Summary
To test whether mechanisms controlling the range of diversity of the developing antibody repertoire in C57BL/6 mice (IgHb) operate similarly to those identified in BALB/c mice (IgHa), we compared the sequences of VH7183-containing H-chain transcripts from sorted adult bone marrow C57BL/6 B-cell subsets with those previously obtained from BALB/c mice. Patterns of VDJ gene segment utilization and CDR-H3 amino acid composition, charge, and average length in C57BL/6 pro-B cells were similar, although not identical, to BALB/c pro-B cells. However, C57BL/6 mature, recirculating B cells failed to demonstrate the reduction in the use of VH81X and the narrowing in the range of variance of CDR-H3 hydrophobicity that characterizes B-cell maturation in BALB/c mice. To further test the ability of the C57BL/6 strain to discard B cells expressing highly charged CDR-H3s, we introduced a mutant IgHa DH allele that forces use of arginine, asparagine and histidine. Unlike BALB/c mice, C57BL/6 mice congenic for the charged DH maintained normal numbers of mature, recirculating B cells that were enriched for charged CDR-H3s. Together; these findings indicate that the mature C57BL/6 B-cell pool permits expression of immunoglobulins with antigen binding sites that are typically discarded during late stage bone marrow B-cell development in BALB/c mice.
Keywords: Antibodies, B cells, Repertoire Development, Rodent
Introduction
The ability to create a diverse immunoglobulin repertoire permits the immune system to produce specific responses to a broad range of ancient and novel antigens [1, 2]. Each individual immunoglobulin is produced by a complex series of V(D)J gene rearrangement events. V(D)J rearrangement is hierarchical, typically beginning with heavy (H) chain DH→JH joining followed by VH→DJH and then light (L) chain VL→JL recombination. B-cell development is marked by passage through successive checkpoints for function. Early checkpoints test the structure of the immunoglobulin products, whereas later ones evaluate antigen-binding properties. The site at which immunoglobulin typically binds antigen is created by the juxtaposition of three hypervariable loops from the H chain and three from the L chain. Of these six loops, termed complementary determining regions (CDRs) [3], the most diverse is CDR-H3 because it is created de novo by V(D)J gene recombination and N addition [1, 2, 4]. CDR-H3 is located at the center of the antigen-binding site where it often plays a critical role in defining antibody specificity [5–7].
In order to gain insight into the mechanisms used to regulate the formation of the antibody repertoire [8]; we previously analyzed the pattern of CDR-H3 repertoire development in the bone marrow of BALB/c mice. We found that constraints on length, amino acid composition and hydrophobicity could readily be identified in pro-B cells and reflected germline sequence imposed constraints on VDJ diversity. Passage through successive checkpoint stages appeared to accentuate these constraints, with enhancement of amino acid preferences and a decrease in the variance of the distribution of lengths and average hydrophobicities.
Although many classic studies of the immune response have been performed using BALB/c mice [9, 10], the sequencing of the C57BL/6 genome and the creation of multiple gene-altered C57BL/6 variants has made it a favored strain for immunologic studies. In part, this preference for the use of C57BL/6 mice also reflects its seemingly reduced resistance to the production of anti-dsDNA antibodies when certain autoimmune susceptibility alleles are introduced [11, 12]. One notable characteristic of these pathogenic anti-dsDNA autoantibodies is the frequent presence of arginine in their antigen binding sites [13].
By evaluating the composition of VH7183-containing H chain transcripts as a function of B-cell development in the bone marrow, we sought to test whether the natural (germline) and somatic (clonal selection) mechanisms used to regulate the composition of the BALB/c antibody repertoire, which is the product of the IgHa H chain allele, were operating to the same extent and outcome in C57BL/6 mice, which carry the IgHb H chain allele. C57BL/6 IgHb differs from BALB/c IgHa in VH, DH and JH gene numbers and sequences [14].
Our comparative study revealed that the constraints on initial VDJ gene segment utilization, amino acid composition, charge, and average CDR-H3 length as observed in C57BL/6 pro-B cells were similar, although not identical, to the constraints introduced by germline VDJ sequence in BALB/c pro-B cells. However, examination of the mature, recirculating B-cell pool in C57BL/6 wild-type and DH-altered mice suggests that the somatic mechanisms of clonal selection that act to focus the repertoire by reducing the variance in CDR-H3 length and hydrophobicity in BALB/c mice appear to operate differently in C57BL/6 mice, permitting increased expression of antigen binding sites enriched for hydrophobic and charged CDR-H3s, including those enriched for arginine residues.
Results
Isolation of B-cell subsets and cloning of H-chain transcripts
We used a combination of the schemes of Melchers [15] and Hardy [16] to sort bone marrow B lineage cells into progenitor (B), early (C) and late (D) precursor, immature (E), and mature (F) B-cell fractions. We then sequenced and analyzed the composition of cloned VH7183DJCμ transcripts expressed in these cells, with a focus on CDR-H3. We chose to study mRNA transcripts as most representative of the expressed, and thus functional, Ig repertoire. We focused on the VH7183 family because it represents a manageable component of the active repertoire, because we and others had previously established patterns of VH7183 utilization during ontogeny and development in BALB/c mice, and because VH7183 gene segments have been shown to be components of antibodies with both self and non-self reactivities (reviewed in [8]).
A total of 577 unique, in-frame, open transcript sequences were obtained including 72 from B (pro-B), 133 from C (early pre-B), 75 from D (late pre-B), 78 from E (immature B), and 219 from F (mature, recirculating B).
Failure to counter select use of the VH81X gene segment in C57BL/6 mature B-cells
The C57BL/6 mouse genome contains only nine VH7183 family gene segments with open reading frames (Figure 1), or ~half that of the BALB/c mouse genome. Of these nine, only seven were identified in our sample of bone marrow transcripts (Figure 2). As in the case of BALB/c mice, the usage of the C57BL/6 VH81X (IGHV05-2, IMGT) gene segment declined four-fold during early B-cell development (28% in B versus 7% in D, p=0.0008). However, unlike BALB/c mice where there was a further five-fold late stage reduction between fractions D or E to F (p<0.02), in C57BL/6 mice the prevalence of VH81X usage did not change between fractions D, E and F (Figure 2).
Figure 1. A comparison of the complement of VH7183, DH and JH gene segments in C57BL/6 and BALB/c mice.
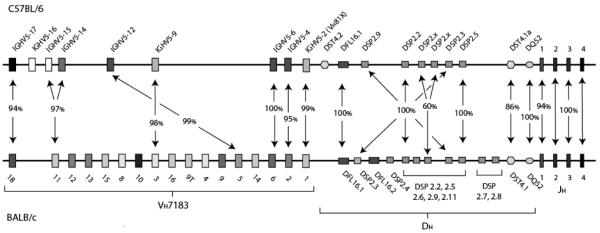
The germline complement of BALB/c VH7183 is based on the report of Feeney and colleagues [30]. The germline complement of DH gene segments is depicted as reported by Kurosawa and Tonegawa [31] and modified by Feeney and Riblet [32]. The precise order of DSP2.2, 2.5, 2.6, 2.9 and 2.11; 2.7 and 2.8 are undetermined [31]. The order and content of the C57BL/6 locus is based on Genbank accession number AC090887 [33]. The closest homologues by amino acid sequence are linked by arrows. Also shown is the percent of amino acid sequence identity between the homologues.
Figure 2. VH 7183 gene segment usage during B-cell development in C57BL/6 8 week bone marrow.
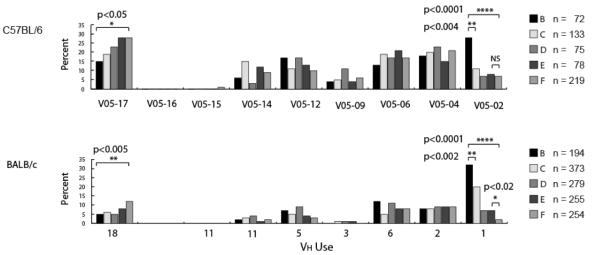
VH gene segments are arranged according to their position relative to the JH locus in the genome. IGHV05-2 (VH81X), the most JH proximal functional gene segment, is at the right. The percent of unique, in-frame sequences using the specified VH gene segment in adult C57BL/6 bone marrow from Hardy fractions B and C Melchers equivalents and from Hardy fractions D, E, and F (Table 1) are displayed and compared with BALB/c bone marrow. The data are obtained from 2 sorts with 2 mice each for a total of 4 mice. Cells from each mouse were not pooled. The number of unique sequences from each fraction is noted with 577 total unique sequences reported. Differences were evaluated using Student's t test. Significance is illustrated as *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p < 0.0001.
Increasing dominance of IGHV05-17 with development
The most JH distal VH, IGHV05-17 (IMGT), exhibited a doubling of usage in the transition from pro B cell to immature B cell and beyond (B→ F, p<0.05), ultimately contributing to almost one-third of the VH7183-containing transcripts from the mature B-cell pool (Figure 2). The closest BALB/c VH7183 homologue to IGHV05-17, VH7183.18, exhibited a similar increase in usage with development, but contributed to only 10% of the final repertoire. Use of the remaining five C57BL/6 VH gene segments did not vary statistically with development, also following the same pattern as their BALB/c homologues (Figure 1). However, the VH gene segment most commonly used in BALB/c mice at all stages of development, VH7183.10, has no C57BL/6 VH7183 homologue; and thus its structure and binding activity is missing in C57BL/6 mice.
Increased use of the DH DFL16.1 gene segment in C57BL/6 mice
Significant differences in the complement of DH gene segments were observed between C57BL/6 and BALB/c mice. The C57BL/6 genome has only one DFL family gene segment, two DST family gene segments and six DSP family gene segments; whereas the BALB/c genome has two DFL family gene segments, one DST family gene segment and nine DSP family gene segments. Both strains of mice had a single DQ52 gene segment that was conserved in sequence. In total, therefore, the C57BL/6 genome contains three fewer functional D gene segments than the BALB/c genome (Figure 1).
If DH usage were primarily a function of gene number, one might expect C57BL/6 mice to halve their use of the DFL family and double the use of DST family. However, while use of the DST family did increase, use of the single DFL gene segment in C57BL/6 mice increased to match the combined usage of the two DFL gene segments in BALB/c mice. Indeed, use of this single DFL gene segment matched the usage of the combined complement of all six C57BL/6 DSP gene segments (Figure 3). The percentage of sequences whose DH progenitor could not be identified (NoD) due to exonucleolytic nibbling of the D and N addition was also more prominent in C57BL/6 fraction B, when compared to BALB/c fraction B (p<0.02). However, the usage of the developmentally regulated DQ52 gene segment in these young adult C57BL/6 mice was essentially the same as in BALB/c mice (Figure 3).
Figure 3. DH, JH, and DH reading frame usage during B-cell development in C57BL/6 vs BALB/c 8-week bone marrow.
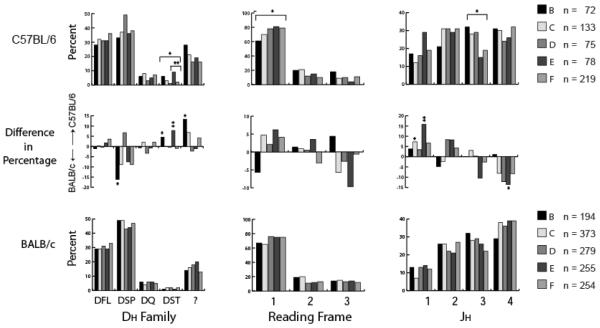
The percent of C57BL/6 sequences using members of the specified DH family (top left); the percent of sequences using DSP or DFL DH gene segment family members in reading frames 1, 2, or 3 (top middle); and percent of sequences using JH1, 2, 3, or 4 displayed by the specified developmental stage (top right) are shown. An equivalent analysis of sequences cloned from 8-week-old BABL/c bone marrow is displayed (bottom, [20]). The divergence in the percentage of DH gene family, DSP and DFL (top), reading frame (middle), and JH between C57BL/6 and BALB/c bone marrow (right) is displayed (middle row). The sequences are drawn from the data set reported in Figure 2. Differences were evaluated using chi square or Fisher's exact test. Significance values are marked as reported in Figure 2.
A decrease in the use of reading frame 2 in the transition from fractions B/C to D
In previous studies of BALB/c B lineage cells [8], we observed a stair-step increase in the use of RF1, which tends to express neutral amino acids including tyrosine, serine and glycine, versus RF2, which expresses hydrophobic amino acids including valine, among CDR-H3 sequences as B lineage cells transition from the progenitor (fraction B) stage to the late pre-B (fraction D) stage (67% RF1, 19% RF2 versus 76% RF1, 11% RF2; p<0.002) (Figure 3). A similar stair-step shift was observed in C57BL/6 B lineage cells (p<0.01) with reading frame 1 usage increasing from 61% in B to 78% in D and reading frame 2 decreasing from 20% to 12% respectively. Thus, both the genetic and somatic mechanisms regulating reading frame choice appeared to be operating similarly in the developing B-cells of these two mouse strains.
A more random use of JH in developing C57BL/6 B-cells
A directional rank order of JH utilization is commonly observed in developing BALB/c B-cells, with increasing usage among JH gene segments that are increasingly distal to the DH locus. This rank order was much less apparent in developing C57BL/6 B-cells. Use of JH1 appeared increased and use of JH4 decreased when compared with that in BALB/c mice (Figure 3). These differences achieved statistical significance for JH1 in Fractions C and E (p<0.05 and p<0.003 respectively); and for JH4 in Fraction E (p<0.04).
An increase in average CDR-H3 length with B-cell development
A key feature of repertoire development in BALB/c mice is an incremental increase in the average length of CDR-H3 with B lineage maturation. A similar increase, statistically indistinguishable from that of BALB/c B lineage cells, was observed in C57BL/6 B lineage cells with an average CDR-H3 length of 11.7 ± 0.3 amino acids in fraction B increasing to 12.3 ± 0.2 in fraction F (p=0.05) (Fig. 4a).
Figure 4. Average CDR-H3 length and CDR-H3 loop charge as a function of B-cell development in the bone marrow of adult C57BL/6 mice compared with that of adult BALB/c mice.
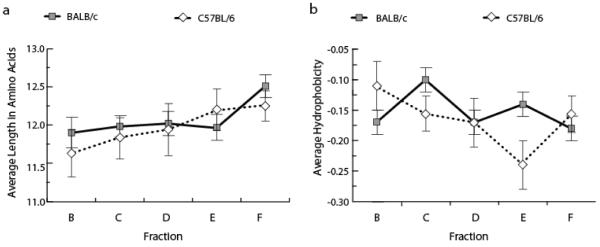
(A) Average CDR-H3 length and (B) average CDR-H3 loop hydrophobicity of the 577 VH7183DJCμ transcripts from C57BL/6 and 1355 BALB/c adult bone marrow transcripts [20] by developmental stage. The sequences are drawn from the data set reported in Figure 2. Data are shown as mean + SEM of the sequences obtained from 4 mice pooled from 2 sorts with 2 mice each. Chi square or Fisher's exact test was used for statistical analysis.
In BALB/c B lineage cells [8], the increase in length from fraction B to fraction F reflected, in part, a reduction in the prevalence of sequences whose CDR-H3 length was less than nine amino acids (Fig. 5). Due to the larger number of sequences available for analysis, this phenomenon was best observed in a comparison between fraction C and F. Of the 192 sequences in fraction C, 24 encoded CDR-H3 of eight amino acids or less (13%); whereas only three of 109 sequences (3%) were eight amino acids or less in fraction F (p < 0.01) [8]. This also led to a significant narrowing in the variance of the distribution of lengths (p = 0.01, Levene's test).
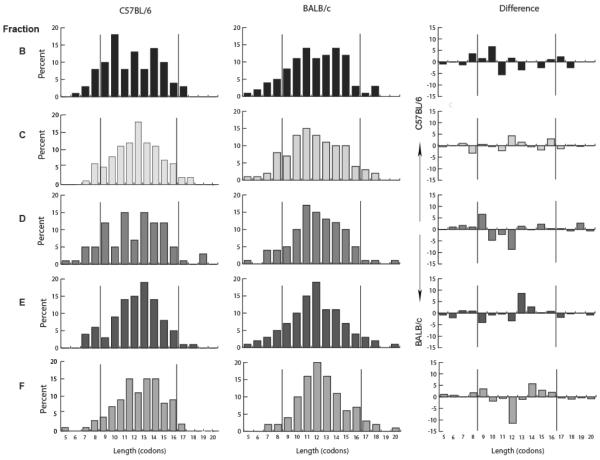
Figure 5. Distribution of CDR-H3 lengths as a function of B-cell development in the bone marrow of adult C57BL/6 as compared with adult BALB/c mice.
Distribution of CDR-H3 lengths in VH7183DJCμ transcripts from adult C57BL/6 bone marrow is shown (left). Distribution of CDR-H3 lengths in VH7183 DJCμ transcripts from adult BALB/c bone marrow as a function of B-cell development [20] is shown (middle). Divergence in the distribution of CDR-H3 length between C57BL/6 and BALB/c bone marrow B lineage cells is displayed (right). To facilitate visualization of the change in variance of the distribution, the vertical lines mark the preferred range of lengths in the bone marrow fraction F. The sequences are drawn from the data set reported in Figure 2. Chi square or Fisher's exact test was used for statistical analysis.
In C57BL/6 B lineage cells, we did not observe a narrowing of the variance in CDR-H3 length with development (p=0.55, Levene's test). Among 133 C57BL/6 fraction C sequences, 16 (12%) were eight amino acids or less; and among 219 fraction F 19 (9%) of sequences exhibit a similar range of short lengths (p=0.81).
A closer examination revealed that the greatest single contributor to the increase in lengths in CDR-H3s of the more mature C57BL/6 B lineage populations was the increase in the use of the single DFL gene segment, DFL16.1, with B-cell development (DFL16.1 is six nucleotides longer than DSP and DST gene segments and 12 nucleotides longer than DQ52). Although there were some slight differences in the extent of N addition and in terminal DH nibbling, none of these achieved statistical significance. In contrast, in BALB/c B lineage cells the increase in the distribution of lengths between fraction B and fraction F reflected increased use of JH4, which is longer. This increase in JH4 usage did not occur in C57BL/6 B lineage cells.
Increased use of serine and asparagine in CDR-H3 loops due to alterations in DH content and usage
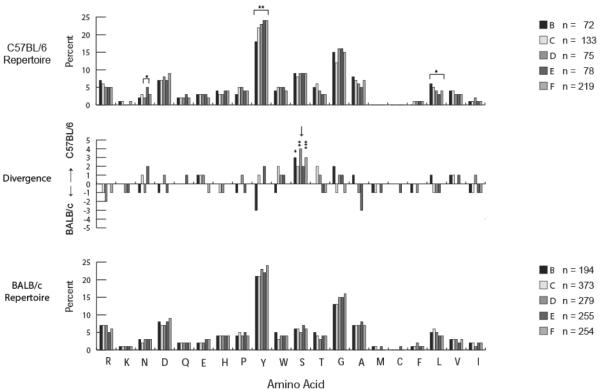
C57BL/6 B lineage cells demonstrated the same preference for tyrosine and glycine in CDR-H3 loops as BALB/c cells (Figure 6); and the use of tyrosine and glycine increased with maturation as in BALB/c bone marrow. However, the C57BL/6 CDR-H3 loop amino acid repertoire differed from the BALB/c repertoire in its increased use of serine and of asparagine. For example, serine contributed to 10% of the total amino acids in C57BL/6 fraction F CDR-H3 loops versus only 6% in BALB/c fraction F CDR-H3 (p=0.0002) [8]. Use of serine in C57BL/6 B lineage cells was also increased in fractions B (p<0.03) and D (p<0.002). These changes reflected the increased use of the DFL16.1 gene segment [17] and the contribution of a variant DSP gene segment, DSP2.x, which is not present in the BALB/c genome. None of the DSP sequences in the BALB/c genome encode serine in RF1, with DSP2.11 in the BALB/c genome, the closest homologue to DSP2.x in the C57BL/6 genome, reading Tyr Tyr Arg Tyr Asp, in RF1. In the C57BL/6 genome, RF1 of DSP 2.x reads Tyr Tyr Ser Asn Tyr, increasing the use of both serine and asparagine.
Figure 6. Amino acid usage as a function of B-cell development in the bone marrow of C57BL/6 as compared with that of BALB/c mice.
The percentage of amino acid use in the CDR-H3 loop of open reading frame, unique sequences from C57BL/6 (top) and BALB/c (bottom) [20]. The divergence in percentage of amino acid use in CDR-H3 between the strains is shown (middle). The sequences are drawn from the data set reported in Figure 2. Chi square or Fisher's exact test was used for statistical analysis. Significance values are marked as reported in Figure 2. Arrows point to features of particular interest.
Failure to reduce the variance of CDR-H3 hydrophobicity with development
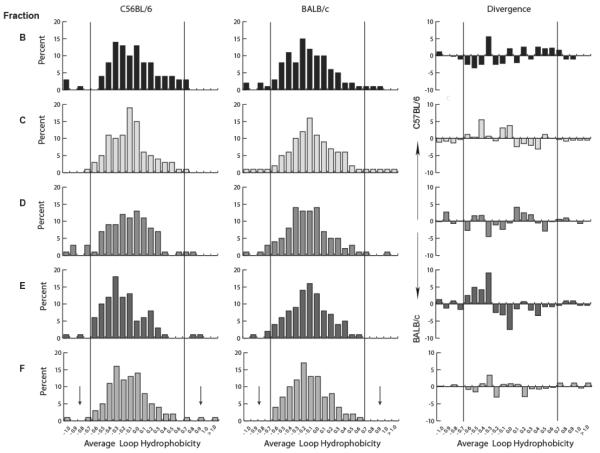
A second prominent feature of repertoire development in BALB/c B lineage cells is the slow, progressive reduction in the variance of average hydrophobicities of the repertoire with development [8]. This shift in variance in the BALB/c CDR-H3 repertoire is most apparent in a comparison between fractions C and F (p<0.01, Levene's test) (Figure 4b). This shift reflects, in part, a decrease in the prevalence of both highly hydrophobic and highly charged sequences among fraction F CDR-H3s when compared to fraction C (Figure 7). For example, 3.8% of BALB/c fraction C CDR-H3 loop sequences were highly hydrophobic (average hydrophobicity >0.6 by Kyte-Doolittle hydrophobicity scale) and 4.6% were highly charged (average hydrophobicity ≤ −0.7); but only 0.39% of fraction F sequences were highly hydrophobic (p=0.006) and 0.39% of fraction F sequences were highly charged when using the same comparison points (p<0.0001) [18].
Figure 7. Distribution of CDR-H3 loop charge as a function of B-cell development in the bone marrow of C57BL/6 as compared with that of BALB/c mice.
Distribution of average CDR-H3 hydrophobicities in VH7183DJCμ transcripts from C57BL/6 bone marrow as a function of B-cell development is shown (left) Distribution of average CDR-H3 hydrophobicities in VH7183DJCμ transcripts from BALB/c bone marrow as a function of B-cell development [20] is shown (middle). Divergence in the distribution of CDR-H3 loop hydrophobicity between C57BL/6 and BALB/c bone marrow B lineage cells is displayed (right). The normalized Kyte-Doolittle hydrophobicity scale [34] has been used to calculate average hydrophobicity. Although this scale ranges from −1.3 to + 1.7, only the range from − 1.0 (charged) to +1.0 (hydrophobic) is shown. Prevalence is reported as the percent of the sequenced population of unique, in-frame, open transcripts from each B lineage fraction. To facilitate visualization of the change in variance of the distribution, the vertical lines mark the preferred range of average hydrophobicity previously observed in wild-type fraction F [8]. The sequences are drawn from the data set reported in Figure 2. Arrows point to features of particular interest.
In contrast, in the developing C57BL/6 CDR-H3 repertoire there was no decrease in variance between fractions C and F (p=0.50, Levene's test) (Figure 4b). Only two of 133 fraction C sequences (1.5%) were highly hydrophobic and five (3.8%) were highly charged; whereas in fraction F seven of 217 CDR-H3 loops (3.2%) were highly hydrophobic (p=0.49) and five of 217 (2.3%) were highly charged (p=0.54). Indeed, the prevalence of highly hydrophobic sequences appeared increased. When compared directly between strains, the increased prevalence of highly hydrophobic CDR-H3s in C57BL/6 mature, recirculating B-cells versus BALB/c mature, recirculating B-cells proved significant (p=0.04). Highly charged CDR-H3 loops were also more prevalent C57BL/6 in mature, recirculating B cells versus BALB/c mature, recirculating B cells, although statistical significance was not achieved with this sample size (p=0.09) (Figure 7). Taken as a whole, the difference between the average charge of all CDR-H3 loops from C57BL/6 Fraction E compared with those from BALB/c Fraction E achieved significance at p=0.02 (Figure 4b), indicating an altered pattern of selection at that developmental stage, as well. Together these findings raised the possibility that the failure of the C57BL/6 mature, recirculating B-cell pool to reduce the variance in average hydrophobicity in the transition from pre-B to mature B-cell stage might reflect greater tolerance or increased survival of developing B cells bearing IgM B-cell receptors with disfavored highly hydrophobic or highly charged CDR-H3s, or both.
Mature B cells forcibly enriched for charged CDR-H3s are better tolerated in C57BL/6 bone marrow
To test the hypothesis that C57BL/6 bone marrow might be more tolerant of producing B cells bearing IgM with charged CDR-H3 loops, including those enriched for arginine, than BALB/c bone marrow; we performed a 22 generation backcrossing into C57BL/6 of an IgHa locus allele, ΔD-iD, which magnifies both the charge and arginine content of the CDR-H3 loops. B-cell progenitors using the ΔD-iD IgHa allele undergo VDJ recombination, pass through all the typical checkpoints of B-cell development and can also undergo class switching. In BALB/c mice, use of the ΔD-iD allele creates a polyclonal repertoire displaying a gradient or more highly charged and arginine-enriched CDR-H3s. These types of antibodies are present in the normal wild-type repertoire, but can be difficult to study due to their very low prevalence [19].
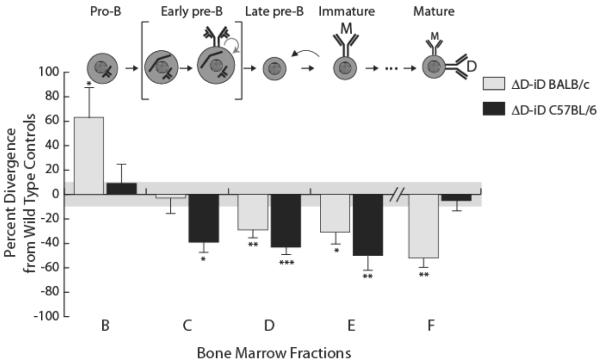
We evaluated the average absolute number of B lineage cells by developmental stage in a cohort of homozygous C57BL/6 ΔD-iD female mice, and compared these numbers with those obtained from a companion cohort of wild type C57BL/6 female littermate controls, as well as to companion historical studies in BALB/c wild-type and ΔD-iD female mice (Figure 8, Supporting Information Figure 1). Among developing C57BL/6 ΔD-iD B cells, a nearly similar number of pro-B (Hardy fraction B-equivalent) cells was followed by a significant decrease of the early pre-B (Hardy fraction C-equivalent) population (p=0.02) when compared to C57BL/6 wild type mice. The late pre-B (fraction D) and immature B (fraction E) compartments had a ~40% and ~50% decrease in numbers when compared to wild type controls (p<0.001 and p=0.002, respectively). This pattern of reduction in cell numbers matched that which we had previously observed at comparable stages of B-cell development on a BALB/c background [19]. However, unlike BALB/c IgHa.ΔD-iD mice where the absolute numbers of mature fraction F B cells in the bone marrow is halved when compared with those of wild-type; in C57BL/6 IgHa.ΔD-iD mice the absolute numbers of fraction F B cells was fully normalized when compared with those from wild-type C57BL/6 control mice (p=0.67) (Table 1).
Figure 8. Divergence in the absolute numbers of B lineage subpopulations from the bone marrow of homozygous ΔD-iD mice relative to their littermate C57BL/6 and BALB/c controls.
Percent loss or gain in homozygous ΔD-iD mice relative to their specific wild type littermate controls in the average absolute number of cells in either Melchers equivalents for bone marrow fractions B and C for C57BL/6 (Table 1) or Hardy fractions B and C [20]; as well as Hardy fractions D, E and F (Table 1). The standard error of the mean of each B lineage subpopulation for the littermate controls averaged approximately 10% of the absolute number of cells in each subpopulation (gray area). Data represent an analysis of 10 mice per group. Student's t test was used for statistical analysis. Error bars depict the standard error of the mean. Significance values are marked as reported in Figure 2.
Table 1.
Cell numbers in bone marrow of normal and mutant C57BL/6 mice
| Fractionc) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Bone Marrowa) | N | Total Cells × 106 | CD19+ × 108 | B × 104 | C × 105 | D × 105 | E × 105 | F × 105 |
| wt | 10 | 17.1 (0.7)b) | 4.0 (0.3) | 2.2 (0.2) | 9.1 (1.1) | 19.9 (1.9) | 5.7 (1.0) | 8.1 (0.6) |
| Δ D-iD | 10 | 12.0 (0.3)***d) | 2.8 (0.1)*** | 2.2 (0.3) | 5.3 (0.4)* | 13.1(0.9)*** | 3.0 (0.4)** | 7.5 (0.6) |
Values shown are cell counts per femur (average cellularity of two femurs collected from each experimental animal) of paired 8-week-old homozygous ΔD-iD (δD-iD) or homozygous wild-type (wt) DH C57BL/6 littermates, with each study representing analysis of the mean of 10 mice each, performed at the same time and conditions.
The mean standard error is provided in parenthesis.
The number of cells in pro B cells which are equivalent to fraction B (B220+ c-KIT+ CD25− BP-1−), early pre B cells equivalent to fraction C (B220+ c-KIT− CD25+ BP-1+), Hardy fractions D (CD19+ CD43− IgM− IgD−), E (CD19+ CD43− IgM+ IgD−), and F (CD19+ CD43− IgMlo IgDhi) was determined from the relative proportion of total cells.
Differences between cell numbers were assessed by Student's t test. Significance is illustrated as
p ≤ 0.05,
p ≤ 0.01
p < 0.0001 versus C57BL/6 wild-type littermates.
In order to distinguish between normalization of mature B-cell numbers due to the enhanced prevalence of B cells bearing IgM with charged, arginine-enriched CDR-H3s versus selection and increased survival for mature B cells that bear IgM with a more neutral CDR-H3 repertoire that could result from DH inversion or increased N addition (potential somatic selection for `normality'); we evaluated 52 in-frame VDJCμ transcripts isolated from C57BL/6 ΔD-iD bone marrow fraction F B cells (Supporting Information Table 2). This permitted direct comparisons between the CDR-H3 loops of fraction F B cells using the same IgHa.ΔD-iD allele, but differing by C57BL/6 versus BALB/c genetic background.
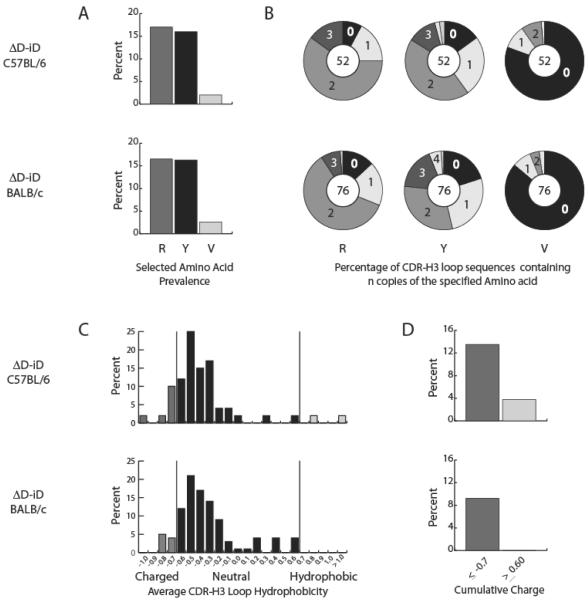
The pattern of reading frame usage, the prevalence of sequences lacking identifiable DH sequence, and the prevalence of N addition was statistically indistinguishable between the IgHa.ΔD-iD repertoires expressed by the two mouse strains. Additionally, both the global prevalence of arginine, tyrosine and valine in CDR-H3 and the relative distribution of CDR-H3 sequences containing one or more of these representative amino acids were statistically indistinguishable (Figure 9A, 9B). The prevalence of neutral CDR-H3 loop sequences did not increase. To the contrary, the prevalence of highly charged and highly hydrophobic CDR-H3 loops in fraction F on the C57BL/6 background proved higher than on the BALB/c background (12.5% vs 9.2% and 3.8% vs 0; respectively) (Figure 9C, 9D). We conclude that the normalization of IgHa.ΔD-iD fraction F B-cell numbers in C57BL/6 mice reflected an increase in the numbers of mature, recirculating cells bearing both highly charged, arginine-enriched CDR-H3 loops and highly hydrophobic CDR-H3 loops (derived from alternative reading frames) when compared with those in BALB/c mice.
Figure 9. Comparison of the usage of selected amino acids and the distribution of hydrophobicity of CDR-H3 in mature, bone marrow B cells from congenic C57BL/6 IgHa.ΔD-iD and BALB/c IgHa.ΔD-iD mice.
(A) The percentage of arginine (R), tyrosine (Y) and valine (V) use in the CDR-H3 loop of open reading frame, unique sequences compared with that of BALB/c congenics. (B) The percentage of CDR-H3 loop sequences containing `n' copies of the specified amino acids in the CDR-H3 loop of open reading frame, unique sequences compared with that of BALB/c congenics. (C) The distribution of charge in the CDR-H3 loop of open reading frame, unique sequences compared with that of BALB/c IgHa.ΔD-iD congenics. (D) The prevalence of highly charged and highly hydrophobic (average Kyte-Doolittle hydrophobicity index value [34] of ≤ −0.700 or > 0.600 respectively). Data shown are from two mice and were performed on the same sorting machine but at different time intervals. The cells from each mouse were not pooled. The original sequences are provided in Supporting Information Table 2.
Discussion
Although the potential diversity of the CDR-H3 component of the immunoglobulin H chain repertoire is astronomical, previous evaluation of the developing repertoire in BALB/c mice has allowed us and others to identify several key elements where there is strong evidence of either developmental or ontological constraints on this diversity (reviewed in [20]). These include differential patterns of V, D and J gene segment usage; the extent of N addition, P junctions and terminal nucleotide loss; the non-random use of DH reading frame; the non-random distribution of amino acids in the CDR-H3 loop; and the close focusing of the distribution of CDR-H3 hydrophobicity. These previous studies in BALB/c mice suggested that the initial constraints regulating CDR-H3 content reflected germline sequence content; i.e., the product of natural selection. Superimposed upon these germline restrictions in diversity were a series of somatic, presumably clonal, selective events that sequentially produced a CDR-H3 repertoire that had undergone `trimming' of apparently `disfavored' sequence content. This process included a reduction in the use of a specific VH gene segment, VH81X; a reduction in the use of very short CDR-H3s; enhanced use of reading frame 1; enhanced use of tyrosine and glycine in the CDR-H3 loop; and a sequential elimination of highly charged or heavily hydrophobic CDR-H3s with development.
The present analysis of immunoglobulin repertoire development in the bone marrow of C57BL/6 mice again demonstrates the effects of germline-imposed restrictions on the range of initial diversity in the H chain repertoire; but would point to significant differences in either the efficiency, the ability or the direction of the late stage somatic, clonal selective events in the bone marrow and the periphery. The end result is a mature, recirculating B-cell repertoire characterized by including IgM BCRs that bear antigen-binding sites that seemed to be not just `disfavored', but commonly `discarded' by the mature, recirculating B cells in BALB/c mice.
At the progenitor B-cell stage, the influence of the germline on the C57BL/6 repertoire is obvious. VH, DH and JH usage in C57BL/6 H-chain transcripts appears to differ from BALB/c H-chain transcripts both due to changes in number as well as the sequence of homologous gene segments. Germline variation appeared to be associated with changes in VDJ rearrangement frequency, although this latter point needs to be confirmed through the analysis of non-functional sequences. Among the features of the C57BL/6 repertoire that most closely matched the BALB/c repertoire were similarities in the initial distribution of N addition, lengths, charge and the usage of 18 of the 20 different potential amino acids. One of the features that varied between the two strains reflected the diminished number of functional VH gene segments, including the absence of the most commonly used VH in the BALB/c genome, VH7183.10. Others included the enhanced use of serine-enriched DFL16.1; the presence of a DSP2.11 homologue, DSP2.x, that encodes serine in RF1; and an increased use of JH1 in place of JH4. Of these changes, the most apparent effect in early B-cell progenitors was on VH content, again due to the absence of many of the VH7183 variant sequences available to BALB/c mice.
The differences between VH and DH gene segment numbers and sequence in the C57BL/6 and BALB/c genomes lead to an alteration in the representation of VH-specific CDR-H1 and –H2 structures and induce a DH-specific enrichment for serine and asparagine in the CDR-H3 loop. It is theoretically possible that the differences in the prevalence of non-neutral CDR-H3s observed in the mature, recirculating B-cell pool reflect the changes in the complement of VH in C57BL/6 B-cells when compared to BALB/c B-cells. However, in previous studies of BALB/c mice we have shown that changes in the global repertoire of CDR-H3 due to changes in DH content had no effect on VH utilization [17, 19, 21]. Thus, this possibility seemed less likely in C57BL/6 mice.
One of the first, critical somatic, clonal selective steps in repertoire development depends on the interaction between the H chain and the surrogate light chain λ5 and VpreB [22, 23]. Successful passage through this checkpoint permits early pre-B fraction C cells to clonally expand and then transition to the late pre-B-cell fraction D stage at which light chain rearrangement occurs. Most of the selective influences that we had observed in developing BALB/c B lineage cells during this transition were also apparent in developing C57BL/6 B lineage cells. This included a decline in the use of VH81X, a decrease in the use of DH RF2 with a compensatory increase in the use of RF1 and a stabilization of average length and average charge [8]. The latter two values in particular were indistinguishable between BALB/c fraction D and C57BL/6 fraction D (Figure 4), suggesting that both mouse strains share similar preference for mechanistic regulation at the step where the interaction between the nascent heavy chain and the surrogate light chain components determine the efficiency of pre BCR formation. For reasons unknown, BALB/c mice carrying the μMT mutation are leaky and can produce some B cells while C57BL/6 mice with the same mutation are not leaky and do not produce B cells suggesting a different timing in the B-cell generation process [24]. Thus it is possible that differences in the timing of Dμ protein or pre-B-cell receptor expression between the two strains could have a downstream effect on repertoire development.
A second selective step is the testing of the reactivity of the nascent IgM in fraction E. Failure at this step can lead to receptor editing, anergy, or cell death, reducing the likelihood of entry or survival of cells bearing `disfavored' IgM in the fraction F pool. Nussensweig and colleagues have clearly demonstrated that this step selects against potentially pathogenic self-reactivity [25]. CDR-H3 sequences obtained from C57BL/6 fraction E cells showed a significant difference in the average hydrophobicity compared to BALB/c fraction E cells suggesting a difference in the intensity or consequences of self-antigen recognition at that stage between the two strains (Figure 4b).
Given the apparent increased susceptibility to the production of anti-dsDNA antibodies by C57BL/6 mice [11], it is remarkable that the reduction in variance of CDR-H3 hydrophobicity that occurs during bone marrow B-cell development in BALB/c mice was much less apparent in C57BL/6 mice. Indeed the mature recirculating B-cell pool in C57BL/6 mice appeared to be retaining both highly hydrophobic and highly charged CDR-H3 sequences. We have previously shown that selection against these types of sequences can be thwarted, to a certain extent, by forcing increased bone marrow production of charged or hydrophobic CDR-H3s [20]. In BALB/c mice, late selective steps appear to ameliorate the effect of the change in the repertoire by reducing the number of B cells that have reached the final maturation step in the bone marrow. This clearly does not occur in C57BL/6 mice, as evidenced by significant increase in hydrophobic CDR-H3 bearing sequences in fraction F B cells as well as the inability of C57BL/6 IgHa.ΔD-iD mice to reduce the numbers of fraction F B cells with highly charged, arginine enriched CDR-H3s when compared with BALB/c IgHa.ΔD-iD mice and wild-type controls (Figures 8 and 9). This apparent inability to efficiently perform late-stage somatic, clonal selection against `disfavored' sequence occurs in parallel with the apparent inability of C57BL/6 wild-type mice to reduce the use of the VH81X gene segment in the transition from fraction E to fraction F. Differences in mechanism could include differences in receptor editing in fraction E, or differences in the consequences of antigen receptor influenced signaling after exposure to antigen in the periphery. These and other mechanisms are currently being studied in our laboratory. Whether or not the difference in the outcome of late stage selection is contributing to the increased propensity of C57BL/6 to produce potentially pathogenic auto-reactive antibodies [26] is unclear. However, as analogous to the comparison of the auto-immune prone C57BL/6 strain to the auto-immune resistant BALB/c strain, previous studies comparing MRL mice to their sister, autoimmune-resistant C3H strain have demonstrated a similar lack of control in the auto-immune prone MRL strain [27]. In either case, it appears that while the C57BL/6 VH7183 repertoire contains reduced diversity of CDR-H1 and CDR-H2 due to decreased numbers of functional VH gene segments, there is increased diversity of CDR-H3 due to altered patterns of somatic selection. This appears to permit mature, recirculating C57BL/6 B-cells to create a subset of antibodies within their repertoire with antigen binding sites that are considerably less common, and potentially even non-existent, in mature, recirculating BALB/c B-cells. The role of these differences in creating a propensity for self-reactivity or other alterations in the immune response is a focus of ongoing investigations in our laboratory.
Materials and Methods
Mice
We obtained bone marrow from C57BL/6 mice with either a wild-type or ΔD-iD [19] DH locus. The latter is the result of gene targeting in the DH locus to force use of a DH gene segment enriched for arginine in reading frame 1. The C57BL/6 mice analyzed represent the progeny of C57BL/6J mice bred in the UAB vivarium. The ΔD-iD DH allele mutation, which had been generated in BALB/c mice [19], was backcrossed onto C57BL/6 mice for 22 generations. Both strains of mice were maintained in a specific pathogen free barrier facility. All experiments with live mice were approved by and performed in compliance with Institutional Animal Care and Use Committee regulations.
Flow cytometry and cell sorting
Flow cytometric analysis and cell sorting of bone marrow mononuclear cells was performed as previously described [8, 17, 19, 28, 29]. Developing B lineage cells were identified on the basis of the surface expression of CD19, CD43, IgM, BP-1, and/or IgD (Supporting information Figure 1). Due to the decreased expression of CD43 on early C57BL/6 B-cell progenitors when compared to BALB/c B-cell progenitors, the scheme of Melchers was used to isolate the equivalent of Hardy fractions B (B220+ cKit+,CD25−, BP-1−) and C (B220+ cKit− CD25+ and BP-1+). .
The following sets of monoclonal antibodies were used: For the equivalent of Hardy fractions B and C, anti-B220 (PerCP) [BD Pharmingen, San Diego, CA], anti-BP-1 (PE) (a gift from JF Kearney), and anti-IgM (Cy5) [Jackson ImmunoResearch], West Grove, PA], anti-cKit (allophycocyanin) [BD Pharmingen, San Diego, CA] and anti-CD25 (FITC) [BD Pharmingen, San Diego, CA]. For Hardy fractions D, E and F, anti-CD19 (SPRD) [Southern Biotech, Birmingham, AL], anti-CD43 (FITC) [BD Pharmingen], anti-IgD (PE) [Southern Biotech, Birmingham, AL], and anti-IgM (Cy-5) [Jackson ImmunoResearch].
Sorting, RNA preparation, RT-PCR and sequencing
Total RNA isolation, VH7183 specific VDJCµ RT-PCR amplification, cloning, sequencing, and sequence analysis was performed as previously described [8, 17, 19]. A listing of the 577 wild-type C57BL/6 VDJCμ unique, in-frame sequences used for analysis in this work is provided in supporting information Table1. A listing of 52 VDJCμ sequences from the congenic C57BL/6 IgHa ΔD-iD mature, recirculating fraction F bone marrow B-cell subset are provided in supporting information Table 2.
Statistical analysis
Differences between populations were assessed where appropriate by Student's t test, two tailed; Fisher's exact test, two tailed; χ2; or Levene's test for the homogeneity of variance. Analysis was performed with JMP version 8 (SAS Institute, Inc., Cary, NC), or with GraphPad Prism 5.03 (GraphPad Software, Inc., La Jolla, CA). Means are accompanied by the standard error of the mean.
Supplementary Material
Acknowledgements
The authors wish to thank Dr. Peter D Burrows for his invaluable advice and support.
This work was supported by NIH AI42732 (HWS), NIH AI48115 (HWS), NIH HD043327 (RLS), and by core facilities supported by NIH G20RR025858, P30 AR48311, P30AI027767, and P30 CA13148.
Footnotes
Conflict of interest The authors declare no financial or commercial conflict of interest.
References
- 1.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 2.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 3.Huetz F, Carlsson L, Tornberg UC, Holmberg D. V-region directed selection in differentiating B lymphocytes. EMBO Journal. 1993;12:1819–1826. doi: 10.1002/j.1460-2075.1993.tb05830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alt FW, Baltimore D. Joining of immunoglobulin heavy chain gene segments: Implications from a chromosome with evidence of three D-J heavy fusions. Proceedings of the National Academy of Sciences, U.S.A. 1982;79:4118–4122. doi: 10.1073/pnas.79.13.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of proteins of immunological interest. U.S.Department of Health and Human Services; Bethesda, Maryland: 1991. [Google Scholar]
- 6.Padlan EA. Anatomy of the antibody molecule. Molecular Immunology. 1994;31:169–217. doi: 10.1016/0161-5890(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 7.Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity. 2000;13:37–45. doi: 10.1016/s1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 8.Ivanov II, Schelonka RL, Zhuang Y, Gartland GL, Zemlin M, Schroeder HW., Jr. Development of the expressed immunoglobulin CDR-H3 repertoire is marked by focusing of constraints in length, amino acid utilization, and charge that are first established in early B cell progenitors. Journal of Immunology. 2005;174:7773–7780. doi: 10.4049/jimmunol.174.12.7773. [DOI] [PubMed] [Google Scholar]
- 9.Manz RA, Lohning M, Cassese G, Thiel A, Radbruch A. Survival of long-lived plasma cells is independent of antigen. International Immunology. 1998;10:1703–1711. doi: 10.1093/intimm/10.11.1703. [DOI] [PubMed] [Google Scholar]
- 10.Teale JM, Kearney JF. Clonotypic analysis of the fetal B cell repertoire: evidence for an early and predominant expression of idiotypes associated with the VH 36–60 family. Journal of Molecular & Cellular Immunology. 1986;2:283–292. [PubMed] [Google Scholar]
- 11.Tarasenko T, Kole HK, Bolland S. A lupus-suppressor BALB/c locus restricts IgG2 autoantibodies without altering intrinsic B cell-tolerance mechanisms. J Immunol. 2008;180:3807–3814. doi: 10.4049/jimmunol.180.6.3807. [DOI] [PubMed] [Google Scholar]
- 12.Sekiguchi DR, Yunk L, Gary D, Charan D, Srivastava B, Allman D, Weigert MG, Prak ET. Development and selection of edited B cells in B6.56R mice. J Immunol. 2006;176:6879–6887. doi: 10.4049/jimmunol.176.11.6879. [DOI] [PubMed] [Google Scholar]
- 13.Kalinina O, Doyle-Cooper CM, Miksanek J, Meng W, Prak EL, Weigert MG. Alternative mechanisms of receptor editing in autoreactive B cells. Proc Natl Acad Sci U S A. 2011;108:7125–7130. doi: 10.1073/pnas.1019389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Retter I, Chevillard C, Scharfe M, Conrad A, Hafner M, Im TH, Ludewig M, et al. Sequence and characterization of the Ig heavy chain constant and partial variable region of the mouse strain 129S1. J Immunol. 2007;179:2419–2427. doi: 10.4049/jimmunol.179.4.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rolink AG, Karasuyama H, Grawunder U, Haasner D, Kudo A, Melchers F. B cell development in mice with a defective lambda 5 gene. European Journal of Immunology. 1993;23:1284–1288. doi: 10.1002/eji.1830230614. [DOI] [PubMed] [Google Scholar]
- 16.Hardy RR, Hayakawa K. B cell development pathways. Annual Review of Immunology. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 17.Schelonka RL, Ivanov II, Jung D, Ippolito GC, Nitschke L, Zhuang Y, Gartland GL, et al. A single DH gene segment is sufficient for B cell development and immune function. Journal of Immunology. 2005;175:6624–6632. doi: 10.4049/jimmunol.175.10.6624. [DOI] [PubMed] [Google Scholar]
- 18.Zemlin M, Schelonka RL, Ippolito GC, Nitschke L, Pelkonen J, Rajewsky K, Schroeder HW., Jr. Forced use of DH RF2 sequence impairs B cell development. In: Kalil J, Cunha-Neto E, Rizzo LV, editors. 13th International Congress of Immunology. Medimond International Proceedings; Bologna: 2007. pp. 507–511. [Google Scholar]
- 19.Ippolito GC, Schelonka RL, Zemlin M, Ivanov II, Kobayashi R, Zemlin C, Gartland GL, et al. Forced usage of positively charged amino acids in immunoglobulin CDR-H3 impairs B cell development and antibody production. Journal of Experimental Medicine. 2006;203:1567–1578. doi: 10.1084/jem.20052217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schroeder HW, Jr., Zemlin M, Khass M, Nguyen HH, Schelonka RL. Genetic control of DH reading frame and its effect on B-cell development and antigen-specifc antibody production. Crit Rev Immunol. 2010;30:327–344. doi: 10.1615/critrevimmunol.v30.i4.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zemlin M, Schelonka RL, Ippolito GC, Zemlin C, Zhuang Y, Gartland GL, Nitschke L, et al. Regulation of repertoire development through genetic control of D H reading frame preference. Journal of Immunology. 2008;181:8416–8424. doi: 10.4049/jimmunol.181.12.8416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burrows PD, Stephan RP, Wang YH, Lassoued K, Zhang Z, Cooper MD. The transient expression of pre-B cell receptors governs B cell development. Seminars in Immunology. 2002;14:343–349. doi: 10.1016/s1044-5323(02)00067-2. [DOI] [PubMed] [Google Scholar]
- 23.Martensson IL, Rolink A, Melchers F, Mundt C, Licence S, Shimizu T. The pre-B cell receptor and its role in proliferation and Ig heavy chain allelic exclusion. Seminars in Immunology. 2002;14:335–342. doi: 10.1016/s1044-5323(02)00066-0. [DOI] [PubMed] [Google Scholar]
- 24.Hasan M, Polic B, Bralic M, Jonjic S, Rajewsky K. Incomplete block of B cell development and immunoglobulin production in mice carrying the muMT mutation on the BALB/c background. Eur J Immunol. 2002;32:3463–3471. doi: 10.1002/1521-4141(200212)32:12<3463::AID-IMMU3463>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 25.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 26.Fukuyama H, Nimmerjahn F, Ravetch JV. The inhibitory Fcgamma receptor modulates autoimmunity by limiting the accumulation of immunoglobulin G+ anti-DNA plasma cells. Nat Immunol. 2005;6:99–106. doi: 10.1038/ni1151. [DOI] [PubMed] [Google Scholar]
- 27.Zemlin M, Ippolito GC, Zemlin C, Link J, Monestier M, Schroeder HW., Jr. Adult lupus-prone MRL/MpJ2+ mice express a primary antibody repertoire that differs in CDR-H3 length distribution and hydrophobicity from that expressed in the C3H parental strain. Molecular Immunology. 2005;42:789–798. doi: 10.1016/j.molimm.2004.07.049. [DOI] [PubMed] [Google Scholar]
- 28.Nobel Lectures . Physiology or Medicine 1901–1921. Elsevier Publishing Company; Amsterdam: 1967. [Google Scholar]
- 29.Schelonka RL, Szymanska E, Vale AM, Zhuang Y, Gartland GL, Schroeder HW., Jr. DH and JH usage in murine fetal liver mirrors that of human fetal liver. Immunogenetics. 2010;62:653–666. doi: 10.1007/s00251-010-0469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chukwuocha RU, Hartman AB, Feeney AJ. Sequences of four new members of the VH7183 gene family in BALB/c mice. Immunogenetics. 1994;40:76–78. doi: 10.1007/BF00163968. [DOI] [PubMed] [Google Scholar]
- 31.Kurosawa Y, Tonegawa S. Organization, structure, and assembly of immunoglobulin heavy chain diversity DNA segments. Journal of Experimental Medicine. 1982;155:201–218. doi: 10.1084/jem.155.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feeney AJ, Riblet R. DST4: a new, and probably the last, functional DH gene in the BALB/c mouse. Immunogenetics. 1993;37:217–221. doi: 10.1007/BF00191888. [DOI] [PubMed] [Google Scholar]
- 33.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2005;33:D34–38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annual Review of Biochemistry. 1984;53:595–623. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.