and damaged high energy-generating intracellular organelles are prominent targets of autophagy in pathologic conditions. Coincidentally, inadequately-folded proteins accumulate and high energy-generating intracellular organelles, such as mitochondria, are damaged by alcohol abuse; these alcohol-induced pathological findings prompted investigation of the role of autophagy in the pathogenesis of alcohol-induced liver damage. Our review summarizes the current knowledge about the role and implications of autophagy in alcohol-induced liver disease.
Keywords: liver, damage, Toll-like receptors, mitophagy, ethanol, mechanisms, mTOR, Atg
Introduction
Alcoholic liver disease (ALD) is a major cause of death in the United States. According to latest surveys, chronic liver disease is the underlying cause of death for ~26,000 persons per year, of which ~46% are associated with alcohol abuse (CDC 1989, Yoon, 2010). Significant progress has been made in recent years toward understanding the mechanisms of alcohol-induced liver injury (Hoek et al. 2004), however little is known about the cellular protective effects against the detriments of ethanol. Several challenging questions in this regard are: how do the host cells cope with the toxic effects of the alcohol and how do they counteract these effects; how do cells recover; at what cost and how long does it take? Recent research on autophagy, initially described in yeast and to a better extent understood in developmental biology, suggests that this self-defense mechanism may account for some of the defense mechanisms mounted by cells after exposure to alcohol. This review summarizes the presentations and the discussions at the symposium entitled “Influence of alcohol abuse on autophagy”, presented at the RSoA 33rd Annual Scientific Meeting on June 27, 2010 in San Antonio, TX, which attempted to identify if there is a role for autophagy in alcohol-induced liver disease.
Autophagy: triggers, mechanisms, markers and outcomes
Autophagy is a genetically programmed, evolutionarily-conserved catabolic process that occurs in response to stress and is manifested as degradation of cellular proteins and damaged organelles in an effort to promote cell survival. Autophagy plays a role in developmental biology (Yang and Klionsky, 2010), in immunity against microbial infections (Kirkegaard et al. 2004), and in a number of human diseases with distinct clinical manifestations but closely-related pathogenesis, such as neurodegenerative diseases, cardiac myopathies and cancer (Edinger and Thompson 2003; Shintani and Klionsky 2004).
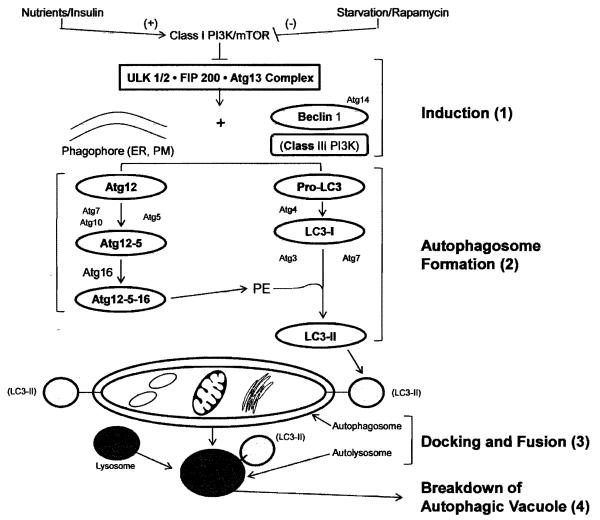
It has been known for over 40 years that cells digest their own contents by autophagy (De Duve, 1966). Among the best understood pathways that lead to autophagy is the one triggered by starvation or ischemia. In oxygen- or nutrient-deprived cells, autophagy involves the mammalian target of rapamycin (mTOR) pathway and undergoes 4 stages of development (Figure 1). Stage 1 involves autophagy induction, which is regulated by mTOR and the downstream ULK/Atg13/FIP200 complex. In stage 2 the autophagosome is formed; this process is regulated by two ubiquitin-like conjugation systems Atg5-12-16 and microtubule light chain-3 (LC3; yeast Atg8) as well as Beclin-1 forms a complex with Atg14, enable the recruitment and conjugation of LC3 with phosphatidylethanolamine (PE) and the formation of an autophagic vacuole (AV). Stage 3 involves docking and fusion of the AV with the lysosome: the cargo-filled autophagosomes enter close proximity with lysosomes and fuse together, forming autolysosomes. Finally, in stage 4 the autophagic vacuole contents are degraded in the autolysosome. In addition to mTOR, several other pathways are involved in induction of autophagy. In yeast, Ras/cAMP-dependent protein kinase (PKA) occurs in well-nourished cells, when Ras1 and Ras2 GTPases are activated and force the adenylyl cyclase to produce cAMP. Cyclic AMP binds to the PKA regulatory subunit (Bcy1) and allows its dissociation from the PKA catalytic subunit Tpk1, Tpk2, and Tpk3, which leads to PKA activation (Thevelein and de Winde 1999). PKA activation prevents the induction of autophagy by nutrient starvation or rapamycin treatment, whereas inactivation of PKA by genetic defects induces autophagy under nutrient-rich conditions without rapamycin (Budovskaya et al. 2004; Schmelzle et al. 2004); the role of PKA-dependent phosphorylation of Atg1, Atg13, Atg18 and Atg21 as a functional link to autophagy is not clear. PKB (Sch9) cooperates with PKA to regulate the induction of autophagy. In mammalian cells, adenosine monophosphate-activated protein (AMP) kinase (AMPK) appears to be required for autophagy. AMP accumulation, caused by excessive energy demands, causes activation of the LKB1-AMPK pathway, which inhibits mTOR by activating TSC1/TSC2 (Hoyer-Hansen and Jaattela 2007) and thus triggers mTOR-dependent autophagy. As a distinct branch, AMPK phosphorylates p27, a cyclin-kinase inhibitor, and thus stabilizes p27 to induce autophagy (Liang et al. 2007). In contrast, the adenosine analogue, adenosine, 5-amino-4-imidazole carboxamide riboside (AICAR), an AMPK activator, strongly suppresses autophagy in hepatocytes (Samari and Seglen, 1998). Recently, it was reported that compound C, an AMPK inhibitor, induces autophagy in cancer cells by AMPK-independent down regulation of Akt/mTOR pathway (Vucicevic et al. 2010). Liver accommodates the mTOR-dependent, mTOR-independent, and probably several yet-to-be-discovered autophagy pathways (Donohue 2009); the effects of alcohol abuse on each of the autophagy pathways (macroautophagy, microautophagy and chaperone-mediated autophagy remain to be determined.
Figure 1. Macroautophagy in mammals.
This diagram was built based on the latest knowledge in the field of autophagy. Only known mammalian counterparts of yeast autophagy factors are included. The mammalian target of rapamycin (mTOR) suppresses autophagy when nutrient supply is adequate or insulin levels are elevated. Under conditions of starvation or rapamycin treatment, mTOR suppression is prevented, causing induction of autophagy (1) by formation of the ULK·FIP200· Atg13 complex, which, in the presence of the phagophore (the autophagosome precursor membrane originating from the endoplasmic reticulum (ER) and/or plasma membrane (PM)), triggers initiation of autophagosome formation (2). This involves the formation of coordinated complexes among beclin-1 and Atg14 and conjugation systems (see text) that enable lipidation of LC3I with phosphatidylethanolamine (PE) to form LC3II. Proteins and organelles are sequestered within the double membrane–bound autophagosome. The latter undergoes fusion with lysosomes (3) to form an autolysosome, which is a degradative organelle. The contents of the autolysosome are acidified and then degraded by lysosomal hydrolases (4). Figure reproduced and modified with permission after Liu, et. al. (2009).
Evidence of autophagy alterations after alcohol exposure
A. Protein degradation after alcohol exposure
Liver enlargement (hepatomegaly) is common in alcoholics and in alcohol-fed laboratory animals. Increased hepatic mass arises from accumulation of both proteins and lipids and each accounts for half the net increase in liver mass (Baraona et al. 1975). Like alcohol-induced fatty liver, the net protein gain likely contributes to the more severe alcohol-induced liver pathologies because some accumulated proteins are damaged by reactive molecules generated from ethanol metabolism, from respiratory leakage of damaged mitochondria, and from ethanol-induced secondary reactions that generate oxidants (Bardag-Gorce et al. 2006). Protein synthetic activity in livers of chronically ethanol-fed rats is normal (Donohue et al. 1987). However, compared with control animals, the rate of hepatic protein degradation in ethanol-fed animals declines by 36-40% (Donohue at al. 1989). Alcohol consumption disrupts the catabolism of long-lived proteins (half-lives ≥ 8 hr) which are mostly degraded in lysosomes. However ethanol administration diminishes the proteolytic capacity of liver lysosomes by alkalinizing their interior (Kharbanda et al.1997) and lowering cathepsin content by disrupting trafficking of these enzymes to lysosomes (Kharbanda et al. 1996). The latter occurs because of an ethanol-induced decrease in the content, of the mannose-6-phosphate receptor (Haorah et al. 2002), which binds to cathepsin precursors for their placement into lysosomes. These same changes in lysosomes likely influence the degradation of autophagosomes during autophagy. These data lead to the hypothesis that ethanol consumption suppresses autophagy (Donohue, 2009). Ethanol consumption inhibits the AMP kinase, an important regulator of catabolic pathways and which, when fully active, suppresses the activity of mTOR, which, in turn negatively regulates autophagosome formation. Because the AMP kinase is inhibited by ethanol metabolism, it is likely that mTOR suppresses autophagy in the alcoholic liver. However, there is evidence of AMPK-independent down regulation of the Akt/mTOR pathway. Additionally, ethanol metabolism disrupts protein and organelle trafficking in hepatocytes (Kharbanda et al. 1996). Because the intracellular movement of autophagic cargo requires a functional microtubule cytoskeleton, ethanol-induced impairment of trafficking probably slows autophagosome-lysosome fusion, thereby decreasing protein and lipid degradation.
B. Evidence of autophagosome induction in model systems of alcohol exposure
Alcohol-exposed VL-17A cells, which metabolize ethanol via alcohol dehydrogenase (ADH) and cytochrome P450 2E1 (CYP2E1) (Donohue 2006), exhibited 1.6- to 1.8-fold enhanced autophagosome content , compared with untreated cells. Ethanol-elicited elevation of autophagosome formation was blocked when ethanol oxidation in these cells was blocked with 4-methyl-pyrazole (4-MP In contrast, autophagosome content was unaffected in HepG2 cells, which do not oxidize ethanol.. The latter two findings clearly indicate that ethanol-induced autophagy depends on the generation of ethanol metabolites.
Similar to VL-17A cells, ethanol-triggered autophagy in primary hepatocytes caused a dose-dependent inhibition of mTOR, indicated by the reduction of both phosphorylated p70 S6 kinase and 4E-binding protein-1, two major downstream targets of mTOR (Ding et al. 2010). 4-MP treatment blocked mTOR inhibition by ethanol. Taken together, these findings support the notion that ethanol oxidation, by generating acetaldehyde and other reactive metabolites thereby suppressed mTOR activity and enhanced autophagosome content (Ding et al. 2010). Further, increased autophagosome content after acute ethanol administration is not associated with changes in long-lived protein degradation but selectively promotes the removal of lipid droplets and damaged mitochondria (Ding et al. 2011). These latter findings underscore the need for further studies to delineate the individual contributions of altered autophagy and proteasome inhibition to hepatic protein accumulation after chronic ethanol consumption.
C. Autophagy induction after alcohol abuse in vivo
-Acute alcohol exposure
In mice, acute gavage of 6 grams ethanol per kg body weight caused ~ 35% enhancement in the hepatic level of LC3 II protein 12 hours after gavage. The rise in autophagosome content coincided with a decline in reduced glutathione (GSH) and a significant elevation in lipid peroxides, both signs of oxidant stress (Donohue et. al., in press). Acute ethanol treatment induced a significant elevation of GFP-LC3 puncta in livers of transgenic GFP-LC3 mice, similarly indicating an enrichment of autophagosomes in the LAMP1-positive heavy membrane fraction, where lysosomes are located (Ding et al. 2010; Ding et al. 2011). Electron microscopic (EM) analysis also indicated an accumulation of autophagosomes in ethanol-exposed mice. This response is suppressed after knockdown of Atg7 expression by its siRNA (Ding et al. 2010). Further, pharmacological and genetic suppression of autophagy enhances ethanol-induced cell death not only in vitro in primary hepatocytes and VL-17A cells but also in intact mouse liver after acute ethanol-binge (Ding et al. 2010).
- Chronic alcohol exposure
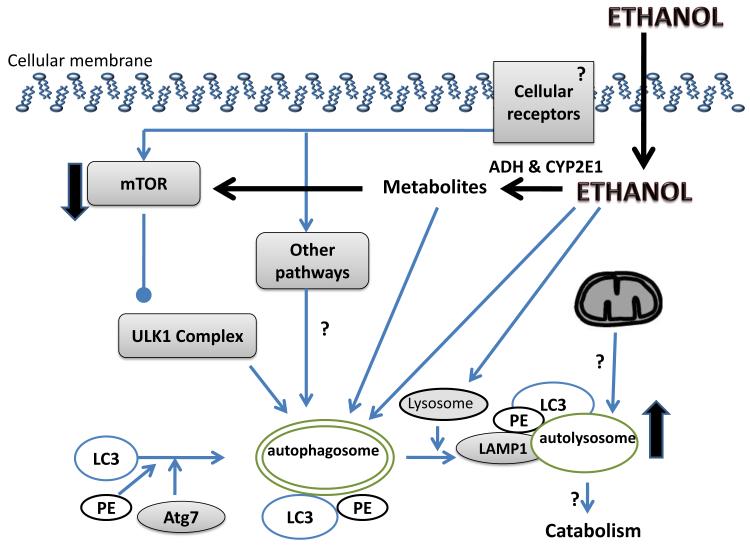
Transgenic GFP-LC3- mice fed control and ethanol-containing liquid diets (Lieber and DeCarli, 1986) for 6 weeks, exhibited hepatomegaly, steatosis and elevated liver protein, suggesting slower catabolism. Alcohol-fed mice had higher serum ALT content, indicating liver injury. Fluorescent microscopy analyses of hepatocytes of ethanol-fed mice revealed four-fold higher green fluorescent autophagosomes than in cells of control animals. These results were confirmed by immunoblot analyses of liver fractions of ethanol-fed mice showing a similar elevation in LC3II levels (Thomes in press 2011; Ding et al. 2010). CYP 2E1 activity and content are elevated in ethanol-fed mice, suggesting enhanced ethanol metabolism, which may have augmented autophagosome content. Hepatic proteasome activity was also suppressed in livers of ethanol-fed mice, thereby contributing to a portion of the net protein gain in the liver. There is evidence that chronic ethanol consumption also inhibits the degradative phase (i.e. autophagosome degradation by lysosomes) of autophagy, which could account, in part, for the rise in autophagosomes (Donohue and Thomes, unpublished). A summary of the known and hypothesized effects of alcohol exposure on autophagy in the liver is depicted in Figure 2.
Figure 2. The proposed mechanism of the alcohol-induced autophagy in the liver.
Cellular receptors trigger the autophagy following the classic 4 stages, as described in the text. Alcohol exposure triggers autophagy. Alcohol and/or its metabolites inhibit the mTOR pathway thus releasing its inhibition of autophagy. The effect of alcohol on protein in the final stages of autophagy, namely autophagosome-lysosome fusion, and on catabolism of cargo proteins is still unclear. The existence of alcohol-induced mitophagy is unknown. The effect of alcohol on receptor-initiated autophagy and their downstream signaling pathways remain largely unexplored. Known effects are depicted with lines and arrows; unknowns are depicted with “?” sign; the up- and down-regulation is depicted by the orientation of the arrows.
Cross talk between cell death and autophagy
Accumulating evidence indicate that autophagy and cell death mutually regulate each other. It is generally agreed that autophagy is a pro-survival mechanism by not only providing nutrients for the cell survival during starvation but also by selectively removing damaged organelles, including damaged mitochondria (Kim et al. 2011b). The relationship between autophagy and multiple types of cell deaths including necroptosis, necrosis, pyroptosis, apoptosis etc, is not fully understood however the interplay between apoptosis and autophagy is best researched to date. Because mitochondria play an essential role in regulating apoptosis by controlling the release of apoptotic factors, removing damaged mitochondria by autophagy (mitophagy) would thus be protective (Kim et al. 2007). Many apoptotic stimuli induce both apoptosis and autophagy at the same time, indicating that the cell’s fate is decided by the balance of autophagy versus the accumulation of damaged mitochondria. It should also be noted that induction of apoptosis can also suppress autophagy by cleaving essential autophagy proteins such as Beclin 1 (Luo et al. 2010, Li et al. 2011). Numerous studies indicate that pharmacologic or genetic inhibition of autophagy greatly enhances cell death (Ding et al. 2007a, Ding et al. 2007b; Komatsu et al. 2005). Liver- specific knockout of Atg7 leads to hepatomegaly and severe liver injury (Komatsu et al. 2005) and suppression of autophagy exacerbates alcohol-induced liver injury by increasing alcohol-induced apoptosis (Ding et al. 2010). Thus, in the absence of autophagy, cell death is significantly increased.
The role of pathogen-derived signals and toll-like receptors (TLRs) in liver autophagy
Toll-like receptors (TLRs) are pathogen-recognition receptors (PRR) that sense pathogen-associated molecular patterns (PAMP). Thirteen TLRs are known so far and each TLR is unique in terms of PAMP recognition and immune responses. TLRs 1,2,4,5, 6 and 11 home to the plasma membrane and recognize mainly pathogen-derived cell wall components, while TLRs 3, 7, 8 and 9 locate in intracellular compartments (endoplasmic reticulum (ER), endosomes, lysosomes and endolysosomes) and recognize nucleic acids.
Endotoxin, (lipopolysaccharide, LPS) of Gr(-) bacteria) levels are elevated in the peripheral circulation of alcohol-fed experimental animals and humans, presumably due to increased translocation from the leaky gut (Keshavarzian et al. 1999). Administration of endotoxin (LPS) to alcohol-fed mice increases their liver damage (Hritz et al. 2008). LPS is recognized by a complex receptor composed of lipopolysaccharide binding protein (LBP), MD-2, CD14 and TLR4. Lack of TLR4 prevents alcohol-induced liver injury and protects against endotoxin-induced liver inflammation (Hritz et al. 2008). Thus, TLR4 plays a key role in the development of alcohol-induced liver disease. LPS engages TLR4 and recruits TIRAP/MyD88 complex for early expression of pro-inflammatory gene products. Alternatively, TLR4 gives preference to TRIF/IRF3 pathway and results in induction of IFNs and late phase NFkB activation. More recent data suggest that TLR4 also triggers autophagy (Xu et al. 2008) in TRIF- , RIP1- and MAPK-dependent, but MyD88- and JNK-independent manner. LPS facilitates autophagic vesicle formation by promoting the incorporation of the PI3-Kinase VPS34 association with the membrane (Xu et al. 2008), and up-regulates LRG47, a GTPase that is involved in initiation of autophagy (MacMicking et al. 2003). Inhibition of TLR4 signaling by TRIF knockdown blocks LPS-induced autophagy (Xu et al. 2008) suggesting that TRIF-dependent TLR4 signaling plays a role in autophagy. However TLR7/8 ligands single-strand (ss)RNA and imiquimod also induce autophagy (Huang et al. 2010) and TLR7/8 function independent of TRIF, thus suggesting an alternative TRIF-independent TLR-triggered autophagy pathway. The common denominator of TLR4 and TLR7/8 receptors is their ability to induce type I IFN production (Kawai and Akira 2010). It is interesting that interferon-producing arm of the TLR4 signaling is key to producing alcohol-induced liver injury (Hritz et al. 2008, Petrasek et al., 2011). Thus the involvement of IFNs and the role of TLRs in alcohol abuse-induced liver autophagy remain to be further researched.
Autophagy and Mitochondria
Alcohol abuse causes functional impairment of mitochondria, including depression of mitochondrial protein synthesis, reduction of mitochondria-specific gene products, dissociation of mitochondrial ribosomes, impaired glutathione uptake, altered oxidative capacity and impaired ATP production (Cunningham et al. 1990). In liver, glucagon released from the pancreas during fasting promotes autophagy, whereas insulin released after feeding suppresses autophagy (Arstila and Trump, 1968). Such autophagy salvages amino acids and fatty acids that are essential for cell survival. Mitochondria are a particularly rich source of protein and lipid and are frequently the target of autophagic sequestration, a process called mitophagy (Lemasters, 2005). Moreover, autophagic elimination of aged and damaged mitochondria appears important to protect cells against mitochondrial release of pro-apoptotic proteins, generation of toxic reactive oxygen species (ROS) and futile hydrolysis of ATP after uncoupling (Lemasters, 2005; Mizushima et al., 2008).
i. Mitochondrial Turnover
In non-proliferating tissues including liver, the half-life of mitochondria is 10 to 25 days (Menzies and Gold, 1971). In this physiological turnover, mitophagy removes mitochondria in balance with biogenesis of new mitochondria. Mitophagy also has an important role in cytoplasmic remodeling. Nonetheless, both inadequate and excess mitophagy appear to promote cell injury and death (Mizushima et al., 2008; Lemasters, 1999). Accordingly, proper regulation of autophagy is fundamental for cellular homeostasis.
ii. Dynamics of Mitophagy
In hepatocytes in the absence of nutrient deprivation, GFP-LC3 is mostly diffuse but some is incorporated into mostly small (0.2-0.3 μm) pre-autophagic structures (PAS) in proximity to mitochondria. Nutrient deprivation and glucagon stimulate autophagy and PAS grow into cups (isolation membranes or phagophores) that envelop and sequester individual mitochondria into autophagosomal vesicles (mitophagosomes), a process that requires phosphatidylinositol 3-kinase (PI3K). Sequestration often occurs coordinately with mitochondrial fission and is completed within 6-7 min. After sequestration, mitochondria depolarize, and intravesicular acidification occurs. Mitochondrial digestion is complete within about 10 min (Kim and Lemasters, 2011a). During nutrient deprivation of hepatocytes, about 85% of autophagic events involve mitochondria.
iii. Organelle Selectivity of Autophagy
Several studies identify proteins that target mitochondria for selective mitophagy. In mammalian cells, Bnip3, parkin and Pink1, are implicated in targeting mitochondria for autophagy (Narendra et al., 2008; Dagda et al., 2009). BNIP3 (Bcl-2/adenovirus E1B 19-kDa-interacting protein 3) is a BH3-only protein that interacts with LC3 and also contributes to cell death through activation of the mitochondrial pathway of apoptosis. PINK1 (PTEN-induced putative kinase 1) and parkin, an E3 ubiquitin ligase, are both proteins whose mutations cause familial forms of Parkinson’s disease. Pink1 appears to recruit Parkin to the outer membrane of depolarized mitochondria; Parkin, in turn, ubiquitinates outer membrane proteins and thus targets mitochondria for autophagic sequestration. Other work identifies the mitochondrial permeability transition (MPT), a phenomenon making the inner membrane non-specifically permeable to solutes up to 1.5 kDa, in targeting mitochondria for autophagy (Elmore et al., 2001; Carreira et al., 2010). Thus, mitophagy in these models shows preference for damaged, dysfunctional mitochondria.
iv. Mitophagy and oxidative stress
Mitochondria are a primary site for formation of ROS. Consequently, mitochondrial DNA (mtDNA) is susceptible to oxidative damage. Since DNA repair mechanisms are limited, damaged mtDNA can accumulate over time, leading to mitochondrial dysfunction and phenotypes of aging (Wallace, 2010). As shown after PicoGreen labeling of mtDNA, mitophagy also degrades mtDNA (Kim and Lemasters, 2011a). Such mitophagy may be important in aging, since caloric restriction and rapamycin, inducers of autophagy, enhance longevity in rodents (Bergamini et al., 2003). Moreover, the process of autophagy declines with age, which may promote accumulation of mtDNA mutations and an age-related decline in mitochondrial function (Bergamini, 2003; Lemasters, 2005),
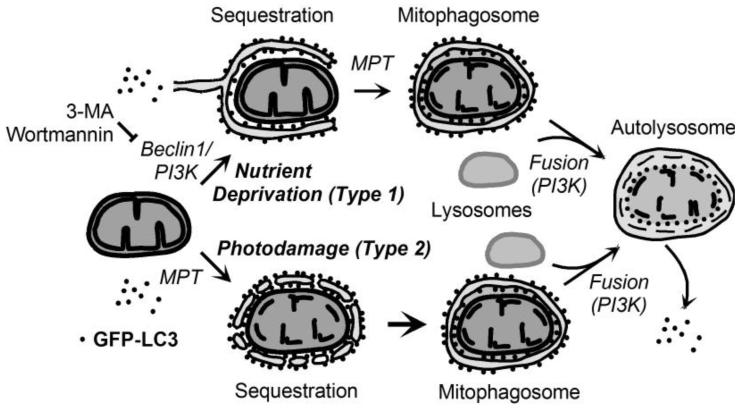
v. Type 1 and Type 2 Mitophagy
Mitophagy has two variants (Figure 3). Type 1 is exemplified by nutrient deprivation-induced mitophagy in which pre-autophagosomal structures (PAS) grow into cup-shaped phagophores that envelop and sequester mitochondria into mitophagosomes often in coordination with mitochondrial fission. Type 2 mitophagy is exemplified by photodamage, when the periphery of damaged mitochondria is decorated with aggregates of GFP-LC3 that then coalesce to sequester individual mitochondria into mitophagosomes. In Type 2 mitophagy, cup-shaped phagophores do not form. In both variants of mitophagy, mitophagosomes form, acidify, fuse with lysosomes and degrade their contents. In Type 1 mitophagy, Class III PI3K/p150 interacts with Beclin1, a homologue of yeast Atg6, to promote autophagosome formation. By contrast, in Type 2 autophagy, PI3K inhibition does not inhibit and actually augments GFP-LC3 association with damaged mitochondria (Kim and Lemasters, 2011b). A noteworthy difference between Type 1 and Type 2 mitophagy is that PI3K inhibition does not block Type 2 mitophagy (Kim and Lemasters, 2011b). Thus enhanced accumulation of mitophagosomes after PI3K inhibition in Type 2 mitophagy suggests that subsequent fusion of mitophagosomes with lysosomes requires PI3K, as shown for the processing of autophagosomes in nutrient deprivation-induced autophagy (Mousavi et al., 2003). Mitophagy, with its Type 1 and 2 characteristics, is largely understudied in alcohol-induced hepatic autophagy.
Figure 3. Scheme of Type 1 and Type 2 mitophagy.
In nutrient deprivation-induced (Type 1) mitophagy, activation of Beclin1/PI3K leads to formation of an GFP-LC3-labeled phagophore, which closes with onset of the MPT to form a mitophagosome. The mitophagosome so formed then fuses with lysosomes in a PI3K-dependent fashion to form an autolysosome where acidic digestion of the entrapped organelle occurs. In photodamage-induced (Type 2) mitophagy, photoirradiation through ROS generation causes MPT onset and mitochondrial depolarization. Membrane vesicles containing GFP-LC3 attach to the depolarized mitochondria and coalesce to form a mitophagosome in a PI3K-independent fashion. The mitophagosome is then processed in a fashion identical to the Type 1 pathway.
Summary and Conclusions
Alcohol up-regulates autophagy, as identified in vitro upon ethanol exposure of cultured hepatoma cells, or in vivo, by analysis of livers after acute and chronic alcohol administration to mice. These recent findings are inconsistent with the earlier hypothesis that ethanol suppresses autophagy (Donohue et al., 2009,) which was largely based on findings of slower protein degradation in livers of ethanol-fed animals owing to disruption of lysosome function and biogenesis (Kharbanda et al. 1996; Kharbanda, 1997; Donohue, 1994; Haorah et al. 2003; Haorah et al. 2002). This apparent disparity can, in part, be explained by the fact that autophagy investigations focused on the proximal marker of autophagy, namely autophagosome formation (stages 1 and 2), while protein degradation occurs during the later stages (3 and 4) of autophagy. In essence it is still possible that both effects occur upon ethanol abuse. Indeed, in alcohol abuse/LPS-induced pancreatitis an inhibition of pancreatic autophagy was reported (Fortunato et al. 2009). So far it is clear that the mechanism(s) by which EtOH induces autophagy is/are dependent on ethanol metabolism and oxidative stress. Yet another layer of complexity upon ethanol abuse is added by the fact that autophagy involves multiple pathways: when macroautophagy fails to function, it is complemented by chaperone-mediated autophagy, and likely microautophagy which act as accessory degradation pathways. The role of TLR-initiated events and the effects of gut-derived endotoxin on alcohol-induced autophagy are yet to be explored. Further research is needed to identify the role of mitophagy in alcoholic liver disease.
Future prospective.
Several immediate questions arise in the field of alcohol research in regards to:
alcohol components (alcohol per se or its metabolites) that cause autophagy in hepatocytes,
mechanistic details of the stimulatory effects of alcohol abuse on autophagy,
outcomes of autophagy in alcohol-induced liver disease,
long-term effects and implications of elevated autophagy in the alcohol-exposed liver.
However, the answers to these questions will largely depend on the progress in the area of basic research of autophagy. Identification of the fundamentals of autophagy is among priorities in this field of research; among them are:
discovery of human homologs of all 31 Atg proteins known so far in yeast and probably more autophagy players,
mechanisms of vesicle formation, sequestration and docking to the lysosomes,
the origin(s) of the lipid membranes of autophagic vesicles,
the nature and the mechanisms of selective types of autophagy including those of energy-producing or energy-demanding organelles.
In-depth studies of autophagy will spearhead our understanding of the molecular mechanism of alcohol-induced liver disease. Most important, the progress in this area will encourage our vision of autophagy-modulating agents for therapeutic purposes.
Acknowledgments
Grant support: Research in Dr Dolganiuc’s laboratory is funded by National Institute of Alcohol Abuse and Alcoholism (NIAAA) grants R21A016571 and R01AA017212. Dr Ding’s work is funded by National Institute of Health (NIH) grants R21AA017421, R01AA02518, P20RR021940, and P20RR016475 from the INBRE program of the National Center for Research Resources. Dr Lemasters’s work is funded by grants R01CA119079, R01DK37034, P01DK59340, R01DK073336 and C06RR015455 from the NIH. The work from Dr Donohue’s laboratory is supported by Dean’s Reviewed Research Grant from the University of Nebraska Medical Center.
References
- Arstila AU, Trump BF. Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration. Am J Pathol. 1968;53(5):687–733. [PMC free article] [PubMed] [Google Scholar]
- Baraona E, Leo MA, Borowsky SA, Lieber CS. Alcoholic hepatomegaly: accumulation of protein in the liver. Science. 1975;190(4216):794–5. doi: 10.1126/science.1198096. [DOI] [PubMed] [Google Scholar]
- Bardag-Gorce F, Li J, French BA, French SW. The effect of ethanol-induced CYP2E1 on proteasome activity: the role of 4-hydroxynonenal. Exp Mol Pathol. 2005;78(2):109–15. doi: 10.1016/j.yexmp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Bergamini E, Cavallini G, Donati A, Gori Z. The anti-ageing effects of caloric restriction may involve stimulation of macroautophagy and lysosomal degradation, and can be intensified pharmacologically. Biomed Pharmacother. 2003;57(5-6):203–8. doi: 10.1016/s0753-3322(03)00048-9. [DOI] [PubMed] [Google Scholar]
- Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem. 2004;279(20):20663–71. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira RS, Lee Y, Ghochani M, Gustafsson AB, Gottlieb RA. Cyclophilin D is required for mitochondrial removal by autophagy in cardiac cells. Autophagy. 2010;19:6. doi: 10.4161/auto.6.4.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC 1989 http://www.cdc.gov/mmwr/preview/mmwrhtml/00018761.htm.
- Cunningham CC, Coleman WB, Spach PI. The effects of chronic ethanol consumption on hepatic mitochondrial energy metabolism. Alcohol Alcohol. 1990;25(2-3):127–36. doi: 10.1093/oxfordjournals.alcalc.a044987. [DOI] [PubMed] [Google Scholar]
- Dagda RK, Cherra SJ, III, Kulich SM, Tandon A, Park D, Chu CT. Loss of pink1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 2009;284(20):13843–55. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Duve C. The significance of lysosomes in pathology and medicine. Proc Inst Med Chic. 1966;26(4):73–6. [PubMed] [Google Scholar]
- Ding WX, Ni HM, Gao W, Hou YF, Melan MA, Chen X, Stolz DB, Shao ZM, Yin XM. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007a;282:4702–10. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007b;171:513–24. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Li M, Chen X, Ni HM, Lin CW, Gao W, Lu B, Stolz DB, Clemens DL, Yin XM. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139(5):1740–52. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Li M, Yin XM. Selective taste of ethanol-induced autophagy for mitochondria and lipid droplets. Autophagy. 2011;7(2):248–249. doi: 10.4161/auto.7.2.14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Manley S, Ni HM. The emerging role of autophagy in alcoholic liver disease. Exp Biol Med (Maywood) 2011;236(5):546–56. doi: 10.1258/ebm.2011.010360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue TM, Jr, McVicker DL, Kharbanda KK, Chaisson ML, Zetterman RK. Ethanol administration alters the proteolytic activity of hepatic lysosomes. Alcohol Clin Exp Res. 1994;18(3):536–41. doi: 10.1111/j.1530-0277.1994.tb00906.x. [DOI] [PubMed] [Google Scholar]
- Donohue TM, Jr, Sorrell MF, Tuma DJ. Hepatic protein synthetic activity in vivo after ethanol administration. Alcohol Clin Exp Res. 1987;11(1):80–6. doi: 10.1111/j.1530-0277.1987.tb01267.x. [DOI] [PubMed] [Google Scholar]
- Donohue TM, Jr, Zetterman RK, Tuma DJ. Effect of chronic ethanol administration on protein catabolism in rat liver. Alcohol Clin Exp Res. 1989;13(1):49–57. doi: 10.1111/j.1530-0277.1989.tb00283.x. [DOI] [PubMed] [Google Scholar]
- Donohue TM, Osna NA, Clemens DL. Recombinant Hep G2 cells that express alcohol dehydrogenase and cytochrome P450 2E1 as a model of ethanol-elicited cytotoxicity. Int J Biochem Cell Biol. 2006;38(1):92–101. doi: 10.1016/j.biocel.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Donohue TM., Jr Autophagy and ethanol-induced liver injury. World J Gastroenterol. 2009;15(10):1178–85. doi: 10.3748/wjg.15.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger AL, Thompson CB. Defective autophagy leads to cancer. Cancer Cell. 2003;4(6):422–4. doi: 10.1016/s1535-6108(03)00306-4. [DOI] [PubMed] [Google Scholar]
- Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- Fortunato F, Bürgers H, Bergmann F, Rieger P, Büchler MW, Kroemer G, Werner J. Impaired autolysosome formation correlates with Lamp-2 depletion: role of apoptosis, autophagy, and necrosis in pancreatitis. Gastroenterology. 2009;137(1):350–60. doi: 10.1053/j.gastro.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Haorah J, MacDonald RG, Stoner JA, Donohue TM., Jr Ethanol consumption decreases the synthesis of the mannose 6-phosphate/insulin-like growth factor II receptor but does not decrease its messenger RNA. Biochem Pharmacol. 2003;65(4):637–48. doi: 10.1016/s0006-2952(02)01605-2. [DOI] [PubMed] [Google Scholar]
- Haorah J, McVicker DL, Byrd JC, MacDonald RG, Donohue TM., Jr Chronic ethanol administration decreases the ligand binding properties and the cellular content of the mannose 6-phosphate/insulin-like growth factor II receptor in rat hepatocytes. Biochem Pharmacol. 2002;63(7):1229–39. doi: 10.1016/s0006-2952(02)00877-8. [DOI] [PubMed] [Google Scholar]
- Hoek JB, Pastorino JG. Cellular signaling mechanisms in alcohol-induced liver damage. Semin Liver Dis. 2004;24(3):257–72. doi: 10.1055/s-2004-832939. [DOI] [PubMed] [Google Scholar]
- Høyer-Hansen M, Jäättelä M. AMP-activated protein kinase: a universal regulator of autophagy? Autophagy. 2007;3(4):381–3. doi: 10.4161/auto.4240. [DOI] [PubMed] [Google Scholar]
- Hritz I, Mandrekar P, Velayudham A, Catalano D, Dolganiuc A, Kodys K, Kurt-Jones E, Szabo G. The critical role of toll-like receptor (TLR) 4 in alcoholic liver disease is independent of the common TLR adapter MyD88. Hepatology. 2008;48(4):1224–31. doi: 10.1002/hep.22470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SW, Liu KT, Chang CC, Chen YJ, Wu CY, Tsai JJ, Lu WC, Wang YT, Liu CM, Shieh JJ. Imiquimod simultaneously induces autophagy and apoptosis in human basal cell carcinoma cells. Br J Dermatol. 2010;163(2):310–20. doi: 10.1111/j.1365-2133.2010.09827.x. [DOI] [PubMed] [Google Scholar]
- Keshavarzian A, Holmes EW, Patel M, Iber F, Fields JZ, Pethkar S. Leaky gut in alcoholic cirrhosis: a possible mechanism for alcohol-induced liver damage. Am J Gastroenterol. 1999;94(1):200–7. doi: 10.1111/j.1572-0241.1999.00797.x. [DOI] [PubMed] [Google Scholar]
- Kharbanda KK, McVicker DL, Zetterman RK, Donohue TM., Jr Ethanol consumption alters trafficking of lysosomal enzymes and affects the processing of procathepsin L in rat liver. Biochim Biophys Acta. 1996;1291(1):45–52. doi: 10.1016/0304-4165(96)00043-8. [DOI] [PubMed] [Google Scholar]
- Kharbanda KK, McVicker DL, Zetterman RK, MacDonald RG, Donohue TM., Jr Flow cytometric analysis of vesicular pH in rat hepatocytes after ethanol administration. Hepatology. 1997;26(4):929–34. doi: 10.1002/hep.510260419. [DOI] [PubMed] [Google Scholar]
- Kim I, Lemasters JJ. Mitochondrial degradation by autophagy (mitophagy) in GFP-LC3 transgenic hepatocytes during nutrient deprivation. Am. J. Physiol Cell Physiol. 2011a;300:C308–C317. doi: 10.1152/ajpcell.00056.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Lemasters JJ. Mitophagy selectively degrades individual damaged mitochondria after photoirradiation. Antioxid.Redox.Signal. 2011b;14(10):1919–28. doi: 10.1089/ars.2010.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462(2):245–53. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol. 2004;2(4):301–14. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang P, Sun Q, Ding WX, Yin XM, Sobol RW, Beer-Stolz D, Yu J, Zhang L. Following cytochrome c release, autophagy is inhibited during chemotherapy-induced apoptosis by caspase-8-mediated cleavage of Beclin-1. Cancer Res. 2011;71(10):3625–34. doi: 10.1158/0008-5472.CAN-10-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, Slingerland JM, Mills GB. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9(2):218–24. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- Liu HY, Han J, Cao SY, Hong T, Zhuo D, Shi J, Liu Z, Cao W. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J. Biol. Chem. 2009;284:31484–92. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Rubinsztein DC. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ. 2010;17:268–77. doi: 10.1038/cdd.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302(5645):654–9. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- Menzies RA, Gold PH. The turnover of mitochondria in a variety of tissues of young adult and aged rats. J. Biol. Chem. 1971;246:2425–2429. [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi SA, Brech A, Berg T, Kjeken R. Phosphoinositide 3-kinase regulates maturation of lysosomes in rat hepatocytes. Biochem. J. 2003;372:861–869. doi: 10.1042/BJ20021136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samari HR, Seglen PO. Inhibition of hepatocytic autophagy by adenosine, aminoimidazole-4-carboxamide riboside, and N6-mercaptopurine riboside. Evidence for involvement of amp-activated protein kinase. J Biol Chem. 1998;273(37):23758–63. doi: 10.1074/jbc.273.37.23758. [DOI] [PubMed] [Google Scholar]
- Schmelzle T, Beck T, Martin DE, Hall MN. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol Cell Biol. 2004;24(1):338–51. doi: 10.1128/MCB.24.1.338-351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306(5698):990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM, de Winde JH. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33(5):904–18. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- Thomes PG, Trambly CS, Thiele GM, Duryee MJ, Fox HS, Haorah J, Donohue TM., Jr Proteasome activity and autophagosome content in liver are reciprocally regulated by ethanol treatment. Biochem Biophys Res Commun. 2011 Nov 28; doi: 10.1016/j.bbrc.2011.11.097. 2011. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Vucicevic L, Misirkic M, Janjetovic K, Vilimanovich U, Sudar E, Isenovic E, Prica M, Harhaji-Trajkovic L, Kravic-Stevovic T, Bumbasirevic V, Trajkovic V. Compound C induces protective autophagy in cancer cells through AMPK inhibition-independent blockade of Akt/mTOR pathway. Autophagy. 2011;7(1):40–50. doi: 10.4161/auto.7.1.13883. [DOI] [PubMed] [Google Scholar]
- Yoon Y, Yi H. Liver cirrhosis in the United States, 1970-2007 surveillance report #88. 2010 NIAAA homepage. http://pubs.niaaa.nih.gov/publications/surveillance88/Cirr07.htm.
- Xu Y, Liu XD, Gong X, Eissa NT. Signaling pathway of autophagy associated with innate immunity. Autophagy. 2008;4(1):110–2. doi: 10.4161/auto.5225. [DOI] [PubMed] [Google Scholar]