Abstract
The clinical efficacy and safety of a drug is determined by its activity profile across multiple proteins in the proteome. However, designing drugs with a specific multi-target profile is both complex and difficult. Therefore methods to rationally design drugs a priori against profiles of multiple proteins would have immense value in drug discovery. We describe a new approach for the automated design of ligands against profiles of multiple drug targets. The method is demonstrated by the evolution of an approved acetylcholinesterase inhibitor drug into brain penetrable ligands with either specific polypharmacology or exquisite selectivity profiles for G-protein coupled receptors. Overall, 800 ligand-target predictions of prospectively designed ligands were tested experimentally, of which 75% were confirmed correct. We also demonstrate target engagement in vivo. The approach can be a useful source of drug leads where multi-target profiles are required to achieve either selectivity over other drug targets or a desired polypharmacology.
The safety and efficacy of a drug is determined not only by its action on an individual protein but also by its interactions with multiple proteins in the proteome. The promiscuous interaction of a drug with undesired proteins frequently causes toxicity1 and adverse effects2,3. Conversely, the modulation of a single drug target can be therapeutically insufficient, particularly in complex neuropsychiatric conditions, infectious diseases and cancer4-6. Instead, it is frequently necessary for a drug to simultaneously engage two or more targets for therapeutic efficacy7. Psychiatric drugs in particular require multiple activities against several targets to therapeutically modulate complex neuropsychiatric domains including perception, cognition and emotion4. However, designing drugs with a specific multi-target profile – to achieve either exquisite selectivity over other drug targets or a desired polypharmacology – is a complex and exceedingly difficult task for medicinal chemistry8. Accordingly, methods are needed to enable drugs to be designed a priori against several molecular targets simultaneously. Here we describe a solution to the complex problem of designing ligands against multiple drug target profiles by automated design.
From prediction to design
The problem of designing ligands against a multi-target profile involves the parallel optimisation of multiple structure-activity relationships (SAR) within a desired range of physico-chemical properties. The prospect of multi-target drug design has been recently aided by the development of computational methods that show success in predicting the molecular targets of drugs3,9-13 (Supplementary Fig. 1) although such approaches are not intrinsically design methods.
Drug design can be modelled as an evolutionary process of iterative cycles of exploration and analysis14,15. Adaptive design processes are efficient at solving complex, multi-objective problems. Accordingly, we developed an automated, adaptive design approach to optimise ligands against polypharmacological profiles.
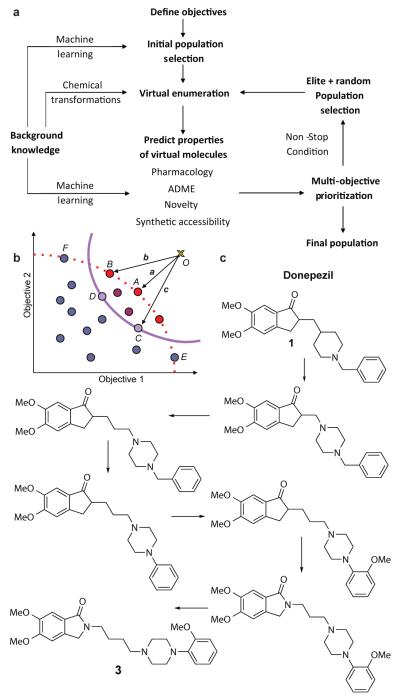
Several de novo drug design methods have been proposed previously16-21. However, of those that have been experimentally tested22-26 only rarely have high affinity ligands been described and these are all against a single molecular-target objective 24,26. In contrast to previous de novo approaches we mimicked the creative process by automated learning of medicinal chemistry design tactics, applying these to the generation of analogues, and then prioritizing them relative to a set of objectives (Fig. 1a) The development of this approach is described below, starting from ‘off-target’ predictions, progressing through ligand design and, finally, the discovery of novel compounds with pre-defined multi-target profiles.
Figure 1. Adaptive drug design.
(a) Closed loop of automated ligand design algorithm by multi-objective evolutionary optimisation. (b) Multi-objective prioritization by vector scalarisation. The multi-target objectives are defined as the co-ordinates of the ideal achievement point, O, (gold cross) and the predicted values of each generated compound (coloured circles) are also defined as a co-ordinates in a multi-dimensional space. The Pareto frontier is displayed as a red dotted line. The multi-objective prioritisation is inverse to the magnitude of vectors (||a|| < ||b|| < ||c||). Compound A is prioritised the highest. Compounds C and D have the same vector length (||c||=||d||) and thus prioritised equally and above the Pareto optimal compounds E and F. (c) Evolution of donepezil (1) (19% inhibition of D2 receptor @ 10μM; dopamine D2 Bayesian score = 25) into dopamine D2 inverse agonist 3 (96% inhibition of D2 receptor @ 10μM; dopamine D2 Bayesian score = 92). The Tanimoto similarity between donepezil and compound 3 is only 0.35.
Evolution of a drug
Sir James Black proposed that, “the most fruitful basis for the discovery of a new drug is to start with an old drug”27. Accordingly, we tested whether the algorithm could automate the evolution of new biological activities, starting from a known drug. Donepezil (compound 1) is an acetylcholinesterase inhibitor approved for cognitive enhancement in Alzheimer’s disease. Bayesian probabilistic activity models9, for 784 molecular targets built from the ChEMBL database28 predicted a moderate likelihood that donepezil possessed D4 dopamine receptor activity and a low chance of D2 dopamine receptor activity (Supplementary Table 1). We found donepezil was a moderately potent D4 inverse agonist (ki=614nM) with minimal D2 activity (Supplementary Fig. 2 and 3 and Supplementary Tables 2 and 3). Donepezil’s D4 inverse agonist activity is intriguing given analyses demonstrating a significant improvement in memory in the Trail Making Test with this drug29 and findings that D4 antagonists can prevent stress-induced cognitive dysfunction in primates30.
We tested our method by evolving the structure of donepezil with the dual objectives of improving D2 activity and achieving blood-brain barrier (BBB) penetration. In our approach the desired multi-objective profile is defined a priori and then expressed as a point in multi-dimensional space termed ‘the ideal achievement point’. In this first example the objectives were simply defined as two target properties and, therefore, the space has two dimensions. Each dimension is defined by a Bayesian score for the predicted activity and a combined score that describe the absorption, distribution, metabolism and excretion (ADME) properties suitable for BBB penetration (D2 score=100, ADME score=50). We next generated alternative chemical structures by a set of structural transformations using donepezil as the starting structure. The population was subsequently enumerated by applying a set of transformations to the parent compound(s) of each generation. In contrast to rules-based or synthetic reaction-based approaches for generating chemical structures16,31-34, we used a knowledge-based approach by mining the medicinal chemistry literature28,35. By deriving structural transformations from medicinal chemistry, we attempted to mimic the creative design process (Supplementary Fig. 4)36. Activity predictions were calculated for each of the enumerated compounds from all Bayesian models. Scores representing the likelihood of CNS penetration and good ADME properties were calculated using the program Stardrop (Optibrium Ltd.) and combined into a single value. The predicted properties of the enumerated structures were then expressed as points in multi-dimensional space. The generated structures were subsequently ranked by the distance (in multi-dimensional space) between the predicted properties for each structure and the ideal achievement point37 (Fig. 1b). Compounds were filtered for novelty, Lipinski’s rule-of-five compliance38 and synthetic accessibility39. The top 10,000 prioritized structures were selected for the next iterative cycle along with 500 random structures from the remaining population. The process was iterated until either a structure close to the objectives was discovered or no further improvements were achieved.
Initially, we evolved a series of isoindoles and prioritized them using our achievement objectives as criteria (Fig. 1c, Supplementary Table 4 and Supplementary Fig. 5). Eight analogues were then synthesized and tested (Fig. 2, Supplementary Figs. 2 and 6 and Supplementary Table 5) with all showing significant D2 affinities (ki’s= 156-1,700 nM; Supplementary Table 3).
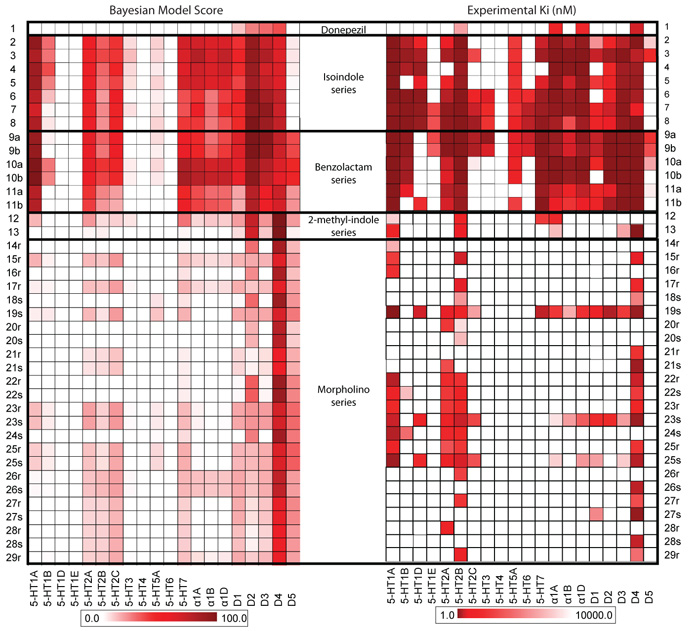
Figure 2. Polypharmacology profiles of designed ligands.
Comparison of the predicted Bayesian and observered polypharmacology profiles for (a) donepezil (1); (b) the isoindole analogues (2-8) – of the 160 ligand-target associations, 100 were correctly predicted by the Bayesian models (p=0.001; probability of success of 0.63 with 95% CI of 0.55 – 0.70); (c) the benzolactam analogues (9a-11b) – of the 160 ligand-target associations, 107 are correctly predicted (p=1.1e−5); (d) the 2,3-dihydro-indol-1-yl analogues (12 and 13); (e) and the morpholino analogues (14-29) – of the 540 ligand-target associations, 437 are correctly predicted (p<2.2e−16). The figure is composed of data in Supplementary Tables 1 and 3. In total, of the 800 predictions in the matrix, on novel, prospectively designed ligands, 599 were experimentally confirmed correct (p<2.2e−16), with a probability of success of 0.75 (95% confidence interval: 0.72 – 0.78).
The second highest ranking compound (3) – chosen from the final population of evolved structures – exhibited the highest D2 receptor affinity (ki=156nM). Thus, we successfully evolved donepezil’s negligible D2 activity into a series of ligands with higher D2 affinities (Fig. 2). Functionally, 3 was a dual D2 inverse agonist/D4 agonist (Supplementary Fig. 3). CNS penetration studies showed that 3 penetrates the brain as predicted, with an in vivo brain/blood ratio (BBR) of 0.5.
Although the evolved compounds were selected for the D2 receptor objective other predicted activities were not selected against. Accordingly, each of the generated compounds had a predicted polypharmacology profile. In general, this set of isoindole analogues was predicted to exhibit promiscuous profiles, with variable activities predicted for multiple serotonergic, adrenergic and dopaminergic receptor subtypes (Supplementary Tables 1 and 5). These predicted promiscuous profiles were subsequently confirmed (Fig. 2; Supplementary Tables 2 and 3), and the predicted multi-target profiles displayed excellent agreement with experimentally determined profiles, thereby implying that the approach can be applied to the de novo design of multi-target agents.
Reducing anti-target activity
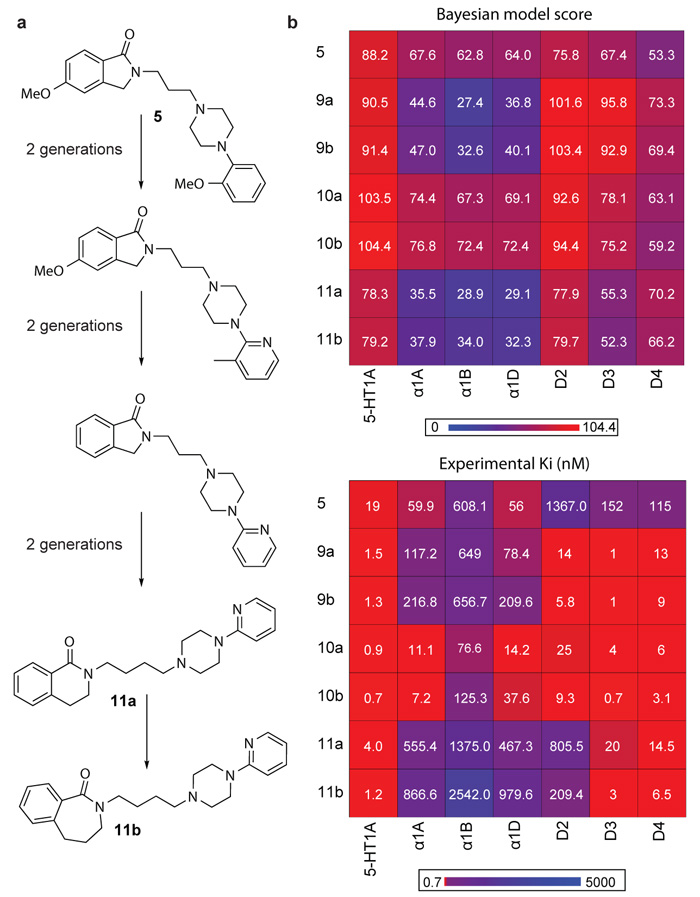
The isoindoles exhibited moderately potent affinities for the α1 adrenoceptors (ki’s = 0.9 nM-3,577 nM; mean ki=277 nM; Supplementary Table 3 and Supplementary Fig. 2). Since α1 adrenoceptor antagonists can induce low blood pressure as a side effect they are considered ‘anti-targets’ to be avoided in drug design40. A common drug design optimisation problem is to reduce such anti-target activity whilst maintaining desired on-target activities. We accordingly evolved the 8 newly-synthesized isoindoles towards a polypharmacological profile (5-HT1A, D2, D3, D4) with selectivity over the three α1 adrenoreceptors anti-targets (α1A, α1B and α1D) whilst maintaining CNS penetration.
To compare evolutionary strategies, the isoindoles were optimised towards polypharmacology objectives with and without highly predicted α1 activities filtered-out at each generation (Fig. 3a, Supplementary Fig. 7, Supplementary Tables 6 and 7). In both optimisations benzolactams, were ranked the highest (compounds 11a and 10a respectively) (Fig. 3b and Supplementary Tables 6 and 7). Six benzolactam analogues were then synthesized based on both sets of objectives (Supplementary Table 8 and Supplementary Fig. 5). Analogues of the benzo-δ-lactam (3,4,-dihydroisoquinolin-1(2H)-one) (9a, 10a and 11a) and benzo-ε-lactam (2,3,4,5-tetrahydro-1-H-benzo[c]azepin-1-one) (9b, 10b and 11b) were synthesized for comparison since both ring systems were highly prioritized.
Figure 3. Reducing> α1 anti-target activity by evolutionary design.
(a) Summary of the evolution of the prioritized benzolactam analogues (compound 11a and 11b) from a parent isoindole analogue (5). The full evolutionary pathway from compound 5 to 11b is show in Supplementary Figure 7. (b) Comparison of polypharmacology profiles for the Bayesian model score and experimental binding affinities (ki) of compound 5 and benzolactam analogues (9a-11b) for the seven target objectives (α1A, α1B, α1D vs 5-HT1A, D2, D3, D4). On average the selectivity ratios for the D2, D3, D4 and 5-HT1A receptors for the synthesized benzolactams over the α1 receptors are 2.8-, 59-, 58- and 312-fold respectively. In comparison, the average selectivity ratios for D2, D3, D4 and 5-HT1A for the isoindole analogues over the α1 receptors are 0.27-, 2.1-, 1- and 14-fold, respectively. With respect to the algorithm prioritization for the benzolactam analogues (Supplementary Table 8) against the multi-target objectives, the order of prioritization matches the experimentally determined order for the analogues (dichloro phenylpiperazine > 2-methoxy phenylpiperazine >2-pyridine piperazine).
Both predicted and observed receptor activity profiles for the synthesized benzolactams are shown in Fig. 3b (Supplementary Tables 1 and 3; Supplementary Fig. 2). The 2-pyridine-piperazine analogues (11a and 11b) have the lowest α1 predictions and, indeed, exhibited the lowest α1 affinities (mean ki=1,131nM). In agreement with the models, 11a and 11b also have the lowest affinity for all dopamine D2-like receptors of the benzolactams tested. The dichloro-phenylpiperazine analogues (9a and 9b) exhibited slighted higher α1 and D2 predictions, which were also confirmed experimentally. In contrast, the 2-methoxy phenylpiperazine analogues (10a and 10b) exhibited potent affinities against the polypharmacology profile 5-HT1A, D2, D3 and D4 receptors but also had the highest α1 predictions and were confirmed as the most potent against the α1 receptors (mean ki=45.3nM) (Fig. 3b). Receptor profiling of the benzolactam series revealed that compared to the isoindoles, from which the compounds were evolved, the benzolactams achieved the objective of an increased polypharmacology profile for 5-HT1A,D2, D3 and D4 over the α1 adrenoceptors (Fig. 3b). The benzolactams are 11-fold more selective with respect to D2 and 25-fold more selective with respect to the polypharmacology profile, compared to the isoindoles. Importantly, the benzolactam 9a penetrated the brain (BBR=5.9), as predicted.
As the benzolactam series was not present in the ChEMBL database (release 1) used to build the Bayesian models, they constituted a novel chemical series for the system to discover41. Intriguingly, benzolactam derivatives have recently been independently synthesized and tested as potent D2/D3 ligands42, however the broader receptor profiles of the compounds were not evaluated. This observation provides additional confidence that the algorithm is capable of generating and prioritising novel chemical structures equivalent to those devised by medicinal chemists.
Potency optimisation
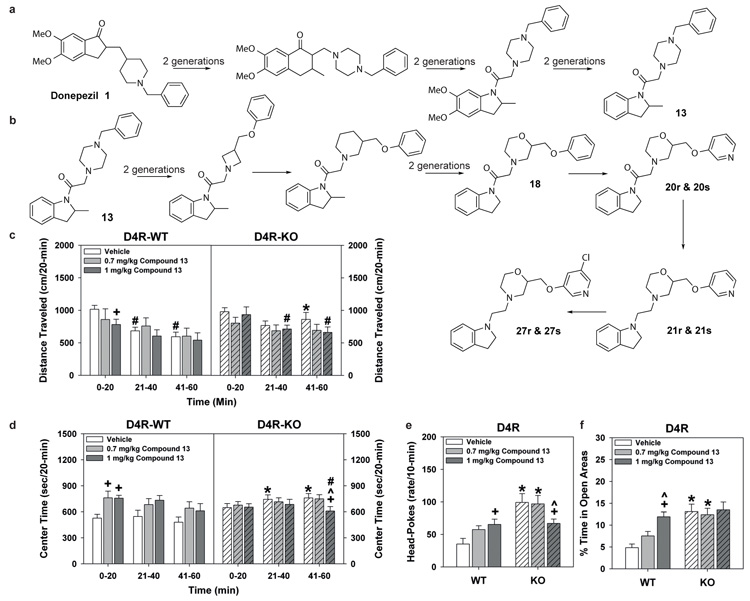
We next explored selectivity in the context of our multi-target objectives and asked whether we could optimise potent, selective, CNS penetrant D4 receptor ligands starting from the chemical structure of donepezil. We executed the optimisation in two stages: we optimised (i) for D4 potency and brain penetrability and (ii) for D4 selectivity. A series of 2-methylindoline derivatives with high predicted D4 activity was evolved from donepezil after six generations (Fig. 4a, Supplementary Fig. 9). Notably, compounds belonging to a 2,3-dihydro-indol-1-yl chemotype were dominant in the prioritized set (Supplementary Table 9). Compounds 12 and 13, which both belong to the 2,3-dihydro-indol-1-yl class, were then selected for testing (Supplementary Figs. 5 & 8). The highest ranking compound, 12, was inactive, while 13, the third highest-ranking design out of the final population, was the most potent D4 ligand amongst all tested compounds (D4 ki=8.9nM). Via optimisation, 13 represents a 69-fold increase in affinity over donepezil. In contrast to the isoindole and benzolactams analogues, 13 is predicted to be a highly selective D4 ligand with 95-fold selectivity over 5-HT2B and weak affinities greater than 1 μM for only 5 other receptors in our panel of GPCRs (Fig. 2). Importantly, and as predicted, 13 is highly CNS penetrant (BBR= 7.5).
Figure 4. Evolution of dopamine D4 ligands from donepezil.
(a) Summary of the evolution of donepezil (1) (dopamine D4 Bayesian score = 26, D4 ki = 614nM) into dopamine D4 inverse agonist 13 (dopamine D4 Bayesian score = 112, D4 ki = 8.9nM). The Tanimoto similarity between donepezil and compound 13 is only 0.26. The full evolutionary pathway from donepezil to 13 is show in Supplementary Figure 9 (b) Summary of the evolution of selective novel dopamine D4 ligands. Compound 13 is further evolvSed by selection for novelty and D4 selectivity into the morpholino analogues 18, 20 (r and s), 21 (r and s) and 27 (r and s). The full evolutionary pathway from 13 to 27 is show in Supplementary Figure 11 (c-f) Behavioral analysis of a novel dopamine D4 ligand in D4 receptor (D4R) knockout (KO) mice. (c) Distance travelled in the open field over 60 min by D4R animals. Mice were given (i.p.) vehicle or 0.7 or 1 mg/kg compound 13 and tested immediately over 60 min. (d) Time spent in the centre zone in the open field by D4R animals. (e) The numbers of head-pokes in the hole-board test in D4R mice. Animals were injected with vehicle or 0.7 or 1 mg/kg compound 13 and were tested 30 min later over 10 min. (f) Percent time in the open areas in the zero maze in D4R (h) mice. Animals were administered vehicle or compound 13 and tested 30 min later for 5 min. N=8-14 D4R mice/treatment-condition. *p<0.05, WT versus D4R-KO mice; +p<0.05, comparisons within genotype to the vehicle; ^p<0.05, comparisons between 0.7 and 1 mg/kg compound 13; ^p<0.05, compared to the 0-20 min time-point.
To verify that the predicted properties of selectivity, potency, and CNS penetration resulted in D4 receptor activity in vivo, we evaluated 13 on behaviour in wild-type (WT) and D4 receptor knockout (D4R-KO) mice (Fig. 4c-f), as well as in proprotein convertase 7 (PC7) KO mice (Supplementary Fig. 10a-d) that display a similar D4R-KO phenotype. Although open field locomotor activity declined in vehicle-treated D4R-WT animals (Fig. 4c), it remained high and showed little habituation in D4R-KO mice (Supplementary Information). While 0.7 mg/kg 13 was without significant effect in either genotype, the 1 mg/kg dose reduced locomotion at 0-20 min in D4R-WT animals. The same dose exerted no effect in D4R-KO mice. In the centre zone, vehicle-treated D4R-KO mice spent more time in this area at 21-60 min than D4R-WT controls (Fig. 4d). Centre time in D4R-WT animals was enhanced at 0-20 min with both doses of 13; in D4R-KO mice it was attenuated at 41-60 min with the 1 mg/kg dose. In the hole-board test, head-poking was increased in vehicle-treated D4R-KO animals compared to D4R-WT controls (Fig. 4e). In D4R-WT mice 13 augmented head-pokes in a dose-dependent fashion; only the 1 mg/kg dose attenuated head-poking in mutants. In the zero maze open-area time was increased in vehicle-treated D4R-KO mice relative to D4R-WT controls (Fig. 4f). One mg/kg compound 13 selectively increased D4R-WT open-area times to levels of the D4R-KO mice. With regards to PC7 mice, PC7-WT and D4R-WT responses were similar (Supplementary Fig. 8a-d). Although behaviors in PC7-KO vehicle controls essentially phenocopied those in vehicle-treated D4R-KO mice, 13 normalized PC7-KO responses to those of PC7-WT controls. By comparison, D4R-KO mice were largely unresponsive to 13 – demonstrating high D4R selectivity. Nonetheless, 13 did not appear to be absolutely selective since in D4R-KO animals the 1mg/kg dose attenuated head-poking in the hole-board test, implying some possible off-target actions at increased doses.
Automated Invention
Using 13, we further expanded our objective to evolve ligands that (i) were highly selective for dopamine D4 (ii) were CNS penetrant and (iii) were a novel chemotype. The evolution of 13 against these objectives resulted in the design of novel morpholino compounds (Fig. 4b, Supplementary Fig. 11) (Supplementary Tables 10 and 11). Compounds with the novel isoindol-1-yl-ethyl-morpholino backbone were prominent in the prioritised final generation population of 10,000 structures (ranked 5th, 6th and 9th of the top 10 compounds in Supplementary Table 10), while most known D4 ligands are 1,4-disubstituted aromatic piperidines and piperazines (1,4-DAPs). However 1,4-DAPs are rather promiscuous substructures common in ligands for many biogenic amine GPCRs43. Therefore the isoindol-1-yl-ethyl-morpholino analogues represent a novel D4 chemotype.
A library of 24 morpholino analogues (compounds 14-29) was then synthesized and profiled against our GPCR panel (Supplementary Table 11; Supplementary Fig 5). Individual R and S morpholino enantiomers were synthesized and assayed separately (chirality designated by suffix). To further reduce complexity, direct analogues with and without the carbonyl oxygen were synthesized (e.g. 20 and 21), as this atom was predicted not to be essential to the overall D4 selectivity profile (Fig. 2).
The assays confirmed the predictions that the new morpholino compounds are generally highly selective for the D4 receptor over the tested receptors (Fig 2). Seventeen of the compounds had affinities for the dopamine D4 receptor, ranging from ki=90nM (Compound 27s) to ki=5,526nM (18s) with eight exhibiting affinities less than 1 μM for D4 (compounds 15r, 19s, 21s, 22r, 23s, 26s, 27s and 28s). Compounds containing the ethanone linker-group were generally less active compared to those with the ethyl linker. For compounds with the ethyl linker, the S enantiomer was more potent than the R enantiomer. Functional assays of an exemplar compound (22r) indicated inverse agonism at D4 (Supplementary Figure 3).
In agreement with the design objectives, the morpholinos displayed exquisite selectivity for the dopamine D4 receptor. Excluding the dopamine receptors, low positive Bayesian scores were observed for 8 of compounds against 5-HT1A, 16 of the compounds against the 5-HT2A/B/C receptors, and almost all had very low scores for 5-HT7 serotonin receptors. The off-target trends were confirmed when the compounds were profiled (Fig. 2). The morpholino compounds, on average, bound to 3.4 targets (including D4 at ki’s <10μM) compared to 15.8 targets for the isoindole and benzolactam compounds. Seven of the active compounds had off-target activities for only 1 of the 20 receptors tested. Compound 26s is the most selective compound with no measured affinity for any other screened receptor. Four compounds possessed both relatively high affinity (D4 ki<1μM) and two or fewer off-target activities out of the 20 GPCRs profiled (21s, 26s, 27s, 28s).
The morpholino series thus represents a new class of highly selective, brain-penetrant, D4-dopamine receptor ligands. Compounds 27s (D4 ki=90nM; D1 ki=5852nM; BBR=2.0) and 21s (D4 ki=182nM, 5-HT2A ki=3,545nM) qualified as lead compounds that fulfilled all of our design objectives of novelty, high affinity for the dopamine D4 receptor and exquisite selectivity and CNS penetration Clearly, the automated design of a novel class of ligands with a desired multi-target profile demonstrates that the method is able to generate novel, drug-like lead compounds directly by automated design.
Drug design ex scientia
We focused on the polypharmacology of bioaminergic GPCRs as a convenient test case, due to the importance of multi-target profiles at these receptors for a variety of neuropsychiatric indications4. In principle the approach is applicable to all drug target classes limited only by the requirement for sufficient structure-activity (SAR) data to create useful models9-12. To generally extend polypharmacology profiling and hence de novo design it will be necessary to develop inferencing methods to build predictive bioactivity models that integrate all available SAR, protein structure and protein sequence information together and fuse data from diverse scoring functions into predictive frameworks44,45.
De novo, automated compound design against multi-target profiles provides a powerful new approach for discovering new ligands and drug leads and for discovering ligands that satisfy specific multi-target objectives. The method is particularly useful as a new source of leads for polypharmacology profiles.
Online Methods
Datasets
All machine learning and data mining of the medicinal chemistry structure-activity data were conducted on the ChEMBL database (release 1) and a pre-release (StARLite version 31)28. The ChEMBL database contains (release 1) over 440,000 compounds abstracted J. Med Chem. and Bioorg. Med Chem. Letts from 1980 to May 2008. The ChEMBL database is available for download from the EBI.
Chemical Transformations
A database of chemical transformations was derived from systematically comparing sets of analogue compounds in ChEMBL28. Sets of analogues with defined structure-activity-relationships (SAR) were identified in ChEMBL28 usually from individual journal articles. The transformations database was seeded with a set of common chemical transformation derived from medicinal chemistry knowledge. The transformations database was then expanded by identifying novel transformations by systematically applied to each of the structures associated with each of the journal articles in ChEMBL. All the transformations were applied to each compound in a journal article. The resulting set of transformed compounds was compared to the published analogues. Analogues that were not present in the transformed set highlighted potential transformations that were missing from the transformation database, and this was subsequently added to the database. This iterative mining method attempts to regenerate all the reported structures of every medicinal chemistry publication reported in ChEMBL. The chemical transformations were encoded in RXN format. The procedure was implemented in Pipeline Pilot. The current database contains over 700 unique structural transformations.
Bayesian Models
Predictive polypharmacology profiling was undertaken using Bayesian activity models, based on our previously published approach9. The Bayesian method for polypharmacology profiling was chosen as it provided both good performance on noisy datasets and a high speed of calculation51. High confidence models were built using ChEMBL (release 1). Activity data were filtered to keep only activity endpoint points that had either IC50, ki or EC50 values and where the ChEMBL confidence score was at least 7 (protein assignment was direct or homologue). A compound was considered active when the mean activity value was below 10μM. All inactive compounds were assigned to the target ‘none’. Following this procedure 133,061 compounds remained with 215,967 activity endpoints, which were used for model building. Multiple category Laplacian-modified naïve Bayesian models were built with ECFP6 representations52 for 784 targets. For each model the data were split in two for the validation step: compounds were clustered and assigned a cluster number. Clusters with an odd number were assigned to the test set, and the clusters with even number were assigned to the training set. Models were built with the training set, and the test set was scored. The training set was scored using its own model as comparison. Finally a model was built with all data and scored against itself, the training set and whole set should provide similar validation statistics. Statistics on the performance of the models are described in Supplementary Table 12. The results for the model containing all 785 targets the results were very similar to the models for the receptor subsets. Two analyses were used to assess the performance of the different models. The first analysis provides an overall score and does not need to specify a cut-off for distinguishing active from inactive compounds. The area under the Receiver Operating Characteristic (ROC) Curve (AUC) provides an indication of the ability of the model to prioritize active compounds over inactive compounds. The ROC curve is the plot of true positive rate versus false positive rate. However it did not provide information on early enrichment, which was important in studies such as the present one where only the top ranking compounds were considered. Therefore the Boltzmann-Enhanced Discrimination of ROC (BEDROC)53 was used, which solves the early enrichment issue by adding a weight to compounds recognized early. BEDROC was derived from the Robust Initial Enhancement (RIE), and the Sum of log of ranks test (SLR)54 which provided a statistical test to assess which method performs better than random. The percent of active compound retrieved in the top 5% is also calculated (Recall =5%). The second analysis required a cut-off to make the distinction between active and inactive as they varied with the rank of the compounds. For each model, the specificity (true negative rate), sensitivity (true positive rate), false positive rate, false negative rate, precision, F-measure and Matthews Correlation Coefficient (MCC) were calculated at different cut-off values. The cut-off providing the best MCC score was used, as it was shown to provide better performance55 (Supplementary Fig. 12). The quality of the models was assessed using an internal leave-one-out validation: one compound was part of the test set, and was scored using the remaining data as the training set. Then the area under the ROC curve was calculated (Supplementary Fig. 13). A cut-off score to minimize the sum of the percent misclassified for category members and for category non-members was calculated and used to classify compounds in the contingency table.
An All Data Model for dopamine receptors only was built using data from pre release of ChEMBL (StARLite version 31), with similar numbers of compounds and endpoints. The model was built without considering the confidence level of target assignment to gather as much data as possible. This model was used for initial calculation on the evolution of the isoindole series and the 2,3-dihydro-indol-1-yl series. The quality of the models was assessed using the same procedures as described above. The results from the All Data Models and the High Confidence Models were very similar (e.g. D2 model R2=0.998, D4 models R2=0.984).
Profile Prediction Probabilities
The cut-off for a good prediction came from the validation steps for the model. From the test set, the cut-off value providing the best MCC value was used. For 5-HT1E, a cut-off of zero was selected. For each ligand-target association, the probability of success was 0.5 (active or inactive where activity was defined as ki<10μM). To test if the profile predictions were better than random, an exact binomial test was performed using R (version 2.8.1), and the cumulative binomial probability was calculated.
Adaptive Optimisation
The adaptive optimization procedure involves defining a set of x achievement objectives (O1-x), where x is at least one, and an initial parent compound population (PP(G=n) where n=0 for the initial population) of at least one molecule. All of the members of the parent population (PP) are subjected to all possible transformations, so as to maximize the pool of molecules in the transformed population (PT). Each member of the transformed population is scored and ranked using the achievement objectives (i.e. Bayesian predictions, molecular properties).
If a stop condition is not satisfied, prioritized individuals with calculated objective parameters that satisfy the defined thresholds are assigned to an elite population (PE) and those that fail are assigned to a non-elite population (PN), of which n random members form a Random Population (PR). For all the calculations performed PE has a maximum size of 10,000 and PR a maximum size of 500. The new parent population (PP(G=n+1)) is created by merging PE and PR (PP(G=n+1) = PE(G=n) + PR(G=n)). This new population is subjected to another transformation process. This process is repeated until a stop condition is satisfied. The new parent population PP from each generation is also added to a combined pool of all observed parents (PPall).
The population of transformed compounds from the last iteration and the pooled parent population are combined and all duplicates and compounds failing structure valency rules removed to produce a merged population (PM) of unique members (PM= PPall + PT). Properties and parameters (e.g. Bayesian activity models values, physico-chemical properties and predicted ADME properties) are calculated for each individual in PM. Each of the n members of the initial population (PM) is evaluated against the achievement objectives (O1-x). Multi-objective prioritization is performed by describing the calculated parameters for a compound of interest as multi-dimensional coordinates. Pareto ranking is a common method for prioritising multiple criteria17. The Parero frontier maps a surface where all solutions are considered equivalent (non-dominant) – where an increase in one objective leads to a decrease in at least one or more other objectives. However finding a Pareto optimal solution becomes difficult when many objectives are considered56. Instead a vector scalarisation procedure is employed37.
The results are ranked by the magnitude of the vector ||a|| between the multi-dimensional co-ordinates of predicted values of the chemical structure of interest (A) and the defined achievement objective point, (O), with the shortest vector length closest to the ideal in multi-dimensional space:
| (1) |
where the achievement objective point has the coordinates (xO1,…,xOn), and predicted values of the molecule i form the coordinates (xA1,…, xAn). Novelty is assessed by comparing the generated compound with compounds in ChEMBL, either as an exact match or by comparison of the Murcko framework41, depending on whether the objectives are defined in terms of novel compounds or novel chemotypes. Novelty is filtered depending on the goals.
ADME properties and CNS penetration are calculated using previously publishing Gaussian Process models57,58 as implemented in StarDrop (Optibrium Ltd). A synthetic accessibility score, representing historical synthetic knowledge is calculated using a previously published algorithm39. The synthetic accessibility score combine the observation of fragments in ChEMBL and a complexity penalty. A limitation of the ECFP6-Bayesian prediction method is that fingerprints cannot distinguish stereochemistry if the stereochemistry is not encoded in the training data. In the ChEMBL database only 43% of the chiral compounds with chiral centres have their stereochemistry fully defined, thus it is not possible to distinguish between R and S enantiomers. To reduce the complexity of synthesis, compounds with two or more chiral centres are filtered from the final population.
Receptor Profiling
The detailed experimental protocols for the radioligand and functional receptor assays are available on the NIMH PDSP website at http://pdsp.med.unc.edu/UNC-CH%20Protocol%20Book.pdf.
hERG Assay
hERG activity was assayed by the patch clamp method on a PatchXpress platform and by FluxOR Tl+ assays. Assays were performed as previously described47.
Metabolic Stability Assay
Metabolic stability was assessed, generating the in vitro intrinsic clearance (Cli) following incubation of test compound with mouse hepatic microsomes. The assay was performed as previously described48.
Brain Penetration Measurement
Mice were housed under standard conditions: 12 h light/dark cycle and food and water available ad libitum throughout the study. The Drug Discovery Unit at the University of Dundee is dedicated to the humane care, maintenance and use of research animals and maintains compliance with UK Home Office regulations. All experiments were approved by the local ethical review committee. The ratio of test compound between brain and blood (B:B ratio) was assessed following intravenous administration to the female NMRI mouse.
Behavioural Testing
Adult male and female C57BL/6J, D4R-KO mice (Jackson Labs, Bar Harbor, ME), and PC7-KO mice (D. Comstam, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) were used. PC7 mice were crossed with C57BL/6J mice for more than 10 generations (N.G. Seidah and A. Prat, IRCM, Montreal, QC, Canada). Animals were maintained in a humidity- and temperature-controlled room under a 14:10 hr light:dark cycle (lights on 0800 hr). All studies were conducted during the light cycle, between 1000 and 1600 hrs in the following order: zero maze, open field, and hole-board, where tests were separated by at least by 7 days. Animals were assigned to vehicle- [0.1% DMA with 15% 2-hydroxypropyl-β-cyclodextrin (Sigma Aldrich, St. Louis, MO)] and compound 13-treated groups and were maintained in these groups throughout the experiments. Water and laboratory chow were supplied ad libitum. All experiments were conducted with an approved protocol from the Duke University Institutional Animal Care and Use Committee and according to NIH Guide for the Care and Use of Laboratory Animals. To overcome the high intrinsic clearance in mouse hepatic microsomes of compound 13 (Cli=13.8mL/min/g), mice were injected (i.p.) with vehicle, or 0.7 or 1 mg/kg compound 13 and placed immediately into the open field for 60 min as described 49. Activity was monitored as distance traveled and time spent in the center zone. In the hole-board and zero maze tests57,50 mice were given (i.p.) vehicle or compound 13 and tested 30 min later. Hole-board test responses were video-taped for 10 min using high-resolution low-light cameras (Panasonic NA, Secaucus, NJ) and were scored using the TopScan program (Clever Sys Inc., Reston, VA) for the rate of head-pokes into the 16 holes. Zero maze behaviors were video-taped over 5 min and were scored by trained observers blinded to the genotype, sex and treatment-condition of the animals using the Observer XT10 program (Noldus Information Technology, Leesburg, VA) for the percent time in the open areas. All behavioral data are presented as means ±SEM and were analyzed by ANOVA and repeated measures ANOVA followed by Bonferroni corrected pair-wise comparisons (IBM SPSS 20, Chicago, IL). A p<0.05 was considered significant.
Supplementary Material
Acknowledgements
This work is supported by SULSA (HR07019), the BBSRC Doctoral Training Programme, the BBSRC Pathfinder (BB/FOF/PF/15/09) and the BBSRC Follow On Fund schemes (BB/J010510/1) (A.L.H.), the University of Dundee’s Pump Priming Fund for Translational Medical Research (I.H.G. and A.L.H.) and supported by grants from the National Institutes of Health contracts supporting drug discovery and receptor pharmacology (B.L.R.) and the NIH grant MH082441 (W.C.W.). The chemical synthesis benefits from the infrastructure investments from the Wellcome Trust Strategic Award (WT 083481). We thank J. Overington of the European Bioinformatics Institute (EMBL-EBI) for StARlite and ChEMBL. We wish to thank C. Means and T. Rhodes for helping with the open field, hole board and zero maze tests. We also wish to thank C. Elms and J. Zhou for their support in the husbandry and generation of the mice used for behavioral testing. We also wish to thank F.Y. Li of Cleversys Inc for customizing the software configuration for the hole-board tests. Some of the equipment used in the behavioral testing was purchased with a grant from the North Carolina Biotechnology Center. BLR also received support from the Michael Hooker Chair of Pharmacology.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions.
A.L.H. devised the method, developed the algorithm and designed the study. J.B. coded the algorithm and undertook the calculations. G.R.B. developed the databases. A.L.H. and J.B. selected the compounds for synthesis. I.H.G., G.F.R. and K.A. designed the synthetic routes and G.F.R. and K.A. undertook the chemical synthesis. L.A.W. purified and analyzed several of the compounds. B.L.R. and V.S. designed the empirical tests for the synthesized compound predictions, analyzed and interpreted the results and performed the experiments. X.P.H. performed the 5-HT2B functional assays and the hERG assays. M.F.S. conducted the dopamine D2 and D4 functional assays. K.D.R. designed the DMPK studies and analyzed the results. S.N. L.S. and F.R.C.S. carried out the DMPK experiments. For the behavioural experiments, D.B.C. disrupted the PC7 gene in mice. A.P. and N.G.S verified the PC7 deletion in many tissues including brain, and then re-derived into a pure C57BL/6 background. W.C.W. designed the studies; R.M.R. and A.I.S. conducted the experiments and analyzed the results; W.C.W, R.M.R., A.I.S., D.B.C., A.P. and N.G.S. interpreted the findings; W.C.W. and R.M.R. prepared the figures. A.L.H. and B.L.R. wrote the manuscript, I.H.G. wrote the synthetic methods, W.C.W and R.M.R wrote the behavioral section of the manuscript. J.B., V.S., W.C.W. and R.M.R. prepared the figures. All the authors discussed the results and commented on the manuscript.
Competing Financial Interest
A.L.H., J.B. and G.R.B. are shareholders in Ex Scientia Ltd, a University of Dundee spin out company that has licensed the technology.
References
- 1.Hughes JD, et al. Physicochemical drug properties associated with in vivo toxicological outcomes. Bioorg. Med. Chem. Lett. 2008;18:4872–4875. doi: 10.1016/j.bmcl.2008.07.071. [DOI] [PubMed] [Google Scholar]
- 2.Roth BL. Drugs and valvular heart disease. N. Engl. J. Med. 2007;356:6–9. doi: 10.1056/NEJMp068265. [DOI] [PubMed] [Google Scholar]
- 3.Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 4.Roth BL, Sheffler DJ, Kroeze WK. Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nature Rev. Drug Discov. 2004;3:353–359. doi: 10.1038/nrd1346. [DOI] [PubMed] [Google Scholar]
- 5.Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat. Rev. Cancer. 2010;10:130–137. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brötz-Oesterhelt H, Brunner NA. How many modes of action should an antibiotic have? Curr. Opin. Pharmacol. 2008;8:564–573. doi: 10.1016/j.coph.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nature Chem. Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 8.Morphy R, Rankovic Z. Designed Multiple Ligands. An Emerging Drug Discovery Paradigm. J. Med. Chem. 2005;48:6523–6543. doi: 10.1021/jm058225d. [DOI] [PubMed] [Google Scholar]
- 9.Paolini GV, Shapland RHB, van Hoorn WP, Mason JS, Hopkins AL. Global Mapping of Pharmacological Space. Nat. Biotechnol. 2006;24:806–815. doi: 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- 10.Keiser MJ, et al. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 11.Keiser MJ, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vidal D, Mestres J. In Silico Receptorome Screening of Antipsychotic Drugs. Mol. Inf. 2010;29:543–551. doi: 10.1002/minf.201000055. [DOI] [PubMed] [Google Scholar]
- 13.Lounkine E, et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider G, So S-S. Adaptive Systems in Drug Design. Landes Biosciences. 2002 [Google Scholar]
- 15.Schneider G, et al. Voyages to the (un)known: adaptive design of bioactive compounds. Trends Biotechnol. 2009;27:18–26. doi: 10.1016/j.tibtech.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Schneider G, Lee ML, Stahl M, Schneider P. De novo design of molecular architectures by evolutionary assembly of drug-derived building blocks. J. Comput. Aided Mol. Des. 2000;14:487–494. doi: 10.1023/a:1008184403558. [DOI] [PubMed] [Google Scholar]
- 17.Gillet VJ, Willett P, Fleming PJ, Green DV. Designing focused libraries using MoSELECT. J. Mol. Graph. Model. 2002;20:491–498. doi: 10.1016/s1093-3263(01)00150-4. [DOI] [PubMed] [Google Scholar]
- 18.Brown N, McKay B, Gasteiger J. The de novo design of median molecules within a property range of interest. J. Comput. Aided Mol. Des. 2004;18:761–771. doi: 10.1007/s10822-004-6986-2. [DOI] [PubMed] [Google Scholar]
- 19.Nicolaou CA, Brown N, Pattichis CS. Molecular optimization using computational multi-objective methods. Curr Opin Drug Discov Devel. 2007;10:316–324. [PubMed] [Google Scholar]
- 20.Liu Q, Masek B, Smith K, Smith J. Tagged fragment method for evolutionary structure-based de novo lead generation and optimization. J. Med. Chem. 2007;50:5392–5402. doi: 10.1021/jm070750k. [DOI] [PubMed] [Google Scholar]
- 21.Dey F, Caflisch A. Fragment-based de novo ligand design by multiobjective evolutionary optimization. J. Chem. Inf. Model. 2008;48:679–690. doi: 10.1021/ci700424b. [DOI] [PubMed] [Google Scholar]
- 22.Vinkers HM, et al. SYNOPSIS: SYNthesize and OPtimize System in Silico. J. Med. Chem. 2003;46:2765–2773. doi: 10.1021/jm030809x. [DOI] [PubMed] [Google Scholar]
- 23.Heikkilä T, et al. The first de novo designed inhibitors of Plasmodium falciparum dihydroorotate dehydrogenase. Bioorg. Med. Chem. Lett. 2006;16:88–92. doi: 10.1016/j.bmcl.2005.09.045. [DOI] [PubMed] [Google Scholar]
- 24.Roche O, Rodríguez Sarmiento RM. A new class of histamine H3 receptor antagonists derived from ligand based design. Bioorg. Med. Chem. Lett. 2007;17:3670–3675. doi: 10.1016/j.bmcl.2007.04.056. [DOI] [PubMed] [Google Scholar]
- 25.Alig L, et al. Benzodioxoles: novel cannabinoid-1 receptor inverse agonists for the treatment of obesity. J. Med. Chem. 2008;51:2115–2127. doi: 10.1021/jm701487t. [DOI] [PubMed] [Google Scholar]
- 26.Schneider G, et al. Reaction-driven de novo design, synthesis and testing of potential type II kinase inhibitors. Future Med. Chem. 2011;3:415–424. doi: 10.4155/fmc.11.8. [DOI] [PubMed] [Google Scholar]
- 27.Wermuth CG. Selective optimization of side activities: the SOSA approach. Drug Discov. Today. 2006;11:160–164. doi: 10.1016/S1359-6446(05)03686-X. [DOI] [PubMed] [Google Scholar]
- 28.Gaulton A, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr777. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ribeiz SR, et al. Cholinesterase inhibitors as adjunctive therapy in patients with schizophrenia and schizoaffective disorder: a review and meta-analysis of the literature. CNS Drugs. 2010;24:303–317. doi: 10.2165/11530260-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 30.Arnsten AF, Murphy B, Merchant K. The selective dopamine D4 receptor antagonist, PNU-101387G, prevents stress-induced cognitive deficits in monkeys. Neuropsychopharmacology. 2000;23:405–410. doi: 10.1016/S0893-133X(00)00133-0. [DOI] [PubMed] [Google Scholar]
- 31.Gillet V, Johnson AP, Mata P, Sike S, Williams P. SPROUT: a program for structure generation. J. Comput. Aided Mol. Des. 1993;7:127–153. doi: 10.1007/BF00126441. [DOI] [PubMed] [Google Scholar]
- 32.Stahl M, et al. A validation study on the practical use of automated de novo design. J. Comput. Aided Mol. Des. 2002;16:459–478. doi: 10.1023/a:1021242018286. [DOI] [PubMed] [Google Scholar]
- 33.Brown N, McKay B, Gilardoni F, Gasteiger J. A graph-based genetic algorithm and its application to the multiobjective evolution of median molecules. J. Chem. Inf. Comput. Sci. 2004;44:1079–1087. doi: 10.1021/ci034290p. [DOI] [PubMed] [Google Scholar]
- 34.Nicolaou CA, Apostolakis J, Pattichis CS. De novo drug design using multiobjective evolutionary graphs. J. Chem. Inf. Model. 2009;49:295–307. doi: 10.1021/ci800308h. [DOI] [PubMed] [Google Scholar]
- 35.Stewart KD, Shiroda M, James CA. Drug Guru: a computer software program for drug design using medicinal chemistry rules. Bioorg. Med. Chem. 2006;14:7011–7022. doi: 10.1016/j.bmc.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 36.Kryssanov VV, Tamaki H, Kitamura S. Understanding design fundamentals: how synthesis and analysis drive creativity, resulting in emergence. Artif. Intell. Eng. 2001;15:329–342. [Google Scholar]
- 37.Deb K, Sundar J, Udaya Bhaskara Rao N, Chaudhuri S. Reference Point Based Multi-Objective Optimization Using Evolutionary Algorithms. Intnat. J. Comp. Intell. Res. 2006;2:273–286. [Google Scholar]
- 38.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Del. Revs. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 39.Ertl P, Schuffenhauer A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J. Cheminform. 2009;10:8. doi: 10.1186/1758-2946-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fanelli F, De Benedetti PG. In: Antitargets: Prediction and Prevention of Drug Side Effects. Vaz RJ, Klabunde T, editors. Wiley-VCH; 2008. [Google Scholar]
- 41.Bemis GW, Murcko MA. The properties of known drugs. 1 Molecular frameworks. J. Med. Chem. 1996;39:2887–2893. doi: 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- 42.Ortega R, et al. Synthesis, binding affinity and SAR of new benzolactam derivatives as dopamine D3 receptor ligands. Bioorg. Med. Chem. Letts. 2009;19:1773–1778. doi: 10.1016/j.bmcl.2009.01.067. [DOI] [PubMed] [Google Scholar]
- 43.Löber S, Hübner H, Tschammer N, Gmeiner P. Recent advances in the search for D3 and D4 selective drugs: probes, models and candidates. Trends Pharm. Sci. 2011;32:148–157. doi: 10.1016/j.tips.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Martin RE, Green LG, Guba W, Kratochwil N, Christ A. Discovery of the first nonpeptidic, small-molecule, highly selective somatostatin receptor subtype 5 antagonists: a chemogenomics approach. J. Med. Chem. 2007;50:6291–6294. doi: 10.1021/jm701143p. [DOI] [PubMed] [Google Scholar]
- 45.Bender A, et al. Chemogenomic data analysis: prediction of small-molecule targets and the advent of biological fingerprint. Comb. Chem. High Throughput Screen. 2007;10:719–731. doi: 10.2174/138620707782507313. [DOI] [PubMed] [Google Scholar]
- 46.Rogers D, Brown RD, Hahn M. Using extended-connectivity fingerprints with Laplacian-modified Bayesian analysis in high-throughput screening follow-up. J. Biomol. Screen. 2005;10:682–686. doi: 10.1177/1087057105281365. [DOI] [PubMed] [Google Scholar]
- 47.Huang XP, Mangano T, Hufeisen S, Setola V, Roth BL. Identification of human Ether-à-go-go related gene modulators by three screening platforms in an academic drug-discovery setting. Assay Drug Dev. Technol. 2010;8:727–742. doi: 10.1089/adt.2010.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruda GF, et al. Aryl Phosphoramidates of 5-phosho erythronohydroxamic acid, a new class of potent trypanocidal agents. J. Med. Chem. 2010;53:6071–6078. doi: 10.1021/jm1004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pogorelov VM, Rodriguiz RM, Insco ML, Caron MG, Wetsel WC. Novelty seeking and stereotypic activation of behavior in mice with disruption of the Dat1 gene. Neuropsychopharma. 2005;30:1818–1831. doi: 10.1038/sj.npp.1300724. [DOI] [PubMed] [Google Scholar]
- 50.Porton B, et al. Mice lacking synapsin III show abnormalities in explicit memory and conditioned fear. Genes Brain Behav. 2010;9(9):257–268. doi: 10.1111/j.1601-183X.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glick M, Jenkins JL, Nettles JH, Hitchings H, Davies JW. Enrichment of high-throughput screening data with increasing levels of noise using support vector machines, recursive partitioning, and laplacian-modified naive bayesian classifiers. Journal of chemical information and modeling. J. Chem. Inf. Model. 2006;46:193–200. doi: 10.1021/ci050374h. [DOI] [PubMed] [Google Scholar]
- 52.Rogers D, Hahn M. Extended-connectivity fingerprints. J. Chem. Info. Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 53.Truchon JF, Bayly CI. Evaluating virtual screening methods: good and bad metrics for the “early recognition” problem. J. Chem. Info. Model. 2007;47:488–508. doi: 10.1021/ci600426e. [DOI] [PubMed] [Google Scholar]
- 54.Zhao W, Hevener KE, White SW, Lee RE, Boyett JM. A statistical framework to evaluate virtual screening. BMC Bioinformatics. 2009;10:225. doi: 10.1186/1471-2105-10-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cannon EO, Nigsch F, Mitchell JB. A novel hybrid ultrafast shape descriptor method for use in virtual screening. Chem. Cent. J. 2008;2:3. doi: 10.1186/1752-153X-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Corne DW, Knowles JD. Proceedings of the 9th annual conference on Genetic and evolutionary computation.pp. 773–780. [Google Scholar]
- 57.Obrezanova O, Csanyi G, Gola JM, Segall MD. Gaussian processes: A method for automatic QSAR modelling of ADME properties. J. Chem. Info. Model. 2007;47:1847–1857. doi: 10.1021/ci7000633. [DOI] [PubMed] [Google Scholar]
- 58.Obrezanova O, Gola JMR, Champness EJ, Segall MD. Automatic QSAR modeling of ADME properties: blood-brain barrier penetration and aqueous solubility J. Comput. Aided Mol. Des. 2008;22:431–440. doi: 10.1007/s10822-008-9193-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.