Abstract
Vertebrate axis specification is an evolutionarily conserved developmental process that relies on asymmetric activation of Wnt signaling and subsequent organizer formation on the future dorsal side of the embryo. Although roles of Wnt signaling during organizer formation have been studied extensively, it is unclear how the Wnt pathway is asymmetrically activated. In Xenopus and zebrafish, the Wnt pathway is triggered by dorsal determinants, which are translocated from the vegetal pole to the future dorsal side of the embryo shortly after fertilization. The transport of dorsal determinants requires a unique microtubule network formed in the vegetal cortex shortly after fertilization. However, molecular mechanisms governing the formation of vegetal cortical microtubule arrays are not fully understood. Here we report that Dead-End 1 (Dnd1), an RNA-binding protein required for primordial germ cell development during later stages of embryogenesis, is essential for Xenopus axis specification. We show that knockdown of maternal Dnd1 specifically interferes with the formation of vegetal cortical microtubules. This, in turn, impairs translocation of dorsal determinants, the initiation of Wnt signaling, organizer formation, and ultimately results in ventralized embryos. Furthermore, we found that Dnd1 binds to a uridine-rich sequence in the 3′-UTR of trim36, a vegetally localized maternal RNA essential for vegetal cortical microtubule assembly. Dnd1 anchors trim36 to the vegetal cortex in the egg, promoting high concentrations of Trim36 protein there. Our work thus demonstrates a novel and surprising function for Dnd1 during early development and provides an important link between Dnd1, mRNA localization, the microtubule cytoskeleton and axis specification.
Keywords: Dnd1, Axis specification, Cortical rotation, Microtubules, Xenopus
INTRODUCTION
Cell-fate determinants are often asymmetrically localized to specific subcellular compartments. After cell division, daughter cells may acquire distinct fates depending upon the inheritance of these cell-fate determinants. Asymmetric inheritance of determinants is a mechanism widely used during embryonic development and in adult tissue homeostasis (Gönczy, 2008; Knoblich, 2008; Knoblich, 2010). In many cases, however, the molecular mechanisms governing asymmetric localization of cell-fate determinants are poorly understood.
During vertebrate axis specification, organizer formation in embryos requires the asymmetric activation of Wnt signaling. As predicted, inactivation of Wnt signaling results in ventralized embryos (for reviews, see Harland and Gerhart, 1997; Heasman, 1997; Sokol, 1999; Weaver and Kimelman, 2004; Schier and Talbot, 2005; Heasman, 2006; Tam et al., 2006; Rivera-Perez, 2007; White and Heasman, 2008; Langdon and Mullins, 2011; Houston, 2012). In Xenopus and zebrafish, the Wnt pathway is triggered by dorsal determinants localized to the vegetal pole of the egg (Fujisue et al., 1993; Holowacz and Elinson, 1995; Kikkawa et al., 1996; Kageura, 1997) and are probably positive components of the Wnt pathway (Larabell et al., 1996; Darras et al., 1997; Marikawa et al., 1997; Rowning et al., 1997; Marikawa and Elinson, 1999; Miller et al., 1999; Weaver et al., 2003; Tao et al., 2005; Cha et al., 2008). In response to sperm entry, these dorsal determinants are translocated from the vegetal pole to the future dorsal side of the embryo. Transport of dorsal determinants after egg activation relies on the formation of a unique vegetal cortical microtubule network. Initially, a poorly ordered meshwork of microtubules forms in the vegetal cortex. These disordered vegetal cortical microtubules soon become organized into numerous wavy parallel bundles (Elinson and Rowning, 1988; Houliston and Elinson, 1991b; Schroeder and Gard, 1992). Vesicle-like organelles, with which dorsal determinants are likely to be associated, are directionally transported along these microtubule arrays to the future dorsal side of the embryo (Larabell et al., 1997; Rowning et al., 1997; Miller et al., 1999; Weaver et al., 2003). In Xenopus embryos, vegetal cortical microtubules also serve as tracks to drive the rotation of the vegetal cortex (Houliston and Elinson, 1991a; Houliston, 1994; Houliston et al., 1994; Larabell et al., 1996). As a result, the cortex rotates 30 degrees relative to the inner yolky cytoplasm. Assembly of vegetal cortical arrays of microtubules and subsequent translocation of dorsal determinants occur before the beginning of first cleavage. Physical or chemical treatments that interfere with vegetal cortical microtubule assembly impair transport of dorsal determinants and result in ventralized embryos (Gerhart et al., 1989; Darras et al., 1997; Medina et al., 1997; Weaver and Kimelman, 2004; Gerhart, 2010).
The vegetal cortex lacks a conventional microtubule-organizing center. Vegetal cortical microtubules extend from the animal hemisphere, where microtubules are nucleated by the sperm centrosome and egg pronuclear associated components (Houliston and Elinson, 1991b; Schroeder and Gard, 1992). Strikingly, polymerization of microtubules into the vegetal subcortex appears to be regulated by vegetally localized maternal factors through an unknown mechanism. It has been reported that knockdown of maternal Trim36, a vegetally localized E3-ubiquitin ligase, impairs the formation of vegetal cortical microtubules and results in ventralized embryos (Cuykendall and Houston, 2009). Here, we report that Xenopus Dnd1, a vegetally localized RNA-binding protein (Horvay et al., 2006) previously found to function exclusively in the germline (Weidinger et al., 2003; Youngren et al., 2005; Horvay et al., 2006; Aramaki et al., 2007; Kedde et al., 2007; Lam et al., 2007; Zhu et al., 2007; Cook et al., 2009; Koebernick et al., 2010; Cook et al., 2011), binds trim36 directly and anchors it to the vegetal cortex. Knockdown of maternal Dnd1 disrupts vegetal cortical microtubule assembly, which is necessary for the translocation of determinants such as wnt11 to the dorsal side. As a result, maternal Wnt signaling is not initiated, leading to impaired organizer formation and ventralized embryos. We show, however, that the dorsal determinants remain functional as dorsal development can be restored by replacing the function of microtubules in moving dorsal determinants by the action of gravity: tipping Dnd1-depleted embryos 90 degrees relative to the animal-vegetal axis during the first embryonic cell cycle. Our work thus demonstrates a novel and surprising function for Dnd1 during early development and provides an important link between Dnd1, mRNA localization, microtubule formation, and axis specification.
MATERIALS AND METHODS
Oocyte and embryo manipulations
Xenopus oocyte and embryos were obtained and injected as described (Sive et al., 2000). Host transfer experiments were performed as described (Heasman et al., 1991). Briefly, manually defolliculated oocytes were injected with oligonucleotides and/or mRNA as detailed in the text. Injected oocytes were cultured for 24 hours and then treated with 2 nM progesterone. Matured oocytes were stained with vital dyes and transferred into a host female frog to complete ovulation. Transferred eggs were recovered and fertilized.
To knock down dnd1, oocytes were injected with 7.5 ng of a phosphorothioate-modified antisense oligonucleotide (AS-oligo) (5′-C*C*C*TCGATTCAGGCCA*C*T-3′; Integrated DNA Technologies). Residues marked by asterisks were modified. The oligo was HPLC purified, followed by a Na+ salt exchange to remove TEAA. In rescue experiments, we co-injected the AS-oligo with Xenopus tropicalis dnd1 RNA, which lacks the AS-oligo binding sequence. Tipping experiments were performed as described (Cuykendall and Houston, 2009). For experiments in which eggs were activated artificially by needle-pricking, manually defolliculated oocytes were injected with the AS-oligo, cultured for 24 hours, and then incubated in progesterone-containing medium for 16 hours at 18°C to induce maturation. Matured oocytes were pricked in the animal hemisphere with a fine glass needle to achieve egg activation.
Western blot, luciferase assay, RT-PCR, histological analysis and whole-mount in situ hybridization
Western blot analysis was performed as described (Jin et al., 2009). Antibodies employed were anti-β-tubulin (Sigma T5293, 1:1000) and anti-Trim36 (Cuykendall and Houston, 2009) (1:500). The anti-Dnd1 antibody (1:500) was generated by immunizing rabbits with KLH-coupled peptide CLGYIPDFSLGDVTARNAL, followed by affinity purification against the Dnd1 peptide. For western blot analysis of Dnd1 and Trim36, Dnd1 or Trim36 proteins were immunoprecipitated from 50 oocytes or eggs and separated on SDS-PAGE. The TOP-Flash luciferase assay was performed as described (Yang et al., 2002). Histological analysis, whole-mount in situ hybridization, RNA extraction and RT-PCR were performed as described (Sive et al., 2000). RT-PCR primers, including siamois, Xnr3, noggin, chordin and odc (Yang et al., 2003); alas (Shi et al., 2008); collagen type II (Sasai et al., 1996); epidermal keratin and Bicaudal-c (Wessely and De Robertis, 2000); NCAM and muscle actin (Sasai et al., 1995); α-globin (Steinbeisser et al., 1995); emi1 (Graindorge et al., 2006); vg1 (Weeks and Melton, 1987); wnt11 (Schroeder et al., 1999) and trim36 (Cuykendall and Houston, 2009) were as reported. dnd1 primers were: 5′-TGGTAATGCTCCAGTCAGTG-3′ and 5′-TAAGCGAACCCTCGATTCAG-3′.
Immunofluorescence
Dnd1 and Nanos1 double staining were performed as described (Lai et al., 2011). Images were collected using an inverted Zeiss LSM-510 confocal laser scanning microscope. Vegetal cortical microtubule formation was analyzed as described (Schroeder and Gard, 1992). Briefly, eggs were fixed in fixation buffer (80 mM Pipes pH 6.8, 5 mM EGTA, 1 mM MgCl2, 3.7% formaldehyde, 0.25% glutaraldehyde, 0.2% Triton X-100 and 0.5 μM taxol) for 4 hours at room temperature, followed by incubation in 100% methanol overnight. After rehydration, eggs were washed in PBS and incubated overnight in PBS containing 100 mM NaBH4 at room temperature. Eggs were then washed with PBS extensively and bisected using a surgical blade. The vegetal halves were incubated with an anti-β-tubulin antibody (Developmental Studies Hybridoma Bank, 1:100) and then an Alexa 564-conjugated anti-mouse IgG (Molecular Probes). Images were collected using a Leica DMI3000B inverted microscope.
In vitro RNA pull-down assay
Bacterially expressed GST-Dnd1 protein (50 μg) was incubated with yeast tRNA (500 μg) in 500 μl RNA-binding buffer (20 mM HEPES pH 7.9, 150 mM NaCl and 0.05% Triton X-100) at 4°C for 4 hours. RNA (1 μg) was incubated with BSA (1% final concentration) and RNasin (200 U) in 500 μl binding buffer at 4°C for 4 hours. Pre-absorbed GST-Dnd1 and RNA were mixed together and incubated at 4°C for 20 minutes. Glutathione-agarose beads were washed three times with RNA-binding buffer and three times with RNA-washing buffer (20 mM HEPES pH 7.9, 150 mM NaCl and 1% Triton X-100). RNAs bound to beads were extracted with Trizol reagent. Recovered RNAs were used for cDNA synthesis and subsequent real-time PCR. The ratio between pulled down RNA and 5% of RNA input was used to determine binding of specific RNAs by GST-Dnd1.
RESULTS
Maternal Dnd1 protein is vegetally localized
Dnd1 is essential for the development of the germline (Weidinger et al., 2003; Youngren et al., 2005; Horvay et al., 2006; Aramaki et al., 2007; Kedde et al., 2007; Lam et al., 2007; Zhu et al., 2007; Cook et al., 2009; Koebernick et al., 2010; Cook et al., 2011). Interestingly, dnd1 mRNA is vegetally localized in Xenopus oocytes (Horvay et al., 2006), suggesting that maternal Dnd1 may be important for early embryonic development. To investigate the function of maternal Dnd1, we first determined the subcellular localization of endogenous Dnd1 protein. We dissected oocytes into animal and vegetal halves. Endogenous Dnd1 protein was immunoprecipitated and analyzed by western blotting. As shown in Fig. 1A, Dnd1 protein was detected in the vegetal, but not in the animal, halves. Dnd1 could not be detected by immunofluorescence in oocytes or eggs (data not shown), most likely because germ plasm at these stages is finely dispersed at the vegetal pole and Dnd1 expression level is low (supplementary material Fig. S2). At the eight-cell stage, the germ plasm has coalesced and endogenous Dnd1 protein was detected concentrated there, and as expected, colocalized with Nanos1 (Lai et al., 2011; Lai et al., 2012) (Fig. 1B).
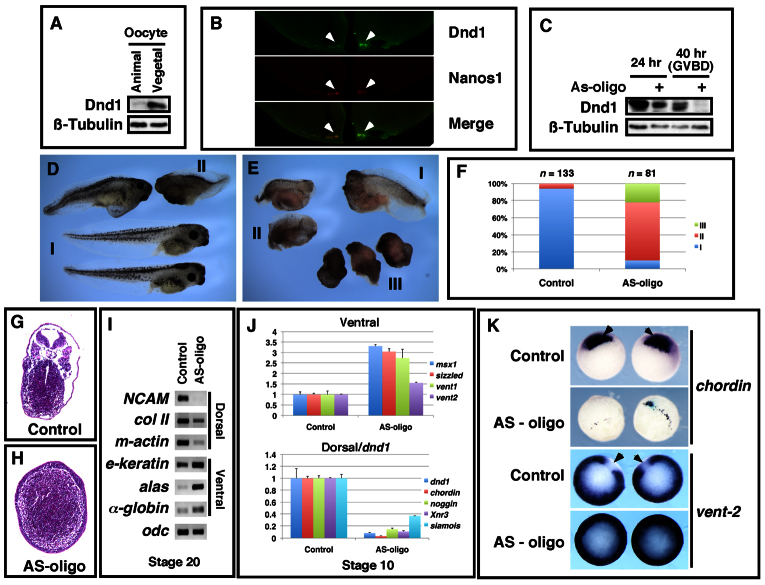
Fig. 1.
Maternal Dnd1 is vegetally localized and required for axis specification. (A) Western blot showing vegetal localization of endogenous maternal Dnd1 protein. Manually bisected animal and vegetal halves of oocytes were analyzed by immunoprecipitation/western blotting. Each group contains 50 animal or vegetal halves. β-tubulin served as the loading control. (B) Immunofluorescence showing colocalization of Dnd1 and Nanos1 in the germ plasm at the vegetal pole at the eight-cell stage (indicated by arrowheads). (C) Dnd1 knockdown efficiency. Oocytes were injected with the AS-oligo against dnd1, cultured for 24 hours, and treated with progesterone to induce germinal vesicle breakdown (GVBD). Uninjected controls and injected oocytes were harvested at 24 hours or 40 hours (after GVBD) after microinjection of the AS-oligo, and analyzed by immunoprecipitation/western blotting. Each group contains 50 oocytes or eggs. β-tubulin served as the loading control. (D,E) Morphology of uninjected controls (D; note rare examples of class II) and AS-oligo-injected host-transfer embryos (E) at the tadpole stage. (F) Summary of the phenotypes of control and AS-oligo-injected host-transfer embryos. (G,H) Cross-section of uninjected (G) and AS-oligo-injected (H) host-transfer embryos at the tadpole stage. H represents the most severe ventralization phenotype. (I) RT-PCR results showing effects of Dnd1 knockdown on the expression of NCAM, type II collagen (col II), muscle actin (m-actin), epidermal keratin (e-keratin), alas and α-globin at stage 20. (J) Real-time RT-PCR showing the expression of Wnt target genes (Xnr3 and siamois), organizer-specific genes (noggin and chordin), and ventral markers (msx1, sizzled, vent1 and vent2), and dnd1 in control and AS-oligo-injected host-transfer embryos (stage 10). All samples were normalized to odc levels. (K) Whole-mount in situ hybridization showing the expression of chordin and vent2 in control and AS-oligo-injected host-transfer embryos at stage 10.5. Arrows point to the dorsalmost region of control embryos, a region positive for chordin expression, but negative for vent2 expression.
Dnd1 is required for dorsal development
We then carried out loss-of-function studies. A phosphorothioate-modified AS-oligo was selected to knock down maternal dnd1 mRNA in Xenopus oocytes (supplementary material Fig. S1). The level of endogenous Dnd1 protein was significantly reduced 24 hours after injection of the AS-oligo (7.5 ng) (Fig. 1C). Using this oligo, we performed host-transfer experiments to generate Dnd1-depleted embryos.
Intriguingly, we observed a strong ventralization phenotype when AS-oligo-injected host transfer embryos reached the tadpole stage (Fig. 1D,E). To simplify our analysis, we divided embryos into three categories using the standard dorsal-anterior index (DAI) (Kao and Elinson, 1988). Embryos falling into category I have head, trunk, and tail structures (DAI 4-5). Category II embryos show an intermediate ventralization phenotype. These embryos developed a trunk and tail, but lacked a head (DAI 2-3). Category III embryos are strongly ventralized, with either no distinguishable structures or a tail-like structure only (DAI 0-1). As shown in Fig. 1D,F, the majority of control embryos (94%) fell into category I. By contrast, AS-oligo-injected embryos were clearly ventralized, with 68% and 22% of embryos falling into categories II and III, respectively (Fig. 1E,F). We picked 10 injected embryos for histological analysis. Most of these embryos exhibited reduced dorsal structures. In the most severe cases (n=2), embryos completely lacked a neural tube, notochord, and somite (Fig. 1H). Injection of AS-oligo significantly decreased the expression of NCAM (neural), collagen II (notochord) and muscle actin (somite) in tailbud stage embryos. By contrast, ventral-specific genes, including epidermal keratin (skin), alas and α-globin (ventral blood island), were all upregulated (Fig. 1I).
To better characterize the phenotypes of Dnd1 knockdown embryos, we analyzed marker gene expression at the beginning of gastrulation (stage 10). Injection of dnd1 AS-oligo markedly decreased the expression of dorsal-specific genes, including siamois, Xnr3, chordin and noggin. The expression of ventral markers, including msx1, sizzled, vent1 and vent2 were upregulated (Fig. 1J). Parallel to the RT-PCR experiments, we performed in situ hybridization analyses. As shown in Fig. 1K, the organizer gene chordin was severely reduced in injected embryos (69%, n=13). The expression of vent2, a direct target of BMP signaling (Henningfeld et al., 2000) that is expressed in the ventral and lateral region in control embryos, was expanded into the most dorsal territory in injected embryos (50%, n=14). These results indicate that injection of dnd1 AS-oligo impairs organizer formation and results in ventralized embryos.
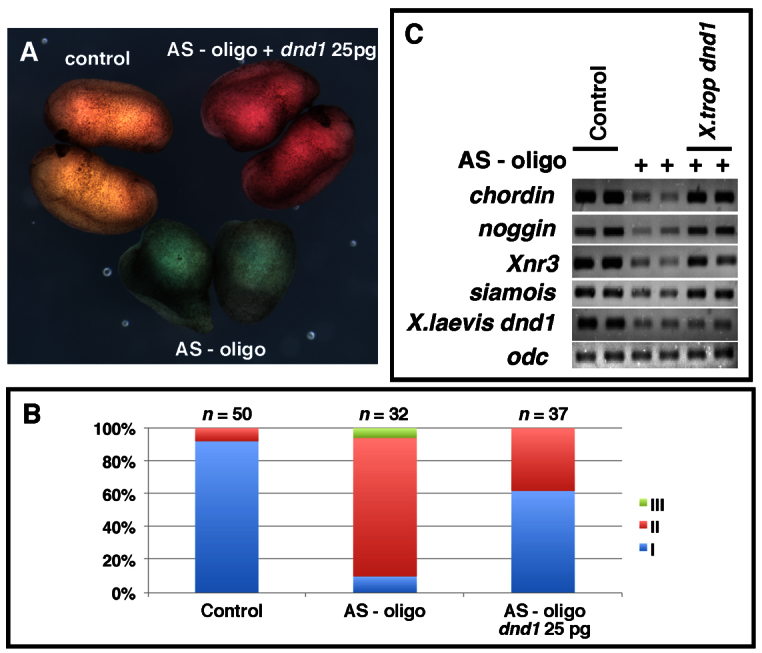
To verify that ventralization caused by injection of dnd1 AS-oligo is due to loss of maternal Dnd1, we performed rescue experiments using Xenopus tropicalis dnd1, which lacks the AS-oligo binding sequence. Consistent with the above observations, injection of dnd1 AS-oligo resulted in ventralization (Fig. 2A,B). Among 32 injected embryos, 6% and 84% fell into category II and III, respectively. Co-injection of dnd1 AS-oligo with dnd1 RNA (25 pg) rescued the ventralization phenotype, with only 38% of embryos now lacking head structures (category II). The rest (62%) of the embryos exhibited a relatively normal morphology. We analyzed marker gene expression at the beginning of gastrulation and found that co-injection of dnd1 RNA rescued the expression of siamois, Xnr3, chordin and noggin in AS-oligo-injected embryos (Fig. 2C). Based on these results, we conclude that maternal Dnd1 is required for dorsal development.
Fig. 2.
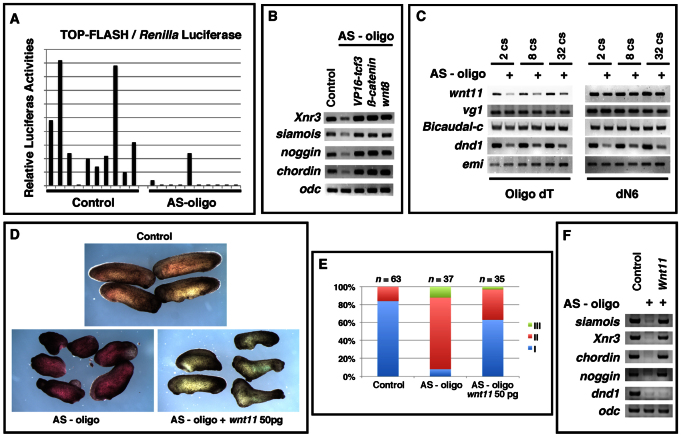
Specificity of Dnd1 knockdown. (A) Morphology of uninjected control (brown), AS-oligo-injected (blue), and AS-oligo- and tropicalis dnd1-injected (red) embryos at stage 25. (B) Summary of experiments shown in A. (C) RT-PCR results showing the expression of noggin, chordin, Xnr3, siamois and dnd1 in control, AS-oligo-injected, and AS-oligo- and tropicalis dnd1-injected (rescued) embryos at stage 10.5.
Dnd1 is required for the initiation of maternal Wnt signaling
During axis specification, several dorsal genes are directly regulated by Wnt signaling. Among these are Xnr3 (McKendry et al., 1997) and siamois (Brannon et al., 1997; Fan et al., 1998), which were significantly downregulated in Dnd1-depleted embryos. This raised the possibility that maternal Wnt signaling is affected by Dnd1 knockdown. To test this hypothesis, we measured the Wnt signaling activity in control and Dnd1 knockdown embryos using TOP-Flash, an artificial Wnt-responsive luciferase reporter (Molenaar et al., 1996). A mixture of the TOP-Flash reporter and pRL-SV40 (a Renilla luciferase control for injection) was injected into control and Dnd1 knockdown embryos. Embryos were harvested at the late blastula stage and subjected to a dual-luciferase assay. The TOP-Flash luciferase activity was detected in control embryos, but not in the majority of Dnd1-depleted embryos (Fig. 3A). This indicates that Wnt signaling is indeed affected by maternal depletion of Dnd1.
Fig. 3.
Maternal Dnd1 is required for the initiation of maternal Wnt signaling during axis specification. (A) TOP-Flash luciferase assay measuring maternal Wnt signaling activities in control and Dnd1-depleted embryos (two embryos each group). Host-transfer embryos were injected with a mixture of TOP-Flash and pRL-SV40 and harvested at stage 9.5. Firefly luciferase activity was normalized to that of Renilla luciferase. (B) RT-PCR showing that overexpression of VP16-tcf3 (10 pg), β-catenin (100 pg) and wnt8 (5 pg) rescued the expression of noggin, chordin, Xnr3 and siamois in Dnd1-depleted embryos at stage 10.5. VP16-tcf3, β-catenin and wnt8 were injected at the four-cell stage into the marginal zone of two blastomeres of Dnd1 knockdown embryos. (C) Depletion of maternal Dnd1 impaired polyadenylation of wnt11. Total RNAs were reverse transcribed with oligo dT (left) and random hexamers (dN6, right). RT-PCR was performed to measure the expression of wnt11, vg1, Bicaudal-c, dnd1 and emi1. (D) Morphology of control (brown, top), AS-oligo-injected (red, lower left), and AS-oligo- and wnt11-injected (50 pg) (blue, lower right) embryos. (E) Summary of the phenotypes observed in the Wnt11 rescue experiments. (F) RT-PCR results showing that overexpression of Wnt11 rescued the expression of noggin, chordin, Xnr3 and siamois in Dnd1 knockdown embryos at stage 10.5.
To determine whether the ventralization phenotype in Dnd1-depleted embryos was caused by defective Wnt signaling, we performed a rescue experiment using Wnt8, β-catenin, and constitutively active TCF3 (VP16-TCF3). We found that overexpression of VP16-TCF3, β-catenin, or Wnt8 rescued the expression of siamois, Xnr3, chordin and noggin in Dnd1 knockdown embryos (Fig. 3B), indicating that the ventralization phenotype in Dnd1-depleted embryos is indeed caused by defective Wnt signaling. The observation that Wnt8 rescues dorsal gene expression in Dnd1 knockdown embryos further suggested that the Wnt pathway, per se, remained intact in Dnd1 knockdown embryos. It seemed likely that Dnd1 is required for the initiation of maternal Wnt signaling.
Elegant studies by Heasman and colleagues have demonstrated that Wnt11 is the key factor that initiates Wnt signaling during Xenopus axis specification (Tao et al., 2005; Kofron et al., 2007; Cha et al., 2008). Shortly after fertilization, maternal wnt11 mRNA is translocated to the future dorsal side (Tao et al., 2005) and becomes polyadenylated (Schroeder et al., 1999). Polyadenylation of wnt11 mRNA correlates with the translation of Wnt11 protein (Schroeder et al., 1999). To understand the mechanism through which maternal Dnd1 regulates the initiation of Wnt signaling during axis specification, we harvested control and Dnd1 knockdown embryos at the 2-, 8- and 32-cell stages and assessed the polyadenylation status of wnt11 mRNA using random primed and oligo(dT) primed cDNAs. Oligo(dT) primed RT-PCR analysis allows preferential detection of polyadenylated mRNA, whereas random-primed RT-PCR analysis measures the total amount of mRNA, regardless of the polyadenylation state. As a control, we also analyzed vg1, a vegetally localized maternal RNA (Weeks and Melton, 1987), Bicaudal-c, a maternal mRNA that undergoes polyadenylation after fertilization (Wessely and De Robertis, 2000), and emi1, a maternal mRNA that is not polyadenylated during early cleavage stages (Graindorge et al., 2006). As shown in Fig. 3C, the total amount of wnt11 mRNA remained unchanged in Dnd1 knockdown embryos, whereas the amount of polyadenylated wnt11 mRNA was reduced. Reduced polyadenylation of wnt11 was not characteristic of a more general defect of Dnd1 knockdown, as vg1, Bicaudal-c and emi1 were unaffected.
Polyadenylation of wnt11 mRNA is important for its translation (Schroeder et al., 1999). Reduced polyadenylation of wnt11 in Dnd1 knockdown embryos suggested that the ventralization phenotype might be caused by the defective biosynthesis of Wnt11. To test this hypothesis, we asked whether overexpression of Wnt11 could rescue the ventralization phenotype in Dnd1-depleted embryos. Consistent with previous results, the majority (92%) of Dnd1-depleted embryos was ventralized, falling into Category II and III. When Wnt11 was overexpressed, only 37% of embryos exhibited ventralized phenotype (Fig. 3D,E). At the gastrula stage (stage 10.5), the expression of siamois, Xnr3, chordin and noggin was rescued by Wnt11 overexpression (Fig. 3F). These results indicate that Dnd1 functions upstream of Wnt11 during axis specification.
Dnd1 is required for vegetal cortical microtubule assembly
As an RNA-binding protein, Dnd1 may bind wnt11 mRNA directly and regulate its polyadenylation. Alternatively, Dnd1 may be involved in early developmental processes that are important for polyadenylation of wnt11. To distinguish between these two possibilities, we first determined whether Dnd1 protein binds wnt11 mRNA using GST-Dnd1 proteins purified from bacteria. We obtained negative results from the in vitro binding assay (Fig. 7C). So we set out to determine whether Dnd1 is involved in early developmental processes important for polyadenylation of wnt11.
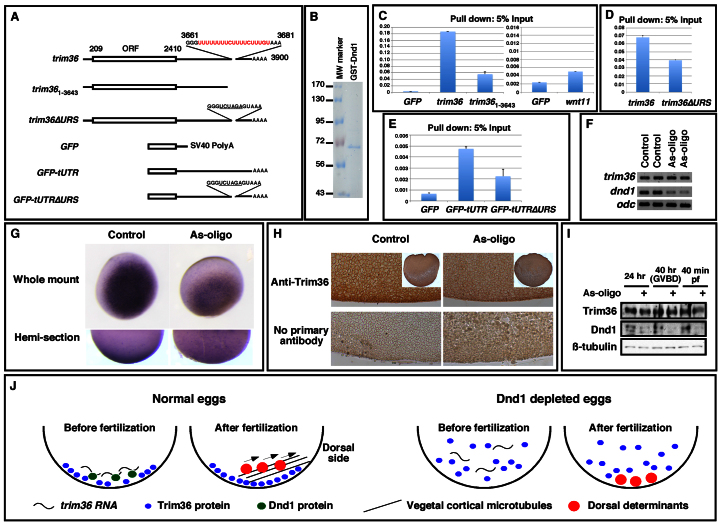
Fig. 7.
Dnd1 binds trim36 directly and anchors trim36 to the vegetal cortex. (A) Schematic of trim36 deletion constructs used in the in vitro pull-down assay. (B) Coomassie Blue staining showing purified GST-Dnd1 protein used in the pull-down assay. (C-E) Real-time RT-PCR showing the ratio between RNAs associated with GST-Dnd1 protein and 5% of RNA input. (C) GST-Dnd1 efficiently pulled down trim36, but not GFP or wnt11. (D) Mutation of the URS in trim36 reduced the binding of trim36 by GST-Dnd1. (E) Fusing the 3′-UTR of trim36 to GFP increased the binding of GST-Dnd1 to GFP RNA. (F) RT-PCR showing that knockdown of Dnd1 had no effect on the level of trim36 mRNA in eggs. (G) In situ hybridization showing that trim36 is no longer localized to the vegetal cortex in dnd1-depleted eggs. (H) Immunohistochemistry showing that knockdown of Dnd1 interfered with the accumulation of Trim36 protein in the vegetal cortex of eggs. Inserts at lower magnification show whole eggs. (I) Immunoprecipitation/western blotting showing that the level of Trim36 protein remained unchanged in Dnd1 depleted eggs. Protein lysates from 50 oocytes or eggs were incubated first with an anti-Trim36 antibody to immunoprecipitate Trim36. Supernatants were then incubated with an anti-Dnd1 antibody to pull down Dnd1. (J) Model of Dnd1 function at the vegetal pole. Dnd1 anchors trim36 within the vegetal cortex resulting in localized translation and vegetal accumulation of Trim36. Trim36 promotes vegetal microtubular array formation required to move and thus activate the dorsal determinants. Without Dnd1, trim36 localization is disrupted, Trim36 protein is diluted, microtubular arrays do not form; dorsal determinants remain inactive at the vegetal pole.
Polyadenylation of wnt11 normally relies on cortical rotation. It has been reported that polyadenylation of wnt11 is sensitive to UV irradiation (Schroeder et al., 1999), a treatment that ventralizes embryos by blocking cortical rotation (Elinson and Rowning, 1988) and the translocation of dorsal determinants from the vegetal pole to the future dorsal side of the embryo (Darras et al., 1997; Medina et al., 1997). We thus assessed vegetal cortical microtubule formation in control and Dnd1 knockdown embryos. At 55 minutes post-fertilization, microtubules were detected in the vegetal cortex in all control embryos, with 38% of them forming disordered microtubules and 62% containing wavy parallel microtubule bundles. The variation in the morphology of vegetal cortical microtubules is likely to be due to a variable delay in fertilization caused by the host-transfer procedure. In contrast to control embryos, most Dnd1 knockdown embryos (73%) completely lacked vegetal cortical microtubules. Only a small percentage of embryos (27%; indicated by arrows) had a few detectable microtubules. Injection of tropicalis dnd1 RNA rescued microtubule formation in the majority of AS-oligo-injected embryos (88%). Among 24 embryos analyzed, 54% formed parallel microtubule arrays, 34% had disordered microtubules, and 12% (not shown) completely lacked microtubules in their vegetal cortex (Fig. 4A). Parallel to these analyses in fertilized eggs, we examined the effect of Dnd1 knockdown on vegetal cortical microtubule formation in eggs artificially activated by needle-pricking. We performed the analysis at 40 minutes post-pricking, when disordered microtubules just begin to polymerize into the vegetal cortex (supplementary material Fig. S4). As shown in Fig. 4B, vegetal cortical microtubules were detected in 84% of control eggs. By contrast, 70% of AS-oligo-injected eggs completely lacked cortical microtubules. Vegetal cortical microtubule formation was rescued by co-injection of tropicalis dnd1 with the AS-oligo (92%).
Fig. 4.
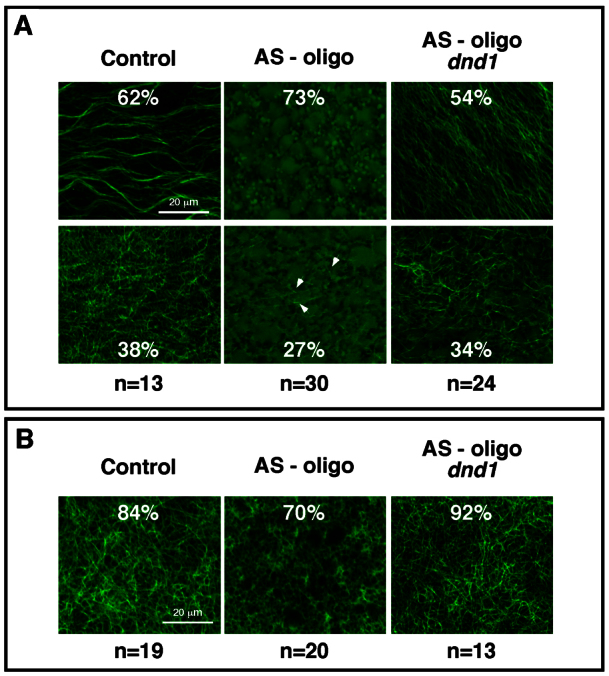
Maternal Dnd1 is required for vegetal cortical microtubule assembly. (A,B) Confocal images showing the formation of vegetal cortical microtubules in fertilized eggs (A, 55 minutes post-fertilization; top and bottom figures) and in artificially activated eggs (B, 40 minutes post-needle pricking). Fixed eggs were bisected and the vegetal halves were stained with an anti-Tubulin antibody. Formation of vegetal cortical microtubules was severely disrupted in all Dnd1 knockdown fertilized eggs. In 27% of Dnd1 knockdown fertilized eggs (A, lower middle), a few microtubules were detected (indicated by arrows). Also note that at 40 minutes post-needle pricking, disordered microtubules just begin to polymerize into the vegetal cortex of activated eggs (supplementary material Fig. S4).
To determine whether Dnd1 is a general regulator of microtubule assembly or controls microtubule formation locally in the vegetal pole of the embryo, we assessed microtubule formation throughout the embryo (40 minutes post-fertilization). We found that microtubule assembly in the animal hemisphere remained unaffected (supplementary material Fig. S3). Cortical microtubule formation was detectably reduced in the marginal zone of Dnd1 knockdown embryos (compare supplementary material Fig. S3C and S3G). In the vegetal pole, cortical microtubule formation was severely disrupted by Dnd1 knockdown (compare supplementary material Fig. S3D and S3H). It appears that Dnd1 is absolutely required for cortical microtubule assembly in the vegetal pole, and to some degree is required for cortical microtubule assembly in the equator of the embryo.
Dorsal determinants remain intact in Dnd1-depleted embryos
During axis specification, vegetal cortical microtubules serve as tracks to transport dorsal determinants from the vegetal pole to the future dorsal side of the embryo (Gerhart et al., 1989; Weaver and Kimelman, 2004; Gerhart, 2010; Houston, 2012). Ventralization caused by defective vegetal cortical microtubule assembly can be rescued by using gravity to force translocation of dorsal determinants from the vegetal pole to the equator of the egg. This can be achieved by centrifuging eggs embedded in gelatin (Black and Gerhart, 1985; Black and Gerhart, 1986), or by tipping embryos 90 degrees relative to the animal-vegetal axis during the first embryonic cell cycle (Scharf and Gerhart, 1980; Scharf and Gerhart, 1983; Cuykendall and Houston, 2009). To determine whether maternal Dnd1 functions in axis specification through controlling vegetal cortical microtubule assembly, we divided Dnd1-depleted embryos into two groups. One group was tipped 90 degrees during the first embryonic cell cycle and the other group was cultured under normal conditions.
As shown in Fig. 5A,B, the majority of Dnd1-depleted embryos cultured under normal conditions were ventralized, with 72% of them falling into category II and III. Tipped embryos, however, exhibited a much milder ventralization phenotype. Formation of a head was observed in 65% of tipped embryos (category I). The remaining 35% of embryos developed trunk structures (category II). At the gastrula stage, the expression of siamois, Xnr3, chordin and noggin, which was reduced in Dnd1-depleted embryos, was rescued by tipping (Fig. 5C). Based on these observations, we conclude that dorsal determinants remained intact in Dnd1-depleted embryos. It appears that maternal Dnd1 functions in axis specification by controlling vegetal cortical microtubule assembly, which in turn supports the translocation of dorsal determinants from the vegetal pole to the dorsal side.
Fig. 5.
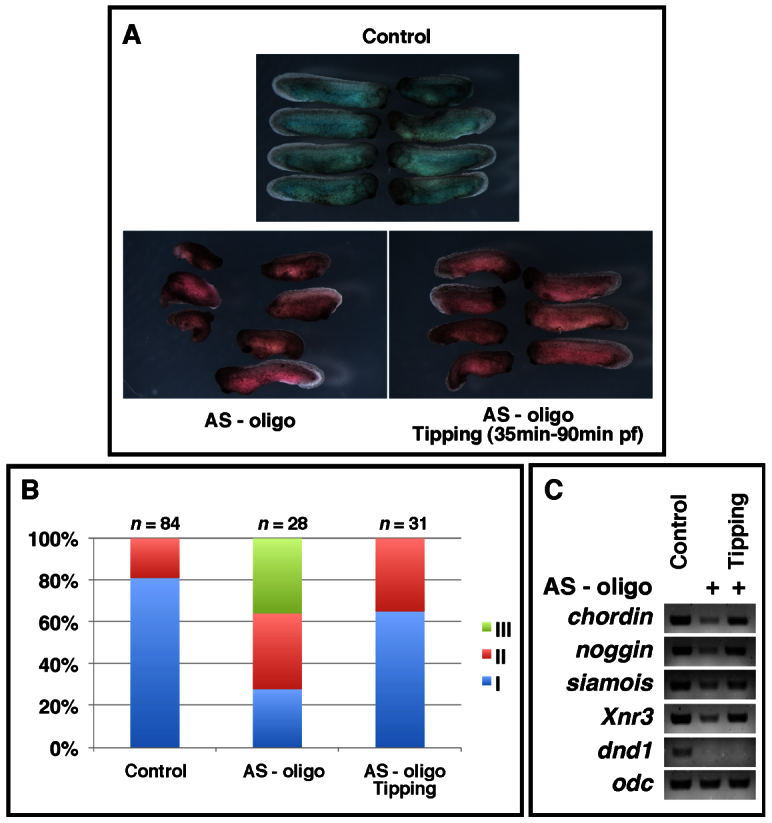
Dorsal determinants remain intact in dnd1-depleted embryos. (A) Morphology of uninjected control embryos (top), dnd1-depleted embryos (AS-oligo, lower left), and dnd1-depleted embryos that were tipped 90 degrees relative to the animal-vegetal axis during the first embryonic cell cycle (lower right). (B) Summary of the phenotypes of the tipping experiment. (C) RT-PCR showing that tipping dnd1-depleted embryos during the first embryonic cell cycle rescued the expression of noggin, chordin, Xnr3 and siamois in Dnd1 knockdown embryos at stage 10.5.
Dnd1 functions as an RNA-binding protein during axis specification
Next, we determined whether Dnd1 acted as an RNA-binding protein to regulate vegetal cortical microtubule assembly and subsequent axis specification. Several amino acid residues in the RNA-recognition motif (RRM) of zebrafish Dnd1 are essential for its function during primordial germ cell (PGC) development. Among these is Y104, which is involved in RRM-RNA interaction (Slanchev et al., 2009). Therefore, we generated Y125C, a mutant form of Xenopus tropicalis Dnd1 that is analogous to the Y104C of zebrafish Dnd1 (Slanchev et al., 2009), and tested the activities of Y125C in rescue experiments.
AS-oligo (7.5 ng) was injected into oocytes, either alone or together with Y125C (25 pg), or with the wild-type dnd1 (25 pg). After oocyte maturation, eggs were activated by needle-pricking. Activated eggs were harvested at 40 minutes post-pricking and subjected to microtubule staining. Consistent with the above observations, vegetal cortical microtubule formation was detected in all control eggs. Depletion of maternal Dnd1 severely interfered with the formation of vegetal cortical microtubules in 86% of eggs. Overexpression of the wild-type Dnd1 rescued vegetal cortical microtubule formation in 88% of eggs. Overexpression of Y125C, however, failed to restore vegetal cortical microtubule formation, with the majority of eggs (90%) lacking detectable microtubule arrays in the vegetal cortex (Fig. 6A).
Fig. 6.
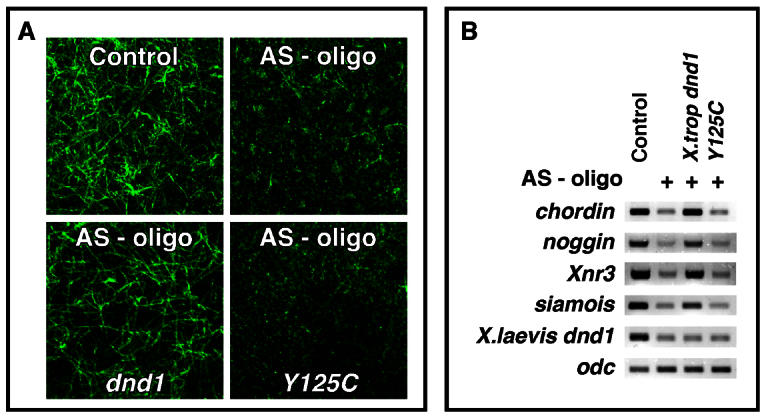
The RNA-binding activity of Dnd1 is required for its function during vegetal cortical microtubule assembly in activated eggs and dorsal development in fertilized eggs. (A) Confocal images showing the formation of vegetal cortical microtubules in uninjected control (upper/left), AS-oligo-injected (upper/right), AS-oligo- and dnd1-injected (lower/left), and AS-oligo- and Y125C-injected (lower/right) eggs (40 minutes post-needle-pricking). Fixed eggs were bisected and the vegetal halves were stained with an anti-Tubulin antibody. (B) RT-PCR showing that overexpression of the wild-type dnd1, but not Y125C, rescued the expression of noggin, chordin, Xnr3 and siamois in Dnd1 knockdown embryos (stage 10.5).
To determine whether Y125C is capable of rescuing dorsal development in Dnd1 knockdown embryos, we performed a host-transfer experiment and analyzed dorsal-specific gene expression when embryos reached stage 10. As shown in Fig. 6B, the expression of siamois, Xnr3, chordin and noggin, which was reduced in Dnd1 knockdown embryos, was rescued by the wild-type dnd1 (25 pg), but not by the same amount of Y125C (25 pg). These results indicate that maternal Dnd1 acts as an RNA-binding protein to control vegetal cortical microtubule formation and subsequent dorsal development.
Dnd1 binds trim36 directly and anchors trim36 to the vegetal cortex
Maternal Trim36 is essential for polymerization of vegetal cortical microtubules during cortical rotation (Cuykendall and Houston, 2009). To gain insight into the mechanism by which Dnd1 regulates vegetal cortical microtubule formation, we performed in vitro RNA pull-down assays to determine whether Dnd1 binds trim36 RNA directly using bacterially expressed GST-Dnd1 protein (Fig. 7B). As shown in Fig. 7C, trim36 RNA, but not GFP or wnt11, was efficiently pulled down by GST-Dnd1, suggesting that Dnd1 binds trim36 directly. Dnd1 binds to uridine-rich sequences (URS) (Kedde et al., 2007). The 3′-UTR of trim36 contains a URS (Fig. 7A). We generated a trim361-3643, which lacks the URS and the 3′-end of trim36. In the pull-down assay, Dnd1 bound to trim361-3643 less efficiently (Fig. 7C). We then constructed a trim36ΔURS, which lacks the URS (Fig. 7A), and assayed the binding of trim36ΔURS by Dnd1. As expected, mutation of the URS in trim36 resulted in a 0.4-fold decrease in Dnd1 binding (Fig. 7D), indicating that the URS is indeed a Dnd1-binding site. In agreement with this view, we found that GFP-tUTR, a GFP construct fused with the wild-type 3′-UTR of trim36, was eightfold more accessible to Dnd1 when compared with GFP with a SV40-polyA signal. By contrast, fusing the mutant form of trim36 3′-UTR to GFP increased the Dnd1 binding only 3.5-fold (Fig. 7E). Collectively, these results demonstrate the URS in the 3′-UTR of trim36 is a Dnd1-binding site. The observation that mutation of the URS only partially reduces the binding of trim36 by Dnd1 argues for the existence of non-uridine-rich Dnd1-binding sites in trim36 RNA. Nevertheless, results from the above experiments allow us to conclude that Dnd1 binds trim36 directly.
To investigate how Dnd1 regulates trim36 in vivo, we first analyzed the total level of trim36 RNA in Dnd1-depleted eggs. RT-PCR analysis revealed that knockdown of Dnd1 had no effect on the abundance of trim36 RNA (Fig. 7F). We then performed in situ hybridization to analyze the localization of trim36. As expected, trim36 was localized to the vegetal cortex in control eggs. In Dnd1 knockdown eggs, however, trim36 was no longer localized to the vegetal cortex (Fig. 7G). This indicates that Dnd1 anchors trim36 mRNA to the vegetal cortex. We consistently found that Trim36 protein was enriched in the vegetal cortex in control eggs. In Dnd1-depleted eggs, however, accumulation of Trim36 protein in the vegetal cortex was not observed (Fig. 7H). Depletion of Dnd1 did not alter the total level of Trim36 protein (Fig. 7I), ruling out the possibility that Dnd1 regulates translation of trim36. Thus, Dnd1 anchors trim36 RNA to the vegetal cortex and is required for accumulation of Trim36 protein in the vegetal cortex.
DISCUSSION
The functions of Dnd1 during PGC development have been studied extensively. Dnd1 is expressed in PGCs (Weidinger et al., 2003; Youngren et al., 2005; Horvay et al., 2006; Aramaki et al., 2007) and is required for their survival and migration (Weidinger et al., 2003; Youngren et al., 2005; Horvay et al., 2006; Cook et al., 2009). In mice with the 129/SvJ background, loss of Dnd1 causes testicular teratomas (Youngren et al., 2005; Lam et al., 2007; Zhu et al., 2007; Cook et al., 2009; Cook et al., 2011). During PGC development, Dnd1 protects its target mRNAs by binding to URSs and blocking microRNAs from their complementary binding sites nearby. In the absence of Dnd1, a subset of PGC-specific mRNAs becomes downregulated in zebrafish embryos (Kedde et al., 2007). In Xenopus, Dnd1 functions together with ElrB1 to protect germ-cell-specific mRNAs from miR-18-mediated degradation during gastrulation (Koebernick et al., 2010). Here, we report a novel and essential function of Dnd1 in Xenopus axis specification, an event that occurs before first cleavage.
Vertebrate axis specification requires asymmetric activation of Wnt signaling and subsequent organizer formation on the future dorsal side of the embryo. In Xenopus, egg activation initiates microtubule dynamics and triggers formation of vegetal cortical microtubular arrays, which serve as tracks to transport dorsal determinants from the vegetal pole to the future dorsal side. As a result, wnt11, which is responsible for the initiation of maternal Wnt signaling, is translocated to the future dorsal side (Tao et al., 2005) and becomes polyadenylated (Schroeder et al., 1999). Dorsally accumulated Wnt11 then activates β-catenin-dependent transcription and ultimately leads to organizer formation. Our studies indicate that maternal Dnd1, localized to the vegetal pole, is responsible for regionally anchoring key regulators of vegetal cortical microtubule formation (see Fig. 7J for our working model). Knockdown of Dnd1 interferes with the vegetal localization of trim36 in oocytes, without affecting other master regulators of early embryonic patterning, such as VegT and wnt11 (supplementary material Fig. S6). After fertilization, the formation of vegetal cortical microtubules was disrupted within the first embryonic cell cycle. Later on, polyadenylation of wnt11, which relies on cortical rotation, was impaired and by the late blastula stage, Wnt signaling activity declined. At the gastrula and tailbud stages, dorsal-specific genes were downregulated in Dnd1 knockdown embryos. This was accompanied by a marked increase in the expression of ventral-specific genes and resulted in morphologically ventralized embryos.
Remarkably, phenotypes caused by maternal depletion of Dnd1 are similar to those observed in embryos lacking maternal Trim36 (Cuykendall and Houston, 2009). Like dnd1, trim36 is localized to the vegetal hemisphere in stage VI oocytes. After the first embryonic cell division, it is restricted to the germ plasm at the vegetal pole (Horvay et al., 2006; Cuykendall and Houston, 2009). When maternal trim36 is depleted, microtubular arrays cannot form in the vegetal cortex and embryos become ventralized (Cuykendall and Houston, 2009). These similarities suggested a link between Dnd1 and Trim36 during the formation of vegetal cortical microtubules.
Indeed, our results demonstrate that trim36 is a target of Dnd1. The URS in the 3′-UTR of trim36 is a Dnd1-binding site. Mutation of this URS reduced the binding of trim36 by Dnd1. Of note, trim36 lacking the URS can still be pulled down by Dnd1 protein, albeit less efficiently. This suggests that in addition to the URS, Dnd1 binds to some non-uridine-rich sequences in trim36. Our analysis further demonstrates that the binding of trim36 by Dnd1 is essential for anchoring trim36 to the vegetal cortex. In agreement with this view, knockdown of Dnd1 disrupted the localization of trim36 in the vegetal cortex, without affecting the overall stability or translation of trim36. Trim36 protein, which is highly enriched in the vegetal cortex in wild-type eggs, is no longer localized in Dnd1-depleted eggs. It seems likely that maternal Dnd1 regulates assembly of vegetal cortical microtubules, at least in part, by anchoring key regulators of vegetal cortical microtubule formation, such as trim36, to the vegetal cortex. Depletion of maternal Dnd1 disrupted vegetal localization of trim36 mRNA. As a result, Trim36 protein was not concentrated vegetally, but became regionally diluted as it dispersed throughout the oocyte and egg. This in turn impaired the formation of vegetal cortical microtubules after fertilization. Consequently, dorsal determinants remained inactive at the vegetal pole (Fig. 7J).
It is worth mentioning that while knockdown of Dnd1 ventralizes embryos, overexpression of Dnd1 had no effect on axis specification (supplementary material Fig. S5). Host-transfer embryos in which Dnd1 was overexpressed showed normal vegetal cortical microtubule formation (supplementary material Fig. S5A), polyadenylation of wnt11 (supplementary material Fig. S5B), and organizer formation (supplementary material Fig. S5C). Nevertheless, these embryos exhibited mild defects in blastopore closure. At the tadpole stage, Dnd1-overexpressed embryos developed normal head and anterior trunk structures, but showed defects in their posterior structures (supplementary material Fig. S5D). It is likely that overexpression of Dnd1, a key regulator of germline development, disrupted endoderm development at the vegetal pole and indirectly affected posterior development.
Previously, Horvay and co-workers reported that Dnd1-depleted embryos lost their PGCs, but were otherwise normal (Horvay et al., 2006). Why do our results differ from what they observed? Dnd1 protein is present in the vegetal pole of fully grown oocytes (Fig. 1A) and anchors trim36 to the vegetal cortex (Fig. 7G). In the study by Horvay et al. (Horvay et al., 2006), a morpholino was injected after fertilization to block further translation of Dnd1. Considering that vegetal cortical microtubule assembly occurs 40 to 70 minutes after egg activation (supplementary material Fig. S4), it is likely that in their experiments, the level of Dnd1 protein was not significantly reduced during this critical period and vegetal localization of Trim36 was not affected. In our experiments, we used an AS-oligo to deplete dnd1 mRNA in the oocyte, followed by oocyte maturation, host transfer, and fertilization. This approach allowed successful knockdown of Dnd1 protein before oocyte maturation (Fig. 1C). It seems likely that Horvay et al. (Horvay et al., 2006) did not observe the early requirement for Dnd1 in cortical microtubule formation because Dnd1 was depleted too late in their experiments.
Interestingly, overexpression of Trim36 cannot rescue vegetal cortical microtubule formation in Dnd1-depleted embryos (not shown). This raises the possibility that in addition to trim36, Dnd1 may regulate other RNAs that are important for vegetal cortical microtubule formation. In the future, it will be important to identify additional targets of Dnd1 and to investigate their roles in controlling microtubule assembly at the vegetal pole. The molecular mechanism through which Trim36 regulates vegetal cortical microtubule formation remains unclear. The vegetal cortex lacks a conventional microtubule-organizing center (Houliston and Elinson, 1991b; Schroeder and Gard, 1992). It is unlikely that Trim36, which is localized to the vegetal cortex, is involved in nucleation of microtubules. However, it has been reported that there is a potent plus-end-specific inhibitor of microtubule assembly in oocytes and a plus-end-specific promoter in activated eggs (Gard and Kirschner, 1987). It is possible that maternal Trim36, an E3 ubiquitin ligase, is involved in the protein degradation step required to remove the putative plus-end-specific inhibitor. Conversely, Trim36 may indirectly control the biosynthesis of the plus-end-specific promoter in the vegetal cortex. Clearly, additional studies will be required to test these possible models.
Supplementary Material
Acknowledgements
We are grateful to Dr P. Klein for insightful discussions and for reading the manuscript. Anti-Tubulin antibody was obtained from the Developmental Studies Hybridoma Bank (DSHB), developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242, USA.
Footnotes
Funding
This work was supported in part by National Institutes of Health (NIH) grants [R01GM093217 to J.Y. and GM33932 to M.L.K.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.094748/-/DC1
References
- Aramaki S., Sato F., Kato T., Soh T., Kato Y., Hattori M. A. (2007). Molecular cloning and expression of dead end homologue in chicken primordial germ cells. Cell Tissue Res. 330, 45-52 [DOI] [PubMed] [Google Scholar]
- Black S. D., Gerhart J. C. (1985). Experimental control of the site of embryonic axis formation in Xenopus laevis eggs centrifuged before first cleavage. Dev. Biol. 108, 310-324 [DOI] [PubMed] [Google Scholar]
- Black S. D., Gerhart J. C. (1986). High-frequency twinning of Xenopus laevis embryos from eggs centrifuged before first cleavage. Dev. Biol. 116, 228-240 [DOI] [PubMed] [Google Scholar]
- Brannon M., Gomperts M., Sumoy L., Moon R. T., Kimelman D. (1997). A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 11, 2359-2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha S. W., Tadjuidje E., Tao Q., Wylie C., Heasman J. (2008). Wnt5a and Wnt11 interact in a maternal Dkk1-regulated fashion to activate both canonical and non-canonical signaling in Xenopus axis formation. Development 135, 3719-3729 [DOI] [PubMed] [Google Scholar]
- Cook M. S., Coveney D., Batchvarov I., Nadeau J. H., Capel B. (2009). BAX-mediated cell death affects early germ cell loss and incidence of testicular teratomas in Dnd1(Ter/Ter) mice. Dev. Biol. 328, 377-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook M. S., Munger S. C., Nadeau J. H., Capel B. (2011). Regulation of male germ cell cycle arrest and differentiation by DND1 is modulated by genetic background. Development 138, 23-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuykendall T. N., Houston D. W. (2009). Vegetally localized Xenopus trim36 regulates cortical rotation and dorsal axis formation. Development 136, 3057-3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darras S., Marikawa Y., Elinson R. P., Lemaire P. (1997). Animal and vegetal pole cells of early Xenopus embryos respond differently to maternal dorsal determinants: implications for the patterning of the organiser. Development 124, 4275-4286 [DOI] [PubMed] [Google Scholar]
- Elinson R. P., Rowning B. (1988). A transient array of parallel microtubules in frog eggs: potential tracks for a cytoplasmic rotation that specifies the dorso-ventral axis. Dev. Biol. 128, 185-197 [DOI] [PubMed] [Google Scholar]
- Fan M. J., Grüning W., Walz G., Sokol S. Y. (1998). Wnt signaling and transcriptional control of Siamois in Xenopus embryos. Proc. Natl. Acad. Sci. USA 95, 5626-5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisue M., Kobayakawa Y., Yamana K. (1993). Occurrence of dorsal axis-inducing activity around the vegetal pole of an uncleaved Xenopus egg and displacement to the equatorial region by cortical rotation. Development 118, 163-170 [DOI] [PubMed] [Google Scholar]
- Gard D. L., Kirschner M. W. (1987). Microtubule assembly in cytoplasmic extracts of Xenopus oocytes and eggs. J. Cell Biol. 105, 2191-2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart J. (2010). Enzymes, embryos, and ancestors. Annu. Rev. Cell Dev. Biol. 26, 1-20 [DOI] [PubMed] [Google Scholar]
- Gerhart J., Danilchik M., Doniach T., Roberts S., Rowning B., Stewart R. (1989). Cortical rotation of the Xenopus egg: consequences for the anteroposterior pattern of embryonic dorsal development. Development 107 Suppl., 37-51 [DOI] [PubMed] [Google Scholar]
- Gönczy P. (2008). Mechanisms of asymmetric cell division: flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 9, 355-366 [DOI] [PubMed] [Google Scholar]
- Graindorge A., Thuret R., Pollet N., Osborne H. B., Audic Y. (2006). Identification of post-transcriptionally regulated Xenopus tropicalis maternal mRNAs by microarray. Nucleic Acids Res. 34, 986-995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland R., Gerhart J. (1997). Formation and function of Spemann's organizer. Annu. Rev. Cell Dev. Biol. 13, 611-667 [DOI] [PubMed] [Google Scholar]
- Heasman J. (1997). Patterning the Xenopus blastula. Development 124, 4179-4191 [DOI] [PubMed] [Google Scholar]
- Heasman J. (2006). Patterning the early Xenopus embryo. Development 133, 1205-1217 [DOI] [PubMed] [Google Scholar]
- Heasman J., Holwill S., Wylie C. C. (1991). Fertilization of cultured Xenopus oocytes and use in studies of maternally inherited molecules. Methods Cell Biol. 36, 213-230 [DOI] [PubMed] [Google Scholar]
- Henningfeld K. A., Rastegar S., Adler G., Knöchel W. (2000). Smad1 and Smad4 are components of the bone morphogenetic protein-4 (BMP-4)-induced transcription complex of the Xvent-2B promoter. J. Biol. Chem. 275, 21827-21835 [DOI] [PubMed] [Google Scholar]
- Holowacz T., Elinson R. P. (1995). Properties of the dorsal activity found in the vegetal cortical cytoplasm of Xenopus eggs. Development 121, 2789-2798 [DOI] [PubMed] [Google Scholar]
- Horvay K., Claussen M., Katzer M., Landgrebe J., Pieler T. (2006). Xenopus Dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Dev. Biol. 291, 1-11 [DOI] [PubMed] [Google Scholar]
- Houliston E. (1994). Microtubule translocation and polymerisation during cortical rotation in Xenopus eggs. Development 120, 1213-1220 [Google Scholar]
- Houliston E., Elinson R. P. (1991a). Evidence for the involvement of microtubules, ER, and kinesin in the cortical rotation of fertilized frog eggs. J. Cell Biol. 114, 1017-1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houliston E., Elinson R. P. (1991b). Patterns of microtubule polymerization relating to cortical rotation in Xenopus laevis eggs. Development 112, 107-117 [DOI] [PubMed] [Google Scholar]
- Houliston E., Le Guellec R., Kress M., Philippe M., Le Guellec K. (1994). The kinesin-related protein Eg5 associates with both interphase and spindle microtubules during Xenopus early development. Dev. Biol. 164, 147-159 [DOI] [PubMed] [Google Scholar]
- Houston D. W. (2012). Cortical rotation and mRNA localization in Xenopus axis formation. Wiley Interdiscip. Rev. Dev. Biol. 1, 371-388 [DOI] [PubMed] [Google Scholar]
- Jin Z., Shi J., Saraf A., Mei W., Zhu G. Z., Strack S., Yang J. (2009). The 48-kDa alternative translation isoform of PP2A:B56epsilon is required for Wnt signaling during midbrain-hindbrain boundary formation. J. Biol. Chem. 284, 7190-7200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageura H. (1997). Activation of dorsal development by contact between the cortical dorsal determinant and the equatorial core cytoplasm in eggs of Xenopus laevis. Development 124, 1543-1551 [DOI] [PubMed] [Google Scholar]
- Kao K. R., Elinson R. P. (1988). The entire mesodermal mantle behaves as Spemann's organizer in dorsoanterior enhanced Xenopus laevis embryos. Dev. Biol. 127, 64-77 [DOI] [PubMed] [Google Scholar]
- Kedde M., Strasser M. J., Boldajipour B., Oude Vrielink J. A., Slanchev K., le Sage C., Nagel R., Voorhoeve P. M., van Duijse J., Ørom U. A., et al. (2007). RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131, 1273-1286 [DOI] [PubMed] [Google Scholar]
- Kikkawa M., Takano K., Shinagawa A. (1996). Location and behavior of dorsal determinants during first cell cycle in Xenopus eggs. Development 122, 3687-3696 [DOI] [PubMed] [Google Scholar]
- Knoblich J. A. (2008). Mechanisms of asymmetric stem cell division. Cell 132, 583-597 [DOI] [PubMed] [Google Scholar]
- Knoblich J. A. (2010). Asymmetric cell division: recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 11, 849-860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koebernick K., Loeber J., Arthur P. K., Tarbashevich K., Pieler T. (2010). Elr-type proteins protect Xenopus Dead end mRNA from miR-18-mediated clearance in the soma. Proc. Natl. Acad. Sci. USA 107, 16148-16153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofron M., Birsoy B., Houston D., Tao Q., Wylie C., Heasman J. (2007). Wnt11/beta-catenin signaling in both oocytes and early embryos acts through LRP6-mediated regulation of axin. Development 134, 503-513 [DOI] [PubMed] [Google Scholar]
- Lai F., Zhou Y., Luo X., Fox J., King M. L. (2011). Nanos1 functions as a translational repressor in the Xenopus germline. Mech. Dev. 128, 153-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F., Singh A., King M. L. (2012). Xenopus Nanos1 is required to prevent endoderm gene expression and apoptosis in primordial germ cells. Development 139, 1476-1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M. Y., Heaney J. D., Youngren K. K., Kawasoe J. H., Nadeau J. H. (2007). Trans-generational epistasis between Dnd1Ter and other modifier genes controls susceptibility to testicular germ cell tumors. Hum. Mol. Genet. 16, 2233-2240 [DOI] [PubMed] [Google Scholar]
- Langdon Y. G., Mullins M. C. (2011). Maternal and zygotic control of zebrafish dorsoventral axial patterning. Annu. Rev. Genet. 45, 357-377 [DOI] [PubMed] [Google Scholar]
- Larabell C. A., Rowning B. A., Wells J., Wu M., Gerhart J. C. (1996). Confocal microscopy analysis of living Xenopus eggs and the mechanism of cortical rotation. Development 122, 1281-1289 [DOI] [PubMed] [Google Scholar]
- Larabell C. A., Torres M., Rowning B. A., Yost C., Miller J. R., Wu M., Kimelman D., Moon R. T. (1997). Establishment of the dorso-ventral axis in Xenopus embryos is presaged by early asymmetries in beta-catenin that are modulated by the Wnt signaling pathway. J. Cell Biol. 136, 1123-1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marikawa Y., Elinson R. P. (1999). Relationship of vegetal cortical dorsal factors in the Xenopus egg with the Wnt/beta-catenin signaling pathway. Mech. Dev. 89, 93-102 [DOI] [PubMed] [Google Scholar]
- Marikawa Y., Li Y., Elinson R. P. (1997). Dorsal determinants in the Xenopus egg are firmly associated with the vegetal cortex and behave like activators of the Wnt pathway. Dev. Biol. 191, 69-79 [DOI] [PubMed] [Google Scholar]
- McKendry R., Hsu S. C., Harland R. M., Grosschedl R. (1997). LEF-1/TCF proteins mediate wnt-inducible transcription from the Xenopus nodal-related 3 promoter. Dev. Biol. 192, 420-431 [DOI] [PubMed] [Google Scholar]
- Medina A., Wendler S. R., Steinbeisser H. (1997). Cortical rotation is required for the correct spatial expression of nr3, sia and gsc in Xenopus embryos. Int. J. Dev. Biol. 41, 741-745 [PubMed] [Google Scholar]
- Miller J. R., Rowning B. A., Larabell C. A., Yang-Snyder J. A., Bates R. L., Moon R. T. (1999). Establishment of the dorsal-ventral axis in Xenopus embryos coincides with the dorsal enrichment of dishevelled that is dependent on cortical rotation. J. Cell Biol. 146, 427-437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar M., van de Wetering M., Oosterwegel M., Peterson-Maduro J., Godsave S., Korinek V., Roose J., Destrée O., Clevers H. (1996). XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 86, 391-399 [DOI] [PubMed] [Google Scholar]
- Rivera-Perez J. A. (2007). Axial specification in mice: ten years of advances and controversies. J. Cell. Physiol. 213, 654-660 [DOI] [PubMed] [Google Scholar]
- Rowning B. A., Wells J., Wu M., Gerhart J. C., Moon R. T., Larabell C. A. (1997). Microtubule-mediated transport of organelles and localization of beta-catenin to the future dorsal side of Xenopus eggs. Proc. Natl. Acad. Sci. USA 94, 1224-1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y., Lu B., Steinbeisser H., De Robertis E. M. (1995). Regulation of neural induction by the Chd and Bmp-4 antagonistic patterning signals in Xenopus. Nature 377, 757 [DOI] [PubMed] [Google Scholar]
- Sasai Y., Lu B., Piccolo S., De Robertis E. M. (1996). Endoderm induction by the organizer-secreted factors chordin and noggin in Xenopus animal caps. EMBO J. 15, 4547-4555 [PMC free article] [PubMed] [Google Scholar]
- Scharf S. R., Gerhart J. C. (1980). Determination of the dorsal-ventral axis in eggs of Xenopus laevis: complete rescue of uv-impaired eggs by oblique orientation before first cleavage. Dev. Biol. 79, 181-198 [DOI] [PubMed] [Google Scholar]
- Scharf S. R., Gerhart J. C. (1983). Axis determination in eggs of Xenopus laevis: a critical period before first cleavage, identified by the common effects of cold, pressure and ultraviolet irradiation. Dev. Biol. 99, 75-87 [DOI] [PubMed] [Google Scholar]
- Schier A. F., Talbot W. S. (2005). Molecular genetics of axis formation in zebrafish. Annu. Rev. Genet. 39, 561-613 [DOI] [PubMed] [Google Scholar]
- Schroeder M. M., Gard D. L. (1992). Organization and regulation of cortical microtubules during the first cell cycle of Xenopus eggs. Development 114, 699-709 [DOI] [PubMed] [Google Scholar]
- Schroeder K. E., Condic M. L., Eisenberg L. M., Yost H. J. (1999). Spatially regulated translation in embryos: asymmetric expression of maternal Wnt-11 along the dorsal-ventral axis in Xenopus. Dev. Biol. 214, 288-297 [DOI] [PubMed] [Google Scholar]
- Shi J., Mei W., Yang J. (2008). Heme metabolism enzymes are dynamically expressed during Xenopus embryonic development. Biocell 32, 259-263 [PubMed] [Google Scholar]
- Sive H., Grainger R., Harland R. (2000) Early Development of Xenopus laevis; A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; [Google Scholar]
- Slanchev K., Stebler J., Goudarzi M., Cojocaru V., Weidinger G., Raz E. (2009). Control of Dead end localization and activity – implications for the function of the protein in antagonizing miRNA function. Mech. Dev. 126, 270-277 [DOI] [PubMed] [Google Scholar]
- Sokol S. Y. (1999). Wnt signaling and dorso-ventral axis specification in vertebrates. Curr. Opin. Genet. Dev. 9, 405-410 [DOI] [PubMed] [Google Scholar]
- Steinbeisser H., Fainsod A., Niehrs C., Sasai Y., De Robertis E. M. (1995). The role of gsc and BMP-4 in dorsal-ventral patterning of the marginal zone in Xenopus: a loss-of-function study using antisense RNA. EMBO J. 14, 5230-5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P. P., Loebel D. A., Tanaka S. S. (2006). Building the mouse gastrula: signals, asymmetry and lineages. Curr. Opin. Genet. Dev. 16, 419-425 [DOI] [PubMed] [Google Scholar]
- Tao Q., Yokota C., Puck H., Kofron M., Birsoy B., Yan D., Asashima M., Wylie C. C., Lin X., Heasman J. (2005). Maternal wnt11 activates the canonical wnt signaling pathway required for axis formation in Xenopus embryos. Cell 120, 857-871 [DOI] [PubMed] [Google Scholar]
- Weaver C., Kimelman D. (2004). Move it or lose it: axis specification in Xenopus. Development 131, 3491-3499 [DOI] [PubMed] [Google Scholar]
- Weaver C., Farr G. H., 3rd, Pan W., Rowning B. A., Wang J., Mao J., Wu D., Li L., Larabell C. A., Kimelman D. (2003). GBP binds kinesin light chain and translocates during cortical rotation in Xenopus eggs. Development 130, 5425-5436 [DOI] [PubMed] [Google Scholar]
- Weeks D. L., Melton D. A. (1987). A maternal mRNA localized to the vegetal hemisphere in Xenopus eggs codes for a growth factor related to TGF-beta. Cell 51, 861-867 [DOI] [PubMed] [Google Scholar]
- Weidinger G., Stebler J., Slanchev K., Dumstrei K., Wise C., Lovell-Badge R., Thisse C., Thisse B., Raz E. (2003). dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr. Biol. 13, 1429-1434 [DOI] [PubMed] [Google Scholar]
- Wessely O., De Robertis E. M. (2000). The Xenopus homologue of Bicaudal-C is a localized maternal mRNA that can induce endoderm formation. Development 127, 2053-2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. A., Heasman J. (2008). Maternal control of pattern formation in Xenopus laevis. J. Exp. Zool. 310B, 73-84 [DOI] [PubMed] [Google Scholar]
- Yang J., Tan C., Darken R. S., Wilson P. A., Klein P. S. (2002). Beta-catenin/Tcf-regulated transcription prior to the midblastula transition. Development 129, 5743-5752 [DOI] [PubMed] [Google Scholar]
- Yang J., Wu J., Tan C., Klein P. S. (2003). PP2A:B56epsilon is required for Wnt/beta-catenin signaling during embryonic development. Development 130, 5569-5578 [DOI] [PubMed] [Google Scholar]
- Youngren K. K., Coveney D., Peng X., Bhattacharya C., Schmidt L. S., Nickerson M. L., Lamb B. T., Deng J. M., Behringer R. R., Capel B., et al. (2005). The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature 435, 360-364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu R., Bhattacharya C., Matin A. (2007). The role of dead-end in germ-cell tumor development. Ann. N. Y. Acad. Sci. 1120, 181-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.