Abstract
The quantitative simultaneous description of both variable region gene usage and antigen specificity of immunoglobulin repertoires is a major goal in immunology. Current quantitative assays are labor intensive and depend on extensive gene expression cloning prior to screening for antigen specificity. Here we describe an alternative method based on high efficiency single B cell cultures coupled with RT-PCR that can be used for rapid characterization of immunoglobulin gene segment usage, clonal size and antigen specificity. This simplified approach should facilitate the study of antibody repertoires expressed by defined B cell subpopulations, the analysis of immune responses to self and nonself-antigens, the development and screening of synthetic antibodies and the accelerated study and screening of neutralizing antibodies to pathogenic threats.
Keywords: B cell repertoire, immunoglobulin genes, antibody specificity, clonal size
1. Introduction
In the fetal liver and then the bone marrow, newly formed B cells are continuously generated from hematopoietic precursors. Progenitor B cells undergo a complex series of somatic DNA rearrangement to create a surface immunoglobulin receptor for antigen. Germline-derived V, D, and J region gene segments can be randomly recombined to generate an enormous diversity of B lymphocytes of greater than 1010 B cell clones, with each B cell clonotype expressing a unique heavy (VHDJH) and light (VLJL) chain gene rearrangement. Mature B cells are destined to a short life unless they encounter antigen and succeed in activating their surface immunoglobulin receptor complex.
Initial studies on the association of antigen specificity with variable region gene usage in the B cell repertoire often depended on the generation of a collection of hybridomas, which were first screened for antigen binding and then analyzed by V gene sequencing (Shlomchik et al., 1990). Although this approach is still in productive use as a means to immortalize specific B cell clones, it introduces an inextricable bias due to its reliance on the non-random process of hybridoma generation, which makes it very difficult to derive quantitative conclusions about the clonal composition of the original repertoire. The advent of single cell sorting and single cell PCR technologies permitted developing of alternative single cell cloning strategies to obtain a more faithful representation of the repertoire of B cells (Brezinschek et al., 1995; Kantor et al. 1997). Recently, this strategy has been buttressed by single cell heavy and light chain gene sequencing and gene expression cloning into recipient cells capable of secreting the cloned immunoglobulin (Tiller et al., 2008). Individual cloned antibodies can then be screened for antigen specificities (Tiller et al., 2008). While this approach avoids the introduction of distortions in clonal sizes created by hybridoma technology, the main obstacle to its widespread usage is the labor-intensive nature of the large scale single cell sequencing, cloning and transfection required to estimate antigen-specific clonal frequencies, especially in unimmunized individuals where that frequency would be expected to be very low. Thus, to date, V gene usage and antigen specificity in the B cell repertoire of naïve mice have only been established for clones that can be detected by antigen-specific screening by flow cytometry. A classic example is the estimation of the number of anti-phosphatidylcholine B cell clones (Seidl et al., 1997) in the circulation. More recently, large scale, screening assays for the isolation of antigen specific plasma cells have been proposed, this approach but depends on very sophisticated technology, thus limiting its application (Jin et al., 2009)
Here, we describe a general approach for the rapid analysis of the B cell repertoire that is based on high efficiency single cell cultures of B lymphocytes simultaneously characterized for VH and VL gene usage, antigen specificity and clonal size. This approach has enabled us to overcome many of the difficulties described above.
2. Material and Methods
2.1. Animals and cells
We chose to use mouse peritoneal B-1a cells as our exemplar for this technique. Peritoneal cells were obtained from female BALB/c WT mice from 8 to 12 weeks of age. Thymocytes used as feeder cells were obtained from 4 week old Sprague–Dawley rats. The bone marrow-derived stromal cell line S17 (Collins and Dorshkind, 1987) was used in most experiments as feeder cells (kindly provided by Dr. Christopher Klug, University of Alabama at Birmingham, USA). All experiments were approved by and performed in compliance with the regulations of the Ethical Committee for Animal Studies of the Instituto de Microbiologia, Universidade Federal do Rio de Janeiro and the IACUC of the University of Alabama at Birmingham.
2.2. Flow cytometry and cell sorting
Single cell suspensions were prepared from the peritoneal cavity by lavage with 10 ml of complete RPMI 1640 medium (GIBCO) (supplemented with 10% heat-inactivated FCS, 2 mM L-glutamine, 1mM sodium pyruvate, 50 μM 2-ME, 100 U penicillin, and 100 μg/ml streptomycin). Cells were washed and resuspended in an appropriate volume of RPMI for counting and staining. The total peritoneal cavity cells from each mouse group were pooled and incubated at 2.5 × 108 cells/mL in RPMI medium containing the followings monoclonal Abs: anti-B220 (RA 3.6B2) Pacific blue (PB) (BD Pharmingen), anti-CD5 PE (53-7.3)(BD Pharmingen), and anti-Mac-1 FITC (M1/70) (BD Pharmingen). Analysis and sorting were then performed on a MoFlo instrument (DakoCytomation) and the cells were collected directly in sterile tubes containing RPMI medium for further use in the cultures. In our studies, we wished to prevent stimulation through the BCR-complex on the B cells prior to their culture, thus anti-CD19 and whole molecule anti-IgM antibodies were avoided. Alternatively, Fab anti-IgM could be used for sort purifying the B cells.
2.3. B cell culture and Limiting Dilution Assay (LDA)
B cell cultures were performed as described previously (Vale et al., 2010) with minor modifications. Sorted B-1a cells were cultured in 250μl of complete RPMI medium in 96 well flat-bottom plates in the presence of 30μg/mL of LPS (Salmonella typhimurium, Sigma-Aldrich), 1.5 μg/ml of CpG (ODN 1828, Invivogen), 1.0 μg/ml of Pam3Cys (Invivogen) and combinations of these TLR ligands. Unless indicated otherwise in the text, all cultures contained 6 × 105 rat thymocytes or 3×103 S17 cells/well for growth support.
The preparation of S17 as feeder cell followed a previously described protocol (Collins and Dorshkind, 1987) with some modifications. Briefly, one day before the start of the LDA cultures, 3 × 103 S17 cells were added per well and incubated overnight at 37°C with 5% CO2. The next day, the S17 culture plates were irradiated at 3000 rads and various numbers of the sorted B-1a cells were added in two different sets of cultures. The first set was used to determine the frequency of IgM secreting clones by ELISA according to the Poisson distribution (Andersson et al., 1977; Taswell, 1981) and contained sorted B-1a cells with 22 replicates for each cell concentration at 18, 6; 2, and 0.66 B cells per well. The second culture set was performed to estimate the clonal frequencies of antigen-specific B cells using an antigen binding assay, with 22 replicates of 10,000; 3,333; 1,111; 370; 123; 41; 14 and 4.6 B cells added per well. To screen for antigen binding, we used the immunoblot protocol described by Nobrega et al. (1998). Culture supernatants for this assay were typically harvested on the 5th day of culture, unless indicated otherwise in the text.
2.4. Single B cell cloning cultures
In parallel to the bulk cultures described above, sorted B-1a cells were diluted to 6.6 cells per mL and cultured on the S17 feeder layer (1×103 S17/well, see Fig. 2C) at a mean concentration of 0.66 cells per well in 96 well round-bottom plates. For the data shown here, 720 replicates per experiment were performed. On the 5th day of culture, 25μl of supernatant were harvested from each well without disturbing the cells on the bottom.
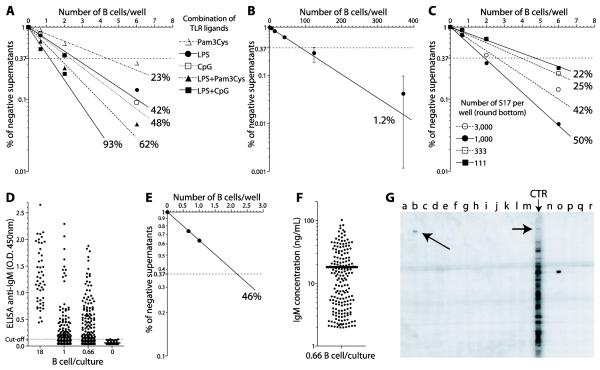
Figure 2. Determination of the frequency of B-1a cells recognizing a 190 kD antigen from mouse brain.
Legend: (A) Sorted B-1a cells were cultivated in variable cell numbers on monolayers of S17 stromal cells in flat-bottom 96 well plates, and the frequencies of cells responding to different mitogens were estimated by limiting dilution analysis. (B) The frequency of Ig secreting B cells producing antibodies to the 190 kD antigen from mouse brain was again calculated by limiting dilution analysis. (C) Sorted B-1a cells were cultivated on variable numbers of S17: 3,000; 1,000; 333 and 111 per well and the percentage of B cells responding to LPS was determined in each condition. (D) B cell cloning cultures, 0.66 cells/well round-bottom 96 well plates, 720 replicates, were performed and 188 scored positive for IgM secretion. (E) Poisson distribution of the cultures containing 18, 1 and 0.66 B-1a cells per well, to double-check the frequency of IgM secreting clones in the B cell cloning cultures. (F) The amount of IgM produced in the B cell cloning cultures (0.66 cell/well) averaged 18 ng/ml. (G) The 188 supernatants from B cell cloning cultures were tested for reactivity with the 190 kD antigen in immunoblots of BALB/c mouse brain ; the reactivities of 16 samples (a-p) are shown. Lane (b) shows a supernatant that scored positive for reactivity with the 190kD antigen (indicated with an arrow); lane (CTR) shows the reactivity profile of a supernatant from a bulk culture of 10,000 B-1a cells and the reactivity with the 190 kD antigen is also indicated. Note that, as expected, the antibodies in these polyclonal B-1a cell cultures recognize many antigens. The supernatant in lane (o) recognizes a 70 kD antigen. Lanes (q-r) contain secondary antibody controls.
2.5. IgM ELISA
To determine the IgM concentration in the supernatants, ELISA was performed as previously described in Vale et al. (2010) using anti mouse IgM-specific reagents (Southern Biotechnology). Standard curves, obtained by using polyclonal, serially 3-fold diluted, mouse IgM (Southern Biotechnology) were used to quantify the IgM.
2.6. One-step RT-PCR amplification of VH and VL genes
B cells cultured with LPS typically divide every 18 hours or less. Within 4 days is possible to observe the colonies of B cells on the S17 monolayer at the bottom of the culture plates by optical microscopy (data not shown). And, by the 6th day of the B cell cloning cultures, approximately 128 cells, all derived from a single precursor, are expected to be present in each well (2). Thus, on day 6, the plates were spun down, the supernatants of each well that previously scored positive for IgM were collected (without disturbing the pellet) and banked stored at 4°C; 10 units of RNAse inhibitor (RNAsin Promega N211A/N211B) was added to the pellet and finally 20μl of cell pellet were harvested from the bottom. Each cell pellet contained B cell plasmacytes. A fraction of the pellet was used directly into the RT-PCR reaction and/or stored at −80°C in order to preserve the mRNA content for further RT-PCR amplifications.
The B cell plasmacytes were processed for one-step RT-PCR. The Ig cDNA was synthesized and amplified in one-step, using 5μl of the sample obtained from the B cell cloning cultures pipetted directly into 20μl of the QIAGEN One-Step RT-PCR Kit master mix, consisting of: 5μl of 5× RT-PCR buffer, 1μl of dNTPs (10mM), 1 μl of RT-PCR Enzyme Mix (Omniscript and Sensiscript Reverse Transcriptases and HotStarTaq DNA Polymerase), 0.25μl of RNAsin (40U/μl, Promega), 0.5μl of each primers (0.5 μl of stock solution that is 10μM for each primer), and RNase-free water sufficient to bring the final volume to 20μl of master mix. Promiscuous primers for VH, Vκ and Vλ were designed according to Kantor et al. (1997) and Seidl et al. (1997): MsVHE = 5′-GAGGTGCAGCTGCAGGAGTCTGG-3′, MsVκM = GAT ATT GTG ATG ACC CAG TCT, and MsVM1 = CAG GCT GTT GTG ACT CAG GAA TCT, respectively. The specific Cμ primer was designed by us: Cμ1 = 5′-GACAGGGGGCTCTCG-3′. The Cκ and Cλ primers, MsCκ1 = 5′-ACA CTC ATT CCT GTT GAA GCT CTT-3′ and MsCλM1 = 5′-GCA GGA GAC AAA CTC TTC TCC ACA-3′, were designed according to Seidl et al. (1997).
Because the current approach involves proliferation and differentiation of B cells induced by LPS, it provides much higher starting amounts of RNA transcripts from plasmacytes than can be obtained from single sorted resting B cells. Thus, a single round of RT-PCR was routinely sufficient to obtain DNA for sequencing. The one-step RT-PCR conditions were as follows: reverse transcription at 50°C for 30 minutes followed by HotStarTaq activation at 94°C for 15 minutes and the First Round PCR performed for 50 cycles at 94°C for 20 sec, 50°C for 30 sec, 72°C for 1 minute and a final extension at 72°C for 10 min. When needed, a second round of amplification was performed using semi-nested PCR using the same promiscuous V primers and internal C region primers: CμIN 5′-G GGA AGA CAT TTG GGA AGG ACT GAC -3′, Cκ M13-MsCκ2 5′-TGT AAA ACG ACG GCC AGT TCT AGA TGG TGG GAA GAT GGA-3′, and Cλ M13-MsCλ2TGT 5′-AAA ACG ACG GCC AGT GAG CTC CTC AGA GGA AGG TGG AAA-3′ (Seidl et al., 1997). In this case, one microliter of the first round PCR reaction was added into 24μl of fresh master mix containing High Fidelity Taq (Invitrogen) polymerase and the appropriate combination of primers for VH or VL. The second round PCR was performed as follows: activation of High Fidelity Taq at 94°C for 3 minutes and 30 cycles at 94°C for 20 sec, 50° C for 30 sec, 68°C for 45 sec and a final extension at 68°C for 5 minutes.
2.7. DNA sequencing and analysis
After PCR, the positive samples were identified by electrophoresis (1.5% agarose gel) and ethidium bromide staining. The cDNAs were purified from the gel using the QIAquick Gel Extraction Kit (QIAGEN), primed with the same promiscuous primers, and sequenced at the University of Alabama at Birmingham Heflin Center Sequencing Core or at FIOCRUZ of Rio de Janeiro, Brazil. Gene segments were assigned according to published germline sequences for the immunoglobulin gene segments as listed in the ImMunoGeneTics database (http://imgt.cines.fr:8104).
2.8. Quantifying clonal sizes
When identical heavy and light chain cDNA sequences were identified in samples from different wells, they were initially scored as repeats of the same clonotype and then confirmed by testing the respective supernatants for matched antigen recognition. When identical heavy and light chain sequences were obtained in two distinct wells, we used controls to check whether this had occurred because of inadvertent contamination during PCR, a potentially serious problem, or if it truly represented a second cell belonging to the same clone. Two sets of controls were used. First, cultures containing B cells that do not recognize the antigen under consideration were amplified in parallel, such that if contamination were a problem, it would likely occur in these samples as well. Second, because the supernatants of all Ig secreting cultures were saved, they could be tested for antigen reactivity to determine if samples possessing identical VH/VL genes coincided for antigen recognition. Should they not coincide, we would surmise that contamination had occurred. However, should they coincide, we would conclude we had validated the existence of a second cell of the same clonotype.
3. Results and Discussion
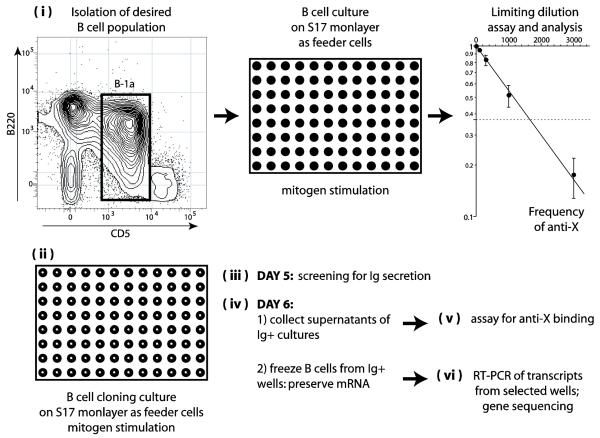
The overall procedure used for the quantitative determination of antigen specific clonal frequencies (anti-X) and V gene usage is schematically depicted in Fig. 1, and each step is described in detail in Section 2. Briefly, (i) we isolated the desired B cell population using flow-cytometry, although magnetic beads and other techniques can also be used. Variable numbers of B cells/well were then cultured on a monolayer of S17 stromal cells and stimulated with polyclonal mitogens. We then (ii) assessed the frequency of B cells responding to the mitogenic stimuli and the frequency of anti-X reactivity using the binding assay of choice (e.g., protein microarray, ELISA, Immunoblot, BIACORE™ system, etc.) and limiting dilution analysis (LDA). A second set of cultures, single B cell cloning cultures on monolayers of S17 feeder cells, was then performed. The number of wells needed for these experiments depends on the frequencies estimated in -i-above (e.g. 500 to 5,000 wells). (iii) On day 5 of culture, 25 μl of supernatants from all culture wells were collected and tested for the presence of total immunoglobulin. (iv) On day 6 of culture, 20 μl were collected from the bottom of the wells that scored positive for Ig secretion in step –iii- and kept frozen with RNAse-inhibitors. These samples contain B cells and plasmacytes that have grown in these wells. (v) The remaining 200 μl of supernatants from the same set of single cell cultures was collected and tested for the presence of antigen specific reactivity (anti-X). Wells scoring positive for anti-X reactivity were identified. (vi) From wells that scored positive for anti-X in step –v-, the frozen 20 μl containing secreting B cells (step –iv-) were thawed and their heavy and light genes were amplified by one step RT-PCR.
Figure 1. Schematic description of the strategy adopted for the quantitative analysis of the immunoglobulin repertoire.
Legend: (i) The desired population of B lymphocytes is isolated and cultivated with mitogens for 7 days in different cell numbers on monolayers of S17 stromal cells in flat-bottom 96 well plates. Supernatants of these cultures are tested for antigen binding and the frequency of secreting B cells producing antibodies to antigen “X” is estimated by limiting dilution analysis. (ii) A large number of replicates of B cell cultures, 0.66 cells/well round-bottom 96 well plates (B cell cloning cultures) are performed, the precise number depending on the frequency of anti-X lymphocytes obtained as above. (iii) On day 5 of culture, 25 microliters of supernatant are collected from each well and screened by ELISA for the presence of secreted immunoglobulin. (iv) On day 6 of culture, 20 microliters are collected from the bottom of the Ig+ wells to obtain B cells and plasmacytes and kept frozen (−80° C) in the presence of RNAse inhibitors. The remaining 200 microliters of supernatants of these wells are also collected for the antigen binding assay. (v) The collected supernatants are screened for binding to antigen “X”. (vi) Frozen samples of B cells and plasmacytes from wells that scored positive for anti-“X” are processed for one step RT-PCR for the amplification of VH and VL genes. Several frozen samples of B cells and plasmacytes from wells that scored negative for anti-“X” are also amplified and serve as controls.
It has been reported that B-1a cells can switch to IgG3 in LPS cultures. However, in bulk culture experiments total IgG was barely detectable by ELISA. Moreover, analysis of day 6 single cell plasmacyte cultures revealed little or no evidence of IgG class switched cells (data not show). Thus, only Cμ primers were used for amplification of IgH chains.
Because antibody-secreting plasmacytes are highly enriched for mRNA transcripts of Ig genes, single round RT-PCR was routinely sufficient to obtain enough material for gene sequencing. Alternatively the first round can be followed by second round of semi-nested PCR. For the amplification of V genes, previously characterized degenerate primers, both for heavy and light chains V genes, were used (Kantor et al., 1997; Seidl et al., 1997). The strategy described above saves the effort of hundreds/thousands of sequencing, cloning and transfection steps because only the cultures scoring positive for the desired antigen binding are processed, together with a defined number of unrelated controls.
Although this rapid methodology has been used in our laboratory to study the immunoglobulin repertoire against antigens such as phosphorylcholine (PC) and constitutive heat shock protein HSC70 (manuscripts in preparation), the method is illustrated here with the cloning of peritoneal B-1a clonotypes recognizing a 190 kD antigen in syngenic mouse brain immunoblots. Many sets of cultures containing varying numbers of mitogen stimulated B-1a cells were prepared for the estimation of the frequency of B-1a cells recognizing this antigen; the supernatants of these cultures were then tested for reactivity with the 190 kD molecule in immunoblots of brain extract.
Polyclonal activation of B lymphocytes with mitogens can be optimized to stimulate up to 100% of B cells, depending on the mouse strain (Vale et al., 2010). In the experiment depicted in Fig. 2, the combination of LPS with CpG stimulated 93% of the B-1a cells to proliferate and secrete IgM (Fig. 2A see also Vale et al., 2010), thus minimizing bias in the repertoire analysis. Fig. 2B demonstrates the Poisson analysis of the frequency of reactivity with the 190 kD antigen among the B-1a cells, which we estimated as 1.2% of the seeded B-1a cells. Once this frequency was calculated, appropriate numbers of B-1a cell cloning cultures were stimulated with LPS in round-bottom plates. To optimize the number of S17 cells in cultures using round-bottom plates, we offset up cultures containing 3,000; 1,000; 333 or 111 S17 cells per well and determined the percentage of B cell responding to LPS in each condition (Fig. 2C). We concluded that for B cell cloning cultures using round-bottom plates, 1×103 S17 cells per well provided efficient growth support yet minimized the accumulation of random cellular debris that might decrease the efficiency of gene amplification by RT-PCR. For this particular experiment we cultured 720 replicates of 0.66 B-1a cells per well on a monolayer of 1×103 S17 cells. We then screened the B cell cloning cultures for IgM production by ELISA. Using the supernatants of cultures containing S17 alone, we were able to determine a cut-off (mean plus 3× standard deviation) above which the B-1a culture supernatants would be considered positive (Fig. 2D). A total of 188 of the single cell cloning cultures averaging 0.66 cell/well scored positive for IgM secretion on day 5. We were able to double-check the frequency of IgM secreting clones according to the Poisson distribution (Fig. 2E). The amount of IgM present in those IgM positive supernatants averaged 18 ng/ml (Fig. 2D), which was sufficient for immunoblot screening for reactivity with the 190 kD antigen and other reactivities (Fig. 2E).
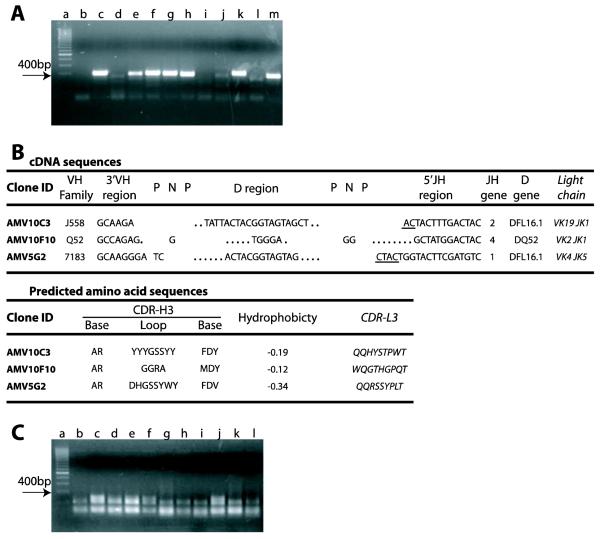
An agarose gel electrophoretic analysis of the products of one-step RT-PCR for the amplification of heavy chain genes from B cell cloning cultures (0.66 cells/well) from a number of wells that scored positive for IgM secretion is presented in Fig 3A. From a total of 188 IgM secreting clones that were obtained from the experiment described in Fig. 2, three cultures scored positive for the recognition of the 190 kD antigen. VH and VL sequencing of B cells from these positive cultures was performed, and the data are presented in Fig. 3B. To detect contamination, samples from cultures of S17 feeder cells alone were added at the two PCR rounds as negative controls. Since we can verify the IgM reactivity for each single clone sequenced, two clones with the same VH/Vκ or VH/Vλ rearrangements but different antigen specificity would be considered contamination products, and both sequences would be discarded from analysis. By contrast, two samples isolated from different cell cultures with identical VH/Vκ or VH/Vλ rearrangements but with the same IgM specificity in the two independent supernatants can be safely considered as isolated from two different cells, it is more likely that they were both derived from the same precursor in vivo. The probable clonal size, in vivo, can be calculated taking into account the frequency of reactivity previously obtained by limiting dilution analysis.
Figure 3. One step RT-PCR amplification of immunoglobulin gene transcripts from B cell cloning cultures.
Legend: (A) Samples containing B cells from cell cloning cultures, 0.66 cells/well, in the presence of S17 stromal cells (3000 cells/well), that scored positive for the secretion of IgM were subjected to one step RT-PCR for the amplification of VH transcripts using a promiscuous VH primer. The final RT-PCR products (~ 400 bp, lanes c-m) were identified by electrophoresis and are indicated in the figure with an arrow. In the gel shown, transcripts from 7 out of 11 IgM+ cultures were amplified (64 % efficiency): lane “a”, DNA ladder; lane “b”, culture with S17 alone. (B) mRNA from samples containing B cells from cloning cultures that scored positive for the recognition of the 190 kD antigen in mouse brain were subjected to one step RT-PCR for the amplification of transcripts of immunoglobulin V genes using a promiscuous primer as described. cDNA sequences: the VH family and the sequence of the heavy chain CDR3 (CDR-H3) are shown with the respective VL and JL genes for each clonotype. Clone ID, clone identifier. VH family, name according to the IMGT classification. 3′VH region, the most 3′ nucleotides of the VH (not including Cys = TGT). P-N-P, palindromic (P) and N region sequence. D region, DH gene sequence. 5′J-Region, the most 5′ nucleotides of the JH (not including Trp = TGG); underlined sequences can be from either of two germline genes. Dots represent nucleotides lost from the germline DH and JH gene segments. JH gene indicates the number of the JH element. DH gene, the DH element used; RF, the DH reading frame used. Light Chain, the Vκ Family and the Jκ usage of the light chain paired with the respective heavy chain is indicated. Predicted amino acid sequences derived from the cDNA sequences: Base, the predicted amino acid sequence of the CDR-H3 base. Loop, the predicted amino acid sequence of the CDR-H3 loop. Hydrophobicity, the average, normalized Kyte-Doolittle hydrophobicity of the CDR-H3 loop. CDR-L3, the predicted amino acid sequence of the light chain CDR-3. (C) A parallel set of cultures were done using rat thymocytes as feeder cells. Samples containing B cells from cell cloning cultures, 0.66 cells/well, that had scored positive for the secretion of IgM were submitted to one step RT-PCR as above. No RT-PCR amplification product could be detected (results representative of 3 different experiments).
On the gel shown in Fig. 3A, 7 out of 11 wells were amplified. This ~70% amplification efficiency has routinely been obtained in several such experiments (data not shown). This efficiency is comparable to the value reported by Kantor et al. (1997) using the same promiscuous VH primer. It is possible that this limitation is due to the VH primer efficiency rather than the method itself. If so, it should be possible to achieve 100% efficiency with suitable mixtures of alternative primers.
Because the cell cloning method we employed was limiting dilution (0.66 cells/well), some positive wells may contain two different clonotypes. However, this is readily detected by sequencing of the RT-PCR products, and we typically discard the products of such wells when they are identified. It is possible that direct automated single cell cloning in 96-well plates by cell sorting could be used to minimize these events, however this step adds another technologic complication to the procedure, limiting its use to laboratories with more sophisticated cell sorting facilities.
It is critical to note that amplification of VH and VL genes was only possible using irradiated S17 stromal cells as feeders cells (Collins and Dorshkind, 1987). Single cell cultures using the classical protocol with young rat thymocytes as feeders cells (600,000/well) resulted in a high percentage of growing clones (Andersson et al., 1977), many of which secreted IgM, but a total failure in RT-PCR amplification (Fig. 3C). Although we have not identified the exact mechanism that prevents the RT-PCR amplification, it is notable that rat thymocytes must be seeded at higher concentration than S17 in order to properly support B cell growth. Thus, one possible explanation for the failure to obtain transcripts when using rat thymocytes could be higher levels of rat thymocyte cellular debris which, along with considerable levels of cell-derived RNAses, may damage the mRNA templates and decrease the efficiency of gene amplification by RT-PCR.
In conclusion, the method described here allows the rapid and quantitative analysis of the immunoglobulin repertoire, facilitating the identification of clonotypes, clonal sizes, and antigen specific frequencies. This new approach should easily lend itself for repertoire studies of natural antibodies, immune responses, and autoimmune diseases in the mouse. Moreover, this quick method could be used to improve the development of synthetic antibodies to be used as targeted therapeutics or laboratory reagents and could also accelerate the screening of neutralizing antibodies. The application of this strategy for studies of human B cell repertoires will depend on the development of an analogous high efficiency single B cell culture system.
HIGHLIGHTS.
▶5 days of single B cell cultures retrieve individual IgM specificities and genes;
▶Paired mRNA/cDNA amplification of VH and VL with promiscuous primers;
▶Limiting dilution analysis of B cell repertoire: frequencies and clonal sizes;
Aknowlegements
The authors wish to thank Drs. John .F. Kearney and Teresa Santiago for their invaluable advice and support. This work was supported in part by CNPq, FAPERJ, and FINEP grants awarded to AN, and by NIH-AI048115 (HWS), AI088498 (HWS), and AI090742 (HWS). AMV was supported in part by a fellowship from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Division of the Brazilian Ministry of Education) and by an exchange-training fellowship from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico - Brazil).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andersson J, Coutinho A, Lernhardt W, Melchers F. Clonal growth and maturation to immunoglobulin secretion in vitro of every growth-inducible B lymphocyte. Cell. 1977;10:27–34. doi: 10.1016/0092-8674(77)90136-2. [DOI] [PubMed] [Google Scholar]
- Brezinschek HP, Brezinschek RI, Lipsky PE. Analysis of the heavy chain repertoire of human peripheral B cells using single-cell polymerase chain reaction. Journal of Immunology. 1995;155:190–202. [PubMed] [Google Scholar]
- Collins LS, Dorshkind K. A stromal cell line from myeloid long-term bone marrow cultures can support myelopoiesis and B lymphopoiesis. Journal of Immunology. 1987;138:1082–1087. [PubMed] [Google Scholar]
- Jin A, Ozawa T, Tajiri K, Obata T, Kondo S, Kinoshita K, Kadowaki S, Takahashi K, Sugiyama T, Kishi H, Muraguchi A. A rapid and efficient single-cell manipulation method for screening antigen-specific antibody-secreting cells from human peripheral blood. Nat Med. 2009;15:1088–92. doi: 10.1038/nm.1966. [DOI] [PubMed] [Google Scholar]
- Kantor AB, Merrill CE, Herzenberg LA, Hillson JL. An unbiased analysis of V(H)-D-J(H) sequences from B-1a, B-1b, and conventional B cells. Journal of Immunology. 1997;158:1175–1186. [PubMed] [Google Scholar]
- Nobrega A, Grandien A, Haury M, Hecker L, Malanch, re E, Coutinho A. Functional diversity and clonal frequencies of reactivity in the available antibody repertoire. European Journal of Immunology. 1998;28:1204–1215. doi: 10.1002/(SICI)1521-4141(199804)28:04<1204::AID-IMMU1204>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Seidl KJ, MacKenzie JD, Wang D, Kantor AB, Kabat EA, Herzenberg LA. Frequent occurrence of identical heavy and light chain Ig rearrangements. International Immunology. 1997;9:689–702. doi: 10.1093/intimm/9.5.689. [DOI] [PubMed] [Google Scholar]
- Shlomchik MJ, Mascelli MA, Shan H, Radic MZ, Pisetsky DS, Marshak-Rothstein A, Weigert MG. Anti-DNA antibodies from autoimmune mice arise by clonal expansion and somatic mutation. Journal of Experimental Medicine. 1990;171:265–297. doi: 10.1084/jem.171.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taswell C. Limiting dilution assays for the determination of immunocompetent cell frequencies. I. Data analysis. Journal of Immunology. 1981;126:1614–1619. [PubMed] [Google Scholar]
- Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–24. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale AM, Hayashi E, Granato A, Schroeder HW, Jr., Bellio M, Nobrega A. Genetic control of the B cell response to LPS: opposing effects in peritoneal versus splenic B cell populations. Immunogenetics. 2010;62:41–8. doi: 10.1007/s00251-009-0404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]