Abstract
The human Y chromosome began to evolve from an autosome hundreds of millions of years ago, acquiring a sex-determining function and undergoing a series of inversions that suppressed crossing over with the X chromosome1,2. Little is known about the Y chromosome’s recent evolution because only the human Y chromosome has been fully sequenced. Prevailing theories hold that Y chromosomes evolve by gene loss, the pace of which slows over time, eventually leading to a paucity of genes, and stasis3,4. These theories have been buttressed by partial sequence data from newly emergent plant and animal Y chromosomes5-8, but they have not been tested in older, highly evolved Y chromosomes like that of humans. We therefore finished sequencing the male-specific region of the Y chromosome (MSY) in our closest living relative, the chimpanzee, achieving levels of accuracy and completion previously reached for the human MSY. We then compared the MSYs of the two species and found that they differ radically in sequence structure and gene content, implying rapid evolution during the past 6 million years. The chimpanzee MSY harbors twice as many massive palindromes as the human MSY, yet it has lost large fractions of the MSY protein-coding genes and gene families present in the last common ancestor. We suggest that the extraordinary divergence of the chimpanzee and human MSYs was driven by four synergistic factors: the MSY’s prominent role in sperm production, genetic hitchhiking effects in the absence of meiotic crossing over, frequent ectopic recombination within the MSY, and species differences in mating behavior. While genetic decay may be the principal dynamic in the evolution of newly emergent Y chromosomes, wholesale renovation is the paramount theme in the ongoing evolution of chimpanzee, human, and perhaps other older MSYs.
This effort required that we first complete the sequencing of the chimpanzee MSY9-11 using large-insert bacterial artificial chromosome (BAC) clones and the iterative mapping and sequencing strategy used to comprehensively sequence the human MSY12,13 (Supplementary Fig. 1 and Supplementary Note 1). This protracted approach is essential because much of the MSY consists of lengthy, highly similar repeat units, or “amplicons,” that cannot be distinguished by conventional mapping methods. BACs deriving from different copies of the same amplicon must be fully sequenced to identify subtle differences that distinguish one amplicon copy from another. These sequence variants are then used to sort other potentially overlapping BACs (Supplementary Fig. 2 and Supplementary File 1). By iterating this process, we assembled and analyzed a tiling path of 219 BAC and 12 fosmid clones from the chimpanzee MSY (Supplementary Fig. 1 and Supplementary Table 1). To avoid polymorphic differences between chimpanzee Y chromosomes that might confound identification of subtle sequence differences between amplicon copies12,13, we intended to sequence the Y chromosome of one male chimpanzee. In fact, all but 17 of the 230 sequenced BAC or fosmid clones were derived from one male.
The resulting euchromatic sequence comprises 25.8 megabases (Mb) in eight contigs, the largest of which spans 10.1 Mb (Supplementary Fig. 1, Supplementary Table 1, and Supplementary File 2). We ordered and oriented the contigs by metaphase fluorescence in situ hybridization (FISH) (Supplementary Fig. 3 and 4) and confirmed amplicon copy numbers by interphase FISH (Supplementary Fig. 5). To test the completeness of our effort, we also analyzed 14 Mb of shotgun sequencing reads from flow-sorted chimpanzee Y chromosomes. This independent sampling of the chromosome confirmed that our sequencing of the chimpanzee MSY was essentially complete (Supplementary Note 2). We estimate that the finished sequence has an error rate of about one nucleotide per Mb.
Our laboratories previously demonstrated that the human MSY euchromatin is largely comprised of two sequence classes: ampliconic and X-degenerate13. We find that the same two sequence classes dominate the chimpanzee MSY euchromatin (Fig. 1A, B), and thus the same was likely true in the common ancestor. The ampliconic segments are composed of large, nearly identical repeat units, most often arrayed as palindromes, and they harbor multi-copy gene families expressed predominantly or exclusively in the testis13 (Supplementary Fig. 6 and 7). By contrast, the X-degenerate segments are dotted with single-copy homologs of X-linked genes. These single-copy MSY genes, most of which are expressed ubiquitously, are surviving relics of ancient autosomes from which the X and Y chromosomes evolved2. Together, the ampliconic and X-degenerate sequences comprise the bulk of the MSY euchromatin in both chimpanzee and human (Fig. 1B). A third sequence class in the human MSY euchromatin – the X-transposed sequences – has no counterpart in the chimpanzee MSY. The presence of these sequences in the human MSY is the result of an X-to-Y transposition that occurred in the human lineage after its divergence from the chimpanzee lineage14.
Figure 1.
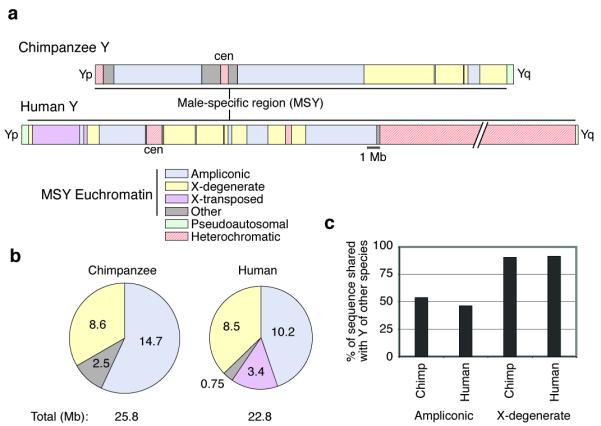
Comparison of chimpanzee and human Y chromosomes. (a) Schematic representations of chromosomes. Yp, short arm. cen, centromere. Yq, long arm. For both chromosomes, male-specific region (MSY) is indicated. Six sequence classes are shown, four of which are MSY euchromatin. (“Other” denotes MSY single-copy sequences that are not X-degenerate or X-transposed.) Chromosomes drawn to scale with exception of large heterochromatic block on human Yq. (b) Sizes (in Mb) of four MSY euchromatin sequence classes in chimpanzee and human. (c) Percentages of ampliconic and X-degenerate sequences present on chimpanzee Y chromosome that are also present on human Y chromosome, and vice versa.
Given that primate sex chromosomes are hundreds of millions of years old2, theories of decelerating decay would predict that the chimpanzee and human MSYs should have changed little since the separation of these two lineages just six million years ago. To test this prediction, we aligned and compared the nucleotide sequences of the chimpanzee and human MSYs (Supplementary File 3). As expected, we found that the degree of similarity between orthologous chimpanzee and human MSY sequences (98.3% nucleotide identity) differs only modestly from that reported when comparing the rest of the chimpanzee and human genomes (98.8%)15. Surprisingly, however, > 30% of chimpanzee MSY sequence has no homologous, alignable counterpart in the human MSY, and vice versa (Supplementary Fig. 8 and Supplementary Note 3). In this respect the MSY differs radically from the remainder of the genome, where < 2% of chimpanzee euchromatic sequence lacks an homologous, alignable counterpart in humans, and vice versa15. We conclude that, since the separation of the chimpanzee and human lineages, sequence gain and loss have been far more concentrated in the MSY than in the balance of the genome. Moreover, the MSY sequences retained in both lineages have been extraordinarily subject to rearrangement: whole-chromosome dot-plot comparison of chimpanzee and human MSYs reveals dramatic differences in gross structure (Fig. 2 and Supplementary Fig. 9), which contrasts starkly with chromosome 21, the only other chromosome comprehensively mapped and sequenced in both species16. Contrary to the decelerating decay theory, the chimpanzee and human MSYs differ dramatically in sequence structure.
Figure 2.
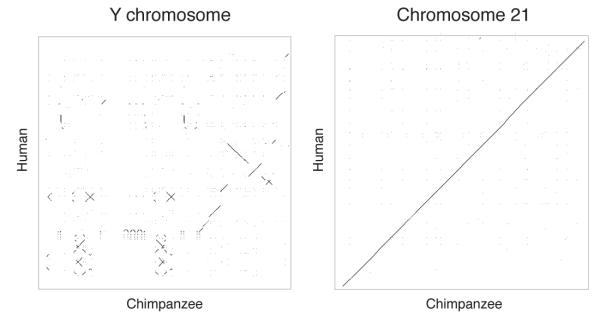
Dot plots of DNA sequence identity between chimpanzee and human Y chromosomes (at left) and chromosomes 21 (at right). Each dot represents 100% chimpanzee-human identity within a 200-bp window. In the Y-chromosome plot, the human chromosome is oriented with short arm to top and long arm to bottom, and the chimpanzee chromosome is oriented with short arm to left and long arm to right. For chromosome 21, which is acrocentric, the plot represents only the long arm.
Have these evolutionary changes involved the ampliconic and X-degenerate regions in equal measure? Previous models of Y-chromosome evolution treated the chromosome as a uniform, homogeneous substrate for evolutionary change1,3,4,17. In fact, the evolution of ampliconic sequences has outpaced that of X-degenerate sequences, and to such a degree that the ampliconic architecture of the common ancestor’s MSY may be difficult to reconstruct even after an outgroup MSY has been sequenced. Comparing the chimpanzee and human MSY sequences, we find that the average length of uninterrupted alignable segments in the ampliconic regions is only one third that in X-degenerate regions: 0.5 Mb vs. 1.5 Mb (Supplementary Fig. 10 and Supplementary Note 3). This reflects extensive rearrangement (Supplementary Fig. 8 and 9) and rampant sequence gain and loss in the ampliconic regions. About half of the chimpanzee ampliconic sequence has no homologous, alignable counterpart in the human MSY, and vice versa, compared to < 10% of the X-degenerate sequence (Fig. 1C).
What molecular mechanisms enabled this wholesale remodeling of ampliconic regions? Although the chimpanzee and human MSYs do not normally participate in meiotic exchange with a partner chromosome, the mirroring of sequences in the ampliconic regions provides ample opportunity for ectopic homologous recombination within the MSY. This recombinational proclivity is well documented in the human MSY, where it has repeatedly given rise to large-scale structural polymorphisms during the past 100,000 years of human history18 as well as to Y-chromosomal anomalies that cause spermatogenic failure and sex reversal in current generations12,19-21. We suggest that ectopic homologous recombination between MSY amplicons has similarly accelerated structural remodeling of the MSY in the chimpanzee and human lineages during the past 6 million years.
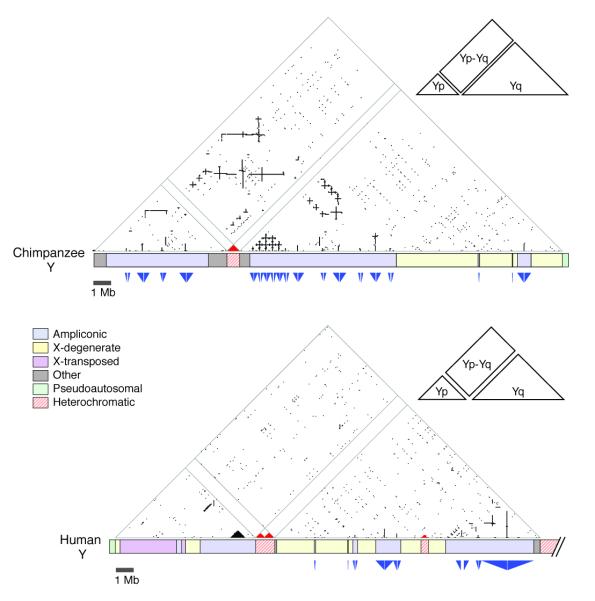
The chimpanzee ampliconic regions are particularly massive (44% larger than in human; Fig. 1B) and architecturally ornate, with 19 palindromes (compared to eight in human) and elaborate mirroring of nucleotide sequences between the short and long arms of the chromosome, a feature not found in the human MSY (Fig. 3, Supplementary Fig. 11). Of the 19 chimpanzee palindromes, only seven are also found in the human MSY; the other 12 are chimpanzee-specific. Unlike the human MSY, nearly all of the chimpanzee MSY palindromes exist in multiple copies (Supplementary Fig. 11), so that each palindrome arm has potential partners for both intra-palindrome and inter-palindrome gene conversion (non-reciprocal transfer)22. This may help explain why arm-to-arm nucleotide sequence divergence in some chimpanzee MSY palindromes (as much as 0.5%; Supplementary Fig. 12 and 13) is more pronounced than in human MSY palindromes (< 0.06%)13.
Figure 3.
Triangular dot plots of DNA sequence identities within euchromatic MSY of chimpanzee (top) and human (bottom). Each dot represents 100% intrachromosomal identity within a 200-bp window. Red dots represent matches between heterochromatic sequences. Direct repeats appear as horizontal lines, inverted repeats as vertical lines, and palindromes as vertical lines that nearly intersect the baseline. Insets indicate that each large triangular plot contains two smaller triangles (one revealing sequence identities within Yp and one revealing identities within Yq) and a rectangle (revealing sequence identities between Yp and Yq). Immediately below plots are schematic representations of chromosomes. Triangles below chromosome schematics denote sizes and locations of palindromes. Gaps between opposed triangles represent the non-duplicated spacers between palindrome arms.
Gene conversion may also account for the relatively low density of retrotransposable elements in ampliconic regions. In the chimpanzee and human MSYs, retrotransposon content is markedly lower in ampliconic than in X-degenerate regions – 41% vs. 63% in both species (p < 0.000001, Z test; Supplementary Fig 1I and Supplementary Table 2). While it is possible that retrotransposons preferentially integrate in X-degenerate sequences, this seems unlikely given the similarity in C+G content and gene density in ampliconic and X-degenerate regions (Supplementary Table 2 and Supplementary Fig. 1H). An alternative explanation is that gene conversion between amplicon copies removes retrotransposons, especially recently integrated ones. Tellingly, an endogenous retrovirus that colonized the chimpanzee genome after the chimpanzee-human split23 is present in 23 copies in the chimpanzee MSY, but only two of these copies are located in ampliconic regions (14.7 Mb) while 21 copies are located in X-degenerate regions (8.6 Mb; p < 0.000001, chi-square test; Supplementary Fig. 14). These findings offer counterpoint to models of unchecked retrotransposon integration as a driving force in Y-chromosome evolution17,24.
Despite the chimpanzee MSY’s elaborate structure, its gene repertoire is considerably smaller and simpler than that of the human MSY (Table 1) as a result of gene loss in the chimpanzee lineage and gene acquisition in the human lineage. For example, we previously discovered that the chimpanzee X-degenerate regions had lost four of 16 genes through inactivating mutations, while the human X-degenerate regions had not lost any genes since the time of the last common ancestor9. We also reported that two X-transposed genes in the human MSY had been acquired since the time of the last common ancestor13.
Table 1.
Comparison of numbers of X-degenerate genes and ampliconic gene families in chimpanzee and human Y chromosomes
| Numbers of intact copies | Difference | |||
|---|---|---|---|---|
| Sequence class | Gene | Human | Chimpanzee | (Human – Chimp) |
| X-degenerate | AMELY | 1 | 1 | - |
| CYorf15A | 1 | 1 | - | |
| CYorf15B | 1 | - | 1 | |
| DDX3Y | 1 | 1 | - | |
| EIF1AY | 1 | 1 | - | |
| KDM5D | 1 | 1 | - | |
| NLGN4Y | 1 | 1 | - | |
| PRKY | 1 | 1 | - | |
| RPS4Y1 | 1 | 1 | - | |
| RPS4Y2 | 1 | 1 | - | |
| SRY | 1 | 1 | - | |
| TBL1Y | 1 | - | 1 | |
| TMSB4Y | 1 | - | 1 | |
| USP9Y | 1 | - | 1 | |
| UTY | 1 | 1 | - | |
| ZFY | 1 | 1 | - | |
| Total | 16 | 12 | 4 | |
| Ampliconic | BPY2 | 3 | 2 | 1 |
| CDY | 4 | 5 | -1 | |
| DAZ | 4 | 4 | - | |
| HSFY | 2 | - | 2 | |
| PRY | 2 | - | 2 | |
| RBMY | 6 | 6 | - | |
| TSPY | ~35 | 6 | 29 | |
| VCY | 2 | 2 | - | |
| XKRY | 2 | - | 2 | |
| Total | 60 | 25 | 35 | |
| X-transposed | TGIF2LY | 1 | - | 1 |
| PCDH11Y | 1 | - | 1 | |
| Grand total (genes) | 78 | 37 | 41 | |
| (53%) | ||||
| Grand total (gene families) | 27 | 18 | 7 | |
| (33%) | ||||
To investigate whether the gene content of the ampliconic regions differs in chimpanzee and human, we searched the chimpanzee MSY sequence for homologs of all known human ampliconic genes, and we assessed their open reading frames, splice sites, and transcriptional activity electronically and experimentally (Supplementary Tables 3-5 and Supplementary Fig. 7). In addition, we searched for novel chimpanzee ampliconic genes using a combination of electronic prediction and shotgun sequencing (>38 Mb total) of chimpanzee testis cDNA. We found no novel chimpanzee ampliconic genes. We did discover that, within the ampliconic regions, three of nine multi-copy, testis-expressed gene families present in human have been mutationally disabled or are simply absent in chimpanzee (Table 1). For example, the chimpanzee MSY contains five loci homologous to the human XKRY gene family, but all five copies share a frameshift mutation that severely truncates the open reading frame and predicted protein (Supplementary Table 3). We confirmed the presence of this disabling mutation in five additional chimpanzees and two bonobos, close relatives of common chimpanzees (data not shown). Similarly, the HSFY and PRY gene families are well represented in the human MSY but absent from the chimpanzee MSY. While it is unclear whether the PRY family was gained in the human lineage or lost in the chimpanzee lineage, the presence of HSFY in the cat25, rhesus macaque and bull MSYs (H.S., personal communication) leads us to conclude that this gene family was deleted outright in the chimpanzee lineage.
In aggregate, the consequence of gene loss and gain in, respectively, the chimpanzee and human lineages is that the chimpanzee MSY contains only two thirds as many distinct genes or gene families as the human MSY, and only half as many protein-coding transcription units (Table 1). By contrast, in the remainder of the genome, comparison of chimpanzee draft sequence with human reference sequence suggests that the gene content of the two species differs by < 1% (ref. 15). Indeed, at six million years of separation, the difference in MSY gene content in chimpanzee and human is more comparable to the difference in autosomal gene content in chicken and human, at 310 million years of separation26.
We have conducted the first comprehensive comparison of Y chromosomes from two species, providing empirical insight into Y-chromosome evolution and a test of decelerating-decay theories. These theories elegantly account for the degeneration observed in neo-Y chromosomes recently evolved from autosomes3-8. However, they did not predict and cannot account for the rapid divergence of the older, highly evolved chimpanzee and human MSYs described here. Instead, remodelling and regeneration have dominated chimpanzee and human MSY evolution during the past six million years. We suggest that this renovation, involving both architecture and genetic repertoire, was propelled by a combination of factors acting in synergy. Three of these factors distinguished the evolving hominid MSY from the bulk of the genome: 1) the highly disproportionate role of MSY genes – especially ampliconic gene families – in sperm production13, 2) the brisk kinetics of ectopic recombination and resultant structural change in ampliconic regions18, and 3) the absence of crossing over with a homolog, which creates the opportunity for a single advantageous mutation to dictate the MSY’s evolutionary fate (“genetic hitchhiking”)1,3. The evolutionary impact of these three MSY features was likely multiplied by sperm competition, especially in the lineage of the modern chimpanzee, where multiple males mate with the same female at each oestrus27. This heightened sperm competition in the chimpanzee lineage, along with positive selection and hitchhiking effects, may account for greater MSY sequence amplification than in the human MSY and extensive gene loss compared with little or none in the human MSY. In the future, complete Y chromosome sequences from additional species will shed further light on these hypotheses.
Methods
BAC selection and sequencing
The iterative mapping and sequencing strategy12 was employed to assemble a path of sequenced clones selected from the CHORI-251 and RPCI-43 BAC libraries and the CHORI-1251 fosmid library (bacpac.chori.org). The rate of error in the finished sequence was estimated by counting mismatches in overlapping clones.
454 sequencing of flow-sorted Y chromosomes and testis cDNA
Chromosomes were harvested from a lymphoblastoid cell line (Coriell repository number S00600, derived from the same chimpanzee used to construct the CHORI-251 BAC and CHORI-1251 fosmid libraries), prepared as described28, and sorted to enrich for Y chromosomes using an Influx cell sorter. The resulting Y-enriched DNA sample was amplified using the GenomiPhi amplification kit (GE Healthcare) to obtain enough template (>1 ug) for 454 sequencing on a GS20 machine. Chimpanzee testis cDNA was generated from total RNA isolated using the RNeasy kit (Qiagen). The cDNA was normalized using the Trimmer kit (Evrogen) and sequenced on a GS20 (454) machine.
FISH analysis
All assays were performed on the chimpanzee lymphoblastoid cell line S00600. Interphase FISH analysis was performed as previously described29. For each probe set, 200 nuclei were scored. Extended metaphase FISH was performed as previously described30.
Sequence analysis, dot plots, and alignments
Chimpanzee and human gene sequences were aligned using CLUSTAL W with default parameters (www.clustal.org). The search for novel chimpanzee Y-chromosome genes was performed using GenomeScan (genes.mit.edu/genomescan.html). Square dot plot and triangular dot plot analyses were performed using custom Perl codes that are available at, respectively, jura.wi.mit.edu/page/papers/Hughes_et_all_2005/tables/dot_plot.pl and jura.wi.mit.edu/page/Y/azfc/self_dot_plot.pl.
RT-PCR
Total RNAs were isolated from male chimpanzee male tissues (testis, liver, lung, and spleen; Yerkes National Primate Research Center) using the RNeasy kit (Qiagen). RT-PCR primer sequences and product sizes are listed in Supplementary Table 5.
Supplementary Material
Acknowledgements
We thank L. Brown, V. Frazzoni, G. Rogers, and A. Berger for technical assistance; and L. Brown, M. Carmell, G. Dokshin, M. Gill, J. Mueller, K. Romer and Y. Soh for comments on the manuscript. Supported by the National Institutes of Health and the Howard Hughes Medical Institute.
Footnotes
Author Contributions J.F.H., H.S., W.W., S.Rozen, R.K.W., and D.C.P. planned the project. J.F.H., H.S., and T.P. performed BAC mapping and RT-PCR analysis. T.A.G., P.J.M., R.S.F., and S.D.M. were responsible for finished BAC sequencing. J.F.H. and H.S. performed comparative sequence analyses. S.K.D. and S.Repping performed FISH analyses. C.F. and B.J.T. flow-sorted chromosomes. D.P.L. and E.R.M. carried out 454 sequencing. J.F.H. and D.C.P. wrote the paper.
Author Information cDNA sequences of chimpanzee ampliconic genes have been deposited in GenBank (www.ncbi.nlm.nih.gov) under accession numbers AY958084 (BPY2), AY958081 (CDY), AY958083 (DAZ), AY995201/2 (RBMY), AY958082 (TSPY), and AY995203 (XKRY). Chimpanzee testis cDNA sequences have been deposited in GenBank under accession numbers EX654441 through EX654483.
Reprints and permissions information is available at npg.nature.com/reprintsandpermission. The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature.
Supplementary Information accompanies this paper.
References
- 1.Rice WR. Evolution of the Y sex chromosome in animals. Bioscience. 1996;46:331–343. [Google Scholar]
- 2.Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 3.Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachtrog D. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics. 2008;179:1513–1525. doi: 10.1534/genetics.107.084012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filatov DA, Moneger F, Negrutiu I, Charlesworth D. Low variability in a Y-linked plant gene and its implications for Y-chromosome evolution. Nature. 2000;404:388–390. doi: 10.1038/35006057. [DOI] [PubMed] [Google Scholar]
- 6.Bachtrog D, Charlesworth B. Reduced adaptation of a non-recombining neo-Y chromosome. Nature. 2002;416:323–326. doi: 10.1038/416323a. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, et al. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature. 2004;427:348–352. doi: 10.1038/nature02228. [DOI] [PubMed] [Google Scholar]
- 8.Peichel CL, et al. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 2004;14:1416–1424. doi: 10.1016/j.cub.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 9.Hughes JF, et al. Conservation of Y-linked genes during human evolution revealed by comparative sequencing in chimpanzee. Nature. 2005;437:100–103. doi: 10.1038/nature04101. [DOI] [PubMed] [Google Scholar]
- 10.Kuroki Y, et al. Comparative analysis of chimpanzee and human Y chromosomes unveils complex evolutionary pathway. Nature Genet. 2006;38:158–167. doi: 10.1038/ng1729. [DOI] [PubMed] [Google Scholar]
- 11.Hughes JF, et al. Has the chimpanzee Y chromosome been sequenced? Nature Genet. 2006;38:853–854. doi: 10.1038/ng0806-853b. [DOI] [PubMed] [Google Scholar]
- 12.Kuroda-Kawaguchi T, et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nature Genet. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- 13.Skaletsky H, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 14.Page DC, Harper ME, Love J, Botstein D. Occurrence of a transposition from the X-chromosome long arm to the Y-chromosome short arm during human evolution. Nature. 1984;311:119–123. doi: 10.1038/311119a0. [DOI] [PubMed] [Google Scholar]
- 15.The Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe H, et al. DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature. 2004;429:382–388. doi: 10.1038/nature02564. [DOI] [PubMed] [Google Scholar]
- 17.Charlesworth B. The evolution of sex chromosomes. Science. 1991;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- 18.Repping S, et al. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nature Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 19.Repping S, et al. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am. J. Hum. Genet. 2002;71:906–922. doi: 10.1086/342928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Repping S, et al. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nature Genet. 2003;35:247–251. doi: 10.1038/ng1250. [DOI] [PubMed] [Google Scholar]
- 21.Lange J, et al. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell. 2009;138:855–869. doi: 10.1016/j.cell.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rozen S, et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- 23.Yohn CT, et al. Lineage-specific expansions of retroviral insertions within the genomes of African great apes but not humans and orangutans. PLoS Biol. 2005;3:e110. doi: 10.1371/journal.pbio.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinemann S, Steinemann M. Retroelements: tools for sex chromosome evolution. Cytogenet. Genome Res. 2005;110:134–143. doi: 10.1159/000084945. [DOI] [PubMed] [Google Scholar]
- 25.Murphy WJ, et al. Novel gene acquisition on carnivore Y chromosomes. PLoS Genet. 2006;2:e43. doi: 10.1371/journal.pgen.0020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hillier LW, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 27.Dixson AF. Primate Sexuality: Comparative Studies of the Prosimians, Monkeys, Apes and Human Beings. University of Chicago Press; Chicago: 1998. [Google Scholar]
- 28.Mefford HC, et al. Comparative sequencing of a multicopy subtelomeric region containing olfactory receptor genes reveals multiple interactions between non-homologous chromosomes. Hum. Mol. Genet. 2001;10:2363–2372. doi: 10.1093/hmg/10.21.2363. [DOI] [PubMed] [Google Scholar]
- 29.Saxena R, et al. Four DAZ genes in two clusters found in the AZFc region of the human Y chromosome. Genomics. 2000;67:256–267. doi: 10.1006/geno.2000.6260. [DOI] [PubMed] [Google Scholar]
- 30.Yeh RF, Lim LP, Burge CB. Computational inference of homologous gene structures in the human genome. Genome Res. 2001;11:803–816. doi: 10.1101/gr.175701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.