Summary
Iron constitutes a major source of toxicity due to its ability to generate reactive oxygen species that can damage cellular macromolecules. However the precise mechanism by which exposure to high iron concentrations results in cellular toxicity remains unknown. Here we identify sphingolipid synthesis and signaling as a major mediator of iron toxicity in S. cerevisiae. Inhibition of sphingolipid synthesis by myriocin treatment or after overexpression of the negative regulator Orm2p confers resistance to high iron. High iron conditions upregulate sphingolipid synthesis, and increasing sphingolipid levels by inactivating Orm2p exacerbates sensitivity to iron. Toxicity is mediated by sphingolipid signaling, as inactivation of the sphingolipid-activated protein kinases Pkh1p and Ypk1p and of the transcription factor Smp1p also enhances resistance to high iron conditions. These results demonstrate an unexpected connection between sphingolipid flux and iron toxicity, and show that activation of a signal transduction cascade contributes to iron-mediated cellular toxicity.
Introduction
Iron (Fe) is a ubiquitous metal in biological systems. However Fe also represents a potential danger to biological macromolecules due to its ability to generate reactive oxygen species (ROS; Touati, 2000; Valentine et al., 1998; Valko et al., 2005). In most organisms, the present paradigm in Fe toxicity suggests that the major toxic effect of Fe is the result of the exposure of cellular macromolecules to ROS (Touati, 2000; Valko et al., 2005). Defects in Fe metabolism can result in various pathologies in diseases of Fe overload (Hentze et al., 2004), underscoring the importance of limiting iron toxicity. Thus, iron levels must be regulated to meet the demands of cellular metabolism but also to prevent an overabundance that might result in potential damages. The unicellular eukaryote S. cerevisiae has been used as a model system to study Fe transport, mtabolism and toxicity (Philpott, 2006; Van Ho et al., 2002; Chen et al., 2002; Lin et al., 2011). In yeast, exposure of cells to high Fe results in growth defects through inhibition of the cell cycle (Philpott et al., 1998). Some of the protective mechanisms against Fe toxicity include storage of iron in organelles, presumably to limit the exposure of cytosolic or nuclear macromolecules to high iron concentrations (Chen and Kaplan, 2000; Li et al., 2001; Lin et al., 2011). This observation is consistent with the hypothesis that Fe causes toxicity by oxidative damage. However recent work has shown that some of the toxic effects of Fe in yeast are independent of oxidative stress (Lin et al., 2011), suggesting that the major pathway(s) by which Fe hampers cellular growth remains to be identified. The results presented here show that reducing sphingolipid synthesis and signaling is sufficient to allow yeast cells to grow in otherwise toxic iron conditions, demonstrating that sphingolipid signaling contributes to mediating iron toxicity in S. cerevisiae.
Results
Genetic or Chemical Inhibition of Sphingolipid Synthesis Confers Resistance to Iron Toxicity
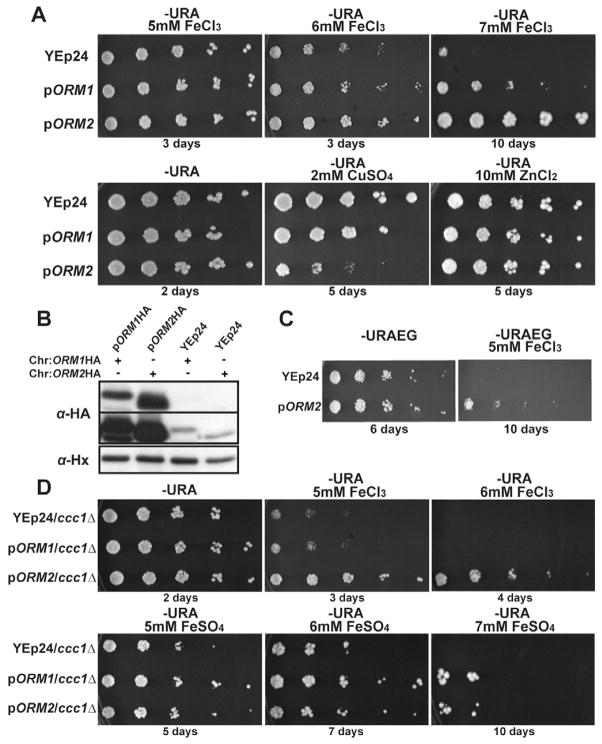
We showed previously that yeast cells lacking the RNase III Rnt1p are hypersensitive to high Fe (Lee et al., 2005). To further understand the molecular basis of Fe toxicity, we performed a high-copy suppressor screen to identify genes, which, when overexpressed, would allow Rnt1p-deficient cells to grow in high Fe. We constructed a genomic library from the rnt1Δ strain in the multi-copy vector YEp24, and performed screens in the presence of high Fe to identify suppressors. We could not reproducibly identify genes that allow rnt1Δ cells to grow in high Fe, so we redirected our efforts to performing this screen in wild-type cells. We isolated a clone exhibiting growth in high Fe conditions, and identified ORM2 as the gene responsible for this effect. We confirmed that an independently cloned version of ORM2 could confer resistance to Fe toxicity when overexpressed in wild-type cells (Fig. 1A). Orm2p and the closely related Orm1p protein were identified as negative regulators of sphingolipid synthesis and mediators of the endoplasmic reticulum (ER) stress response (Breslow et al., 2010; Breslow and Weissman, 2010; Han et al., 2010). We tested the ability of Orm1p to confer resistance to iron toxicity and found that ORM2-overexpressing cells (ORM2ox; pORM2 on the figures) grew faster than ORM1ox in high Fe conditions, suggesting that Orm2p was a more potent high copy suppressor of Fe toxicity than Orm1p. We monitored Orm1p and Orm2p levels in high Fe conditions, both when the proteins were HA-tagged at their respective endogenous loci, and when the same HA-tagged proteins were overexpressed (Fig. 1B). We found that in this overexpression system, Orm2p was expressed at higher levels than Orm1p in high Fe conditions (Fig. 1B), and at levels much higher than the endogenous copy. Thus, the ability of Orm2p to confer better growth in high iron than Orm1p might be linked to higher overexpression levels.
Figure 1. ORM2 confers specific resistance to iron toxicity in a variety of physiological contexts.
A. Strains were grown in SD medium lacking uracil (-URA) supplemented with the indicated metals. These, and all subsequent assays show 10-fold dilution series from left to right. For the bottom panel, plates were incubated at 30°C. B. Cells expressing Orm1p or Orm2p HA-tagged at their chromosomal loci (Chr), transformed with either the YEp24 vector, or with plasmids containing HA-tagged Orm1p or Orm2p (pORM1/2-HA) were grown in 7mM FeCl3, and the levels of Orm1/2p analyzed by immunoblot. Upper panels are two different exposures to detect endogenous and overexpressed Orm1/2p. C. Strains were grown on ethanol-glycerol carbon source lacking uracil (URAEG), without or with 5mM FeCl3. D. Legends as in A.
The resistance of ORM2ox cells to high Fe was specific to this particular stress. In contrast to high iron, ORM2ox cells actually grew slower than wild-type in high copper conditions (Fig. 1A). In addition, we found no growth difference between wild-type and ORM2ox cells in high zinc (Fig. 1A) or oxidative stress conditions (Fig. S1A). We also found no differences in the levels of oxidized proteins between wild-type and ORM2ox cells in high Fe (Fig. S1B), no growth difference in high salt (Fig. S1C), nor in survival after heat-shock (Fig. S1D). Finally, because media containing high Fe require acidic conditions in order for Fe to be soluble, we also tested the growth of wild-type and ORM2ox cells in acidic medium and found no difference (Fig. S1E). Thus, ORM2 overexpression does not confer general resistance to a variety of cellular stresses but seem specific to Fe toxicity.
The genetic screen identifying ORM2 was performed in glucose-containing medium, where oxidative stress is low. We found that overexpressing Orm2p conferred resistance to Fe toxicity in media containing the non-fermentable carbon sources ethanol and glycerol (Fig. 1C). Thus, Orm2p can also suppress Fe toxicity when oxidative stress is increased by respiration. To further investigate the potency of the suppression of Fe toxicity by Orm2p, we overexpressed Orm2p in a strain deficient for the vacuolar Fe importer Ccc1p. Strains lacking Ccc1p are hypersensitive to high Fe because they cannot store iron in the vacuole (Li et al., 2001; Lin et al., 2011). Overexpression of ORM2 was sufficient to rescue the growth of ccc1Δ cells at 6mM FeCl3 (Fig. 1D), showing that Orm2p can also rescue the growth of cells deficient in iron storage. Overexpressing Orm1p also resulted in some suppression, although this effect was more robust when cells were grown in medium containing FeSO4 (Fig. 1D). We do not fully understand why Orm1p seems to be more potent in these conditions, but this effect might be due to differential expression. Collectively, these results show that overexpressing Orm1/2p can suppress Fe toxicity in a variety of physiological contexts and can compensate for defects in vacuolar iron storage. These results also show that the capacity of Orm1/2p to suppress Fe toxicity is not linked to their ability to influence vacuolar iron storage by modulation of the activity of the vacuolar ATPase (Finnigan et al., 2011), since cells lacking Ccc1p cannot store iron in the vacuole. Taken together, these results show that overexpression of Orm2p can suppress Fe toxicity in a variety of physiological contexts.
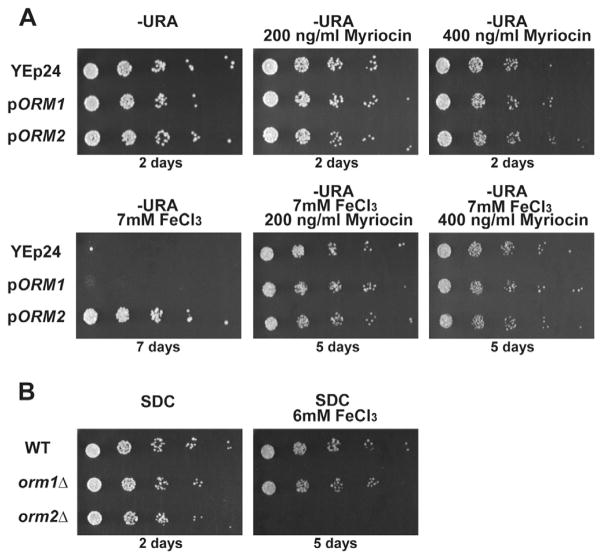
Based on its cellular functions (Breslow et al., 2010; Han et al., 2010), the ability of Orm2p to enhance resistance to Fe toxicity could be attributed to two distinct pathways: (i) control of ER stress and unfolded protein response (UPR), or (ii) inhibition of sphingolipid synthesis. Since metals such as Cd can induce the UPR (Gardarin et al., 2010), we tested whether ORM2 confers resistance to iron toxicity by modulating the UPR, which might have been activated by high Fe. Splicing of the HAC1 mRNA, a hallmark of the UPR (Cox and Walter, 1996), was not triggered by growing cells in high Fe (Fig. S2A), and no differences in HAC1 splicing were found when overexpressing ORM2 (Fig. S2A). These results show that in contrast to Cd stress, high Fe conditions do not trigger the UPR, and that Orm2p is unlikely to mediate resistance to high iron by modulating ER stress. To test whether Orm2p confers resistance to Fe toxicity by inhibiting sphingolipid synthesis, we used the chemical inhibitor myriocin, which modulates sphingolipid synthesis through inhibition of the long-chain base (LCB)-synthesizing enzyme serine palmitoyltransferase (Breslow and Weissman, 2010; Cowart and Obeid, 2007; Dickson et al., 2006). If ORM2 overexpression enhances resistance to iron toxicity by inhibiting sphingolipid synthesis, exposure of cells to sub-toxic concentrations of myriocin was predicted to have a similar protective effect. While myriocin at the concentrations used had no effect on overall growth in normal medium, it allowed wild-type cells to survive in high Fe conditions (Fig. 2A). There seemed to be no cumulative effect of exposing cells to myriocin and overexpressing ORM2, consistent with the hypothesis that these two treatments act on the same pathway.
Figure 2. Modulation of sphingolipid synthesis changes resistance to high iron.
A. Strains were grown on normal medium or with the indicated concentrations of myriocin and FeCl3. B. Strains were grown on normal or 6mM FeCl3 conditions in minimal medium (SDC).
The previous results are consistent with the hypothesis that ORM2 overexpression and myriocin treatment suppress iron toxicity by reducing sphingolipid levels. To test whether an increase of sphingolipid flux is toxic when combined with high Fe concentrations, we used an orm2Δ strain, since ORM2 inactivation was shown to upregulate sphingolipid synthesis (Breslow et al., 2010; Breslow and Weissman, 2010; Finnigan et al.; Han et al., 2010). As shown in Fig. 2B, cells lacking ORM2 were unable to grow in 6mM Fe, while wild-type cells were still viable. By contrast, the orm1Δ strain did not exhibit any strong growth phenotype (Fig. 2B), suggesting a more prominent function for Orm2p in these conditions. These results show that an increase of sphingolipid synthesis confers hypersensitivity to subtoxic Fe concentrations, further correlating sphingolipid flux and Fe-mediated toxicity.
Orm2p Overexpression does not reduce Cellular Iron Levels
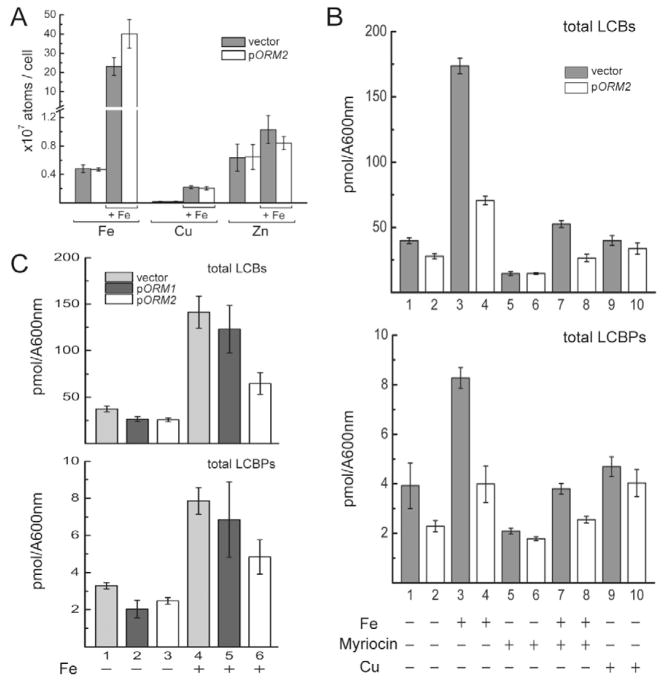
Since sphingolipids are integral components of membranes, the capacity of Orm2p to enhance resistance to Fe toxicity could be linked to a potential function in modulating the activity of a transmembrane Fe transporter and Fe import. To test this hypothesis, we measured the iron content of wild-type and ORM2ox cells by ICP-MS (Fig. 3A). In normal Fe conditions, these strains showed comparable low levels of Fe and of other metals (Fig. 3A). In high Fe conditions, both strains exhibited an increase of intracellular Fe and ORM2ox cells showed slightly higher Fe levels than the wild-type (Fig. 3A). In contrast, the level of other metals such as Zn or Cu was similar in both strains (Fig. 3A), showing that the slightly higher Fe content in ORM2ox cells is specific to this metal. Prior work had shown that sphingolipids regulate the expression of Fet3p (Villa et al., 2009), a multi-copper oxidase involved in Fe transport. We detected similar levels of the FET3 mRNA in wild-type and ORM2ox cells (Fig. S2B) in normal conditions, and could not detect FET3 in high Fe conditions in both strains, suggesting that ORM2 overexpression in high iron does not change the expression of this key mediator of Fe assimilation. Taken together, these results rule out the hypothesis that Orm2p suppresses cellular Fe toxicity by preventing intracellular iron uptake.
Figure 3. High iron conditions result in higher intracellular iron concentrations and sphingolipid LCBs.
A. Intracellular metal content analysis. Strains were grown on SD plates containing a normal concentration of Fe and incubated for 2 days, or on SD+7mM FeCl3 and incubated for 6 days (pORM2) and 11 days (YEp24) before harvesting for ICP-MS measurement. The results shown are the average of 6 samples (triplicate samples from 2 independent experiments), except for pORM2 in normal conditions, which is the average of 5 samples (3 and 2 samples from 2 independent experiments).
B. Sphingolipid analysis in normal, high Fe and high Cu conditions. Strains were grown either on normal medium, 7mM FeCl3, 200ng/ml myriocin, 7mMFeCl3+200ng/ml myriocin or 2mM CuSO4 conditions and incubated at 25°C until colonies reached a similar size before harvesting for LCBs analysis. Triplicate samples for LCBs analysis were prepared as described in experimental procedures. LCB and LCBP levels are indicated as pmol/A600. Statistical analysis is provided in Supplementary Table S1. Evidence for reproducibility of the effect of high iron conditions and ORM2 overexpression on LCB levels is shown in Figure S3.
C. Comparison of the effects of Orm1p and Orm2p overexpression on sphingolipid levels. Procedures were the same as in B, except that cells were transformed with the vector alone (YEp24), or the vector overexpressing Orm1p or Orm2p (pORM1/2) and grown in -URA medium (lanes 1,2, 3) or with -URA + 7mM FeCl3. Shown are the average values of 3 independent biological samples with the standard deviations.
High Iron Conditions increase Sphingolipids Synthesis
Since the previous data showed that decreasing sphingolipid synthesis enhances the capacity to grow in high Fe, we investigated whether high Fe conditions trigger an increase in sphingolipid synthesis. We measured the concentration of sphingolipid long-chain bases (LCBs) and long-chain base phosphates (LCBPs) metabolic intermediates in wild-type and ORM2ox cells, since their level is sensitive to changes in growth rate and environmental stress (Cowart and Obeid, 2007; Dickson et al., 2006). Consistent with previous results (Breslow et al., 2010; Han et al., 2010), overexpression of ORM2 or myriocin treatment significantly reduced LCB but not LCBP levels in normal medium (Fig. 3B; detailed values and statistical analysis shown in Table S1; Fig. S3; Fig. S4). When grown in high Fe, wild-type cells showed a statistically significant increase in total LCBs and LCBPs compared to normal medium (Fig. 3B; Fig. S3). These data support the hypothesis that high Fe causes an accumulation of sphingolipid intermediates. Overexpressing Orm2p or myriocin treatment significantly reduced LCBs in Fe-treated cells (Fig. 3B, Fig. S3), and a stronger reduction was observed when combining Orm2p overexpression and myriocin (Fig. 3B, Lanes 4 and 8). Similar trends were observed for LCBPs (Fig. 3B; Fig. S3). The complete analysis for each species of LCB and LCBP is shown in Fig. S4 and shows the same tendencies as total LCBs and LCBPs. These effects were specific to Fe, as high Cu was found to have no effect on LCBs and LCBPs levels (Fig. 3B). We also compared the ability of Orm1p and Orm2p to modulate sphingolipid synthesis in high Fe conditions when each of these proteins was overexpressed (Fig. 3C). We found that Orm2p overexpression conferred a statistically significant reduction of LCBs and LCBPs in high iron compared to cells transformed with the YEp24 vector (p-values of 7.3 10−5 and, 5.2 10−5 respectively, t-test), while Orm1p overexpression did not lead to a significant effect. These results corroborate the suppression analysis showing that Orm2p is a much more potent suppressor of Fe toxicity than Orm1p (Fig. 1A). Taken together, these results show that the levels of LCBs and LCBPs are specifically increased in high Fe conditions and that the capacity of Orm2p or myriocin to enhance resistance to high Fe is correlated with a reduction in LCB and LCBP levels.
Inactivation of sphingolipid-mediated signaling enhances resistance to high iron
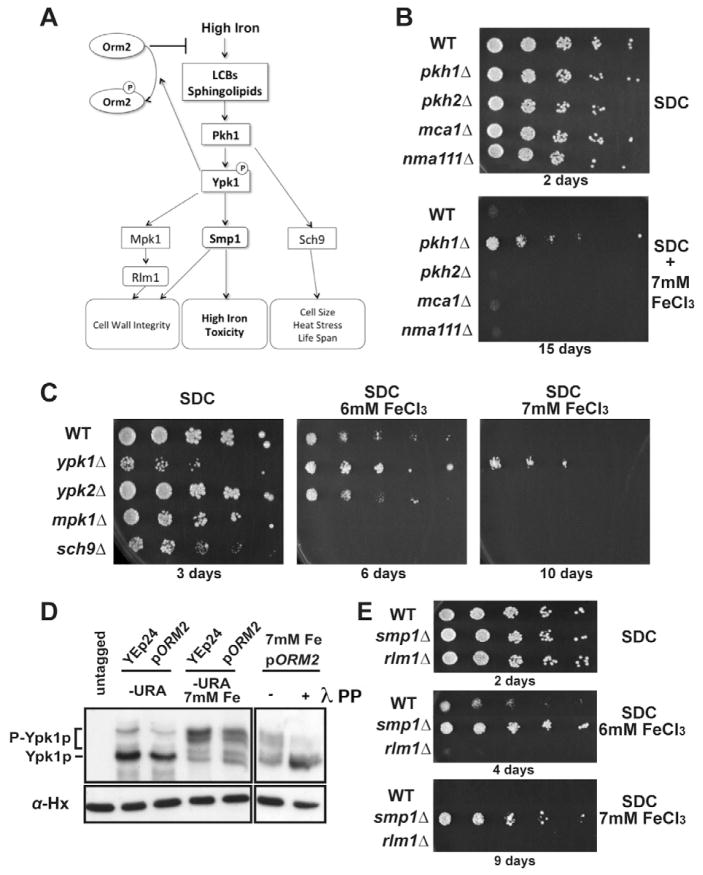
Ceramides have been shown to induce apoptosis in mammals (Colombini, 2010). In addition, previous data have shown that exposure of yeast cells to metals such as Cu and Mn can trigger apoptosis (Liang and Zhou, 2007). Thus, the increase in sphingolipid levels detected in high Fe conditions might potentially cause apoptosis through an accumulation of ceramides. To test this hypothesis, we analyzed a number of mutants in this pathway. Blocking apoptosis by inactivation of the metacaspase Mca1p/Yca1p (Madeo et al., 2002) or of the serine protease Nma111p (Fahrenkrog et al., 2004) did not restore growth in high Fe conditions (Fig. 4B). We conclude that unlike Cu or Mn, high Fe is unlikely to mediate toxicity through apoptosis.
Figure 4. Inactivation of sphingolipid signaling confers resistance to iron toxicity.
A. Yeast sphingolipid-mediated signal transduction pathways. B. Strains were grown on normal minimal medium (SDC) or SDC containing 7mM FeCl3. C. Legends as in Fig. 1A. D. Immunoblot analysis of myc-tagged Ypk1p in normal and high iron conditions in wild-type cells or cells overexpressing Orm2p. SDS-PAGE was performed in the presence of 20 μM Phos-Tag. To demonstrate that the higher bands corresponded to phospho-Ypk1p, the extracts prepared from cells overexpressing Orm2p were treated with lambda (λ) phosphatase (right lane). E. Legends as in 4B.
In addition to triggering apoptosis, sphingolipids are involved in many cellular functions, as structural components of membranes but also as signaling molecules during stress (Breslow and Weissman, 2010; Dickson, 2008; Dickson et al., 2006). In S. cerevisiae their signaling function is primarily mediated by LCBs, which activate the Pkh1p and Pkh2p protein kinases (Fig. 4A; Liu et al., 2005; Roelants et al., 2002), although other sphingolipids can act as signaling molecules as well (Roelants et al., 2010). If the contribution of LCBs to cellular toxicity in high iron conditions is due to their signaling function, we hypothesized that strains deficient in these kinases might also exhibit resistance to high Fe. Indeed, the pkh1Δ strain exhibited increased resistance to high Fe (Fig. 4B), while cells lacking Pkh2p did not show this phenotype, suggesting a specific role for Pkh1p. Pkh1p has been shown to activate several signal transduction cascades (Fig. 4A). To investigate which cascade is responsible for mediating toxicity in high iron conditions, we inactivated the Mpk1p, Sch9p, Ypk1p and Ypk2p kinases and assessed the growth of these mutants in high Fe. Strikingly, inactivation of Ypk1p resulted in resistance to high Fe, while none of the other kinase knockout strains exhibited this phenotype (Fig. 4C). This suppression phenotype is particularly striking considering that the ypk1Δ strain shows a growth defect in normal medium (Fig. 4C, upper panel). We analyzed the expression of some genes known to be controlled by the Pkh1p/Ypk1p kinases, but none of them showed a change in expression in high iron or upon Orm2p overexpression (Fig. S2C). Thus, these kinases control other cellular functions besides the expression of known target genes.
To directly demonstrate that sphingolipid signaling is activated during high Fe conditions, we assessed Ypk1p phosphorylation by immunoblot using a strain expressing myc-tagged Ypk1p (Fig. 4D). In cells shifted to high Fe, we found increased amounts of slower mobility forms of Ypk1p (Fig. 4D). Strikingly, the level of these forms was reduced upon Orm2p overexpression (Fig. 4D). These slower mobility bands correspond to phosphorylated forms of Ypk1p, as their mobility was increased upon treatment of extracts with phosphatase prior to fractionation (Fig. 4D). This result directly shows that increased sphingolipid levels in high iron activate the Pkh1/Ypk1 signal transduction cascade, and that Orm2p overexpression confers resistance to high iron by inhibiting this response and reducing Ypk1p phosphorylation. While this work was being considered, it was shown that Ypk1p can phosphorylate and inactivate Orm2p (Roelants et al., 2011; Sun et al., 2012). Therefore the observation that Ypk1p depletion could rescue iron toxicity might indicate that this effect could be due to Orm2p activation in the absence of Ypk1p-mediated phosphorylation. If this were the case, we would not expect any downstream target of Ypk1p to have any influence on the ability of cells to grow in high iron conditions. However, we found that inactivation of Smp1p also rescued the ability of cells to grown in high iron conditions (Fig. 4E). This result contrasts with the observation that Smp1p inactivation usually results in decreased growth in stress conditions, such as high osmolarity (de Nadal et al., 2003). In contrast, inactivation of another transcription factor, Rlm1p had a negative effect on growth in high iron (Fig. 4E). Taken together, these results show that sphingolipid signaling through Pkh1p, Ypk1p and Smp1p contributes to mediating iron toxicity.
Discussion
In this study, we report the unexpected finding that genetic or chemical inhibition of sphingolipid synthesis allow yeast cells to grow in higher iron conditions, showing that sphingolipid levels are limiting the ability of cells to grow in these conditions. We cannot completely exclude that the increased growth of cells overexpressing Orm2p cells in high iron is due to general effects on cellular viability, or that overexpressing Orm2p can confer resistance to other transition metals. However the effect of Orm2p seem specific to Fe, as overexpressing Orm2p does not confer resistance to Cu or Zn, or to all other stresses tested - indeed, the opposite effect is observed for Cu, since cells overexpressing Orm2p are more sensitive to Cu. A link between Fe levels and sphingolipids was shown previously, as Fe deficiency was found to reduce sphingolipids (Shakoury-Elizeh et al., 2010). This effect might be explained by the inactivation in low iron conditions of sphingolipid biosynthetic enzymes that require Fe as an essential cofactor. In contrast, we found that high Fe conditions increase sphingolipid levels. This effect might be due to a general increase of synthesis of these lipids in response to environmental stress, as shown previously (Liu et al., 2005). However, unlike other cellular stresses such as heat shock, elevated sphingolipid do not confer resistance to high iron conditions, but rather, are toxic. Thus there is a unique relationship between sphingolipid levels and iron-mediated cellular toxicity, as preventing upregulation is sufficient to allow cells to survive in high Fe.
Our data indicate that sphingolipids mediate cellular toxicity by acting as signaling molecules. It was shown recently that sphingolipid signaling affect yeast lifespan (Huang et al., 2012). This effect is mechanistically distinct from those described here, as inactivation of Sch9p increases lifespan (Huang et al., 2012) but decreases the ability of cells to grow in high Fe (Fig. 4C). Thus, sphingolipid signaling can modulate cellular fitness in a variety of mechanisms. Sphingolipid signaling through Pkh1p/Ypk1p was shown to be required for endocytosis (deHart et al., 2002; Friant et al., 2001), and it is known that some divalent cation transporters are endocytosed following exposure to high metal concentrations (Philpott, 2006). However, our data are inconsistent with the idea that these lipids play a role in limiting Fe toxicity by controlling Fe transporters endocytosis, since we found slightly elevated Fe levels in cells overexpressing Orm2p. Instead, sphingolipids play a function in mediating Fe toxicity by triggering a change in cellular properties through the activation of the Pkh1/Ypk1p signal transduction cascade. Although we favor LCBs as regulators of Pkh1-Ypk1 signaling in high Fe, we cannot exclude that other sphingolipids are also involved because their concentration is also affected when serine palmitoyltransferase activity is reduced (Breslow et al. 2010; Huang et al., 2012). This signaling cascade involves the Smp1p transcription factor, as Smp1p inactivation increases resistance to high Fe. Smp1p might mediate downstream events responsible for mediating Fe toxicity, which could include changes in cell cycle events known to be affected in high Fe (Philpott et al., 1998). Alternatively, other molecular events controlled by sphingolipids, such as the activity of aminophospholipid flippases (Roelants et al., 2010) might modify the properties of the membrane or the cell wall and thus make the cells more sensitive to high Fe.
Iron is generally thought to mediate cellular toxicity by virtue of its ability to generate damaging ROS (Touati, 2000; Valko et al., 2005). By contrast, our data show that a significant part of iron toxicity in yeast involves the activation of sphingolipid-signaling. This result is in agreement with the pioneer work from Lin and colleagues, who showed that some of the toxic effects of Fe are independent from oxidative stress in yeast, and therefore from the ability of Fe to generate ROS (Lin et al., 2011). Taken together, these findings provide a paradigm change by showing that Fe toxicity in a model eukaryotic unicellular system is mediated, at least in part, by the activation of a biological signal transduction cascade. It remains to be investigated if a similar signaling pathway is activated in other eukaryotic cells exposed to high iron conditions.
Experimental Procedures
Strains and plasmids
S. cerevisiae strains were derived from BMA64 (Lee et al., 2005). Genomic library construction and screening, and construction of the plasmids used in this study are described in supplemental materials. Cells were grown on synthetic defined (SD) medium plates with or without specific concentrations of added FeCl3 or FeSO4 or of other metals, as described in supplemental materials. Most of the growth assays were performed at 25°C, unless indicated otherwise in the figure legends.
Sphingolipid and Metal Analysis
Preparation of extracts for the analysis of LCB sphingolipids is described in detail in supplemental materials. Analysis of LCB by HPLC was performed as described (Lester and Dickson, 2001). For measurement of metal content by ICP-MS, cells were washed with 1mM EDTA and water after harvesting, before digestion with nitric acid. ICP-MS measurement was performed as described (Kropat et al., 2011).
RNA and Protein Analysis
Protein extracts were prepared as described in supplemental materials. Western blot analysis of strains expressing myc-tagged Ypk1p was performed using anti-myc polyclonal antibody. Western blot analysis of strains expressing Orm1p or Orm2p tagged at the C-terminus with the HA epitope were performed using anti-HA monoclonal antibody. Analysis of Ypk1p phosphorylation by phosphatase treatment and fractionation on SDS-PAGE containing Phos-tag is described in supplemental materials.
Statistical Analysis
Statistical analysis was performed using a two tailed t-test in Excel.
Supplementary Material
Highlights.
Inhibiting sphingolipid synthesis allows yeast to grow in higher iron conditions
Yeast exposed to high iron increase sphingolipid production
High iron conditions activate sphingolipid signaling
Inhibiting Pkh1p/Ypk1p/Smp1p signaling confers relative resistance to iron toxicity.
Acknowledgments
We thank S. Covarrubias for help with the construction of the genomic library, I. Toesca for help with initials screens, R. Lester for advice on sphingolipid analyses and C. Clarke, J. Valentine and J. Torres for discussions. Supported by NIGMS grant GM61518 to G.C, AG024377 grant to R.C.D and DOE-DE-FG02-04ER15529 grant to S.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Weissman JS. Membranes in balance: mechanisms of sphingolipid homeostasis. Mol Cell. 2010;40:267–279. doi: 10.1016/j.molcel.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen OS, Hemenway S, Kaplan J. Genetic analysis of iron citrate toxicity in yeast: implications for mammalian iron homeostasis. Proc Natl Acad Sci U S A. 2002;99:16922–16927. doi: 10.1073/pnas.232392299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen OS, Kaplan J. CCC1 suppresses mitochondrial damage in the yeast model of Friedreich’s ataxia by limiting mitochondrial iron accumulation. J Biol Chem. 2000;275:7626–7632. doi: 10.1074/jbc.275.11.7626. [DOI] [PubMed] [Google Scholar]
- Colombini M. Ceramide channels and their role in mitochondria-mediated apoptosis. Biochim Biophys Acta. 2010;1797:1239–1244. doi: 10.1016/j.bbabio.2010.01.021. [DOI] [PubMed] [Google Scholar]
- Cowart LA, Obeid LM. Yeast sphingolipids: recent developments in understanding biosynthesis, regulation, and function. Biochim Biophys Acta. 2007;1771:421–431. doi: 10.1016/j.bbalip.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- de Nadal E, Casadome L, Posas F. Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol Cell Biol. 2003;23:229–237. doi: 10.1128/MCB.23.1.229-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deHart AK, Schnell JD, Allen DA, Hicke L. The conserved Pkh-Ypk kinase cascade is required for endocytosis in yeast. J Cell Biol. 2002;156:241–248. doi: 10.1083/jcb.200107135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC. Thematic review series: sphingolipids. New insights into sphingolipid metabolism and function in budding yeast. J Lipid Res. 2008;49:909–921. doi: 10.1194/jlr.R800003-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC, Sumanasekera C, Lester RL. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog Lipid Res. 2006;45:447–465. doi: 10.1016/j.plipres.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B, Sauder U, Aebi U. The S. cerevisiae HtrA-like protein Nma111p is a nuclear serine protease that mediates yeast apoptosis. J Cell Sci. 2004;117:115–126. doi: 10.1242/jcs.00848. [DOI] [PubMed] [Google Scholar]
- Finnigan GC, Ryan M, Stevens TH. A Genome-Wide Enhancer Screen Implicates Sphingolipid Composition in Vacuolar ATPase Function in Saccharomyces cerevisiae. Genetics. 2011;187:771–783. doi: 10.1534/genetics.110.125567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friant S, Lombardi R, Schmelzle T, Hall MN, Riezman H. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 2001;20:6783–6792. doi: 10.1093/emboj/20.23.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardarin A, Chedin S, Lagniel G, Aude JC, Godat E, Catty P, Labarre J. Endoplasmic reticulum is a major target of cadmium toxicity in yeast. Mol Microbiol. 2010;76:1034–1048. doi: 10.1111/j.1365-2958.2010.07166.x. [DOI] [PubMed] [Google Scholar]
- Han S, Lone MA, Schneiter R, Chang A. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc Natl Acad Sci U S A. 2010;107:5851–5856. doi: 10.1073/pnas.0911617107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- Huang X, Liu J, Dickson RC. Down-regulating sphingolipid synthesis increases yeast lifespan. PLoS Genet. 2012;8:e1002493. doi: 10.1371/journal.pgen.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J, Hong-Hermesdorf A, Casero D, Ent P, Castruita M, Pellegrini M, Merchant SS, Malasarn D. A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 2011;66:770–780. doi: 10.1111/j.1365-313X.2011.04537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Henras AK, Chanfreau G. Multiple RNA surveillance pathways limit aberrant expression of iron uptake mRNAs and prevent iron toxicity in S. cerevisiae. Mol Cell. 2005;19:39–51. doi: 10.1016/j.molcel.2005.05.021. [DOI] [PubMed] [Google Scholar]
- Lester RL, Dickson RC. High-performance liquid chromatography analysis of molecular species of sphingolipid-related long chain bases and long chain base phosphates in Saccharomyces cerevisiae after derivatization with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate. Anal Biochem. 2001;298:283–292. doi: 10.1006/abio.2001.5368. [DOI] [PubMed] [Google Scholar]
- Li L, Chen OS, McVey Ward D, Kaplan J. CCC1 is a transporter that mediates vacuolar iron storage in yeast. J Biol Chem. 2001;276:29515–29519. doi: 10.1074/jbc.M103944200. [DOI] [PubMed] [Google Scholar]
- Liang Q, Zhou B. Copper and manganese induce yeast apoptosis via different pathways. Mol Biol Cell. 2007;18:4741–4749. doi: 10.1091/mbc.E07-05-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Li L, Jia X, Ward DM, Kaplan J. Genetic and biochemical analysis of high iron toxicity in yeast: iron toxicity is due to the accumulation of cytosolic iron and occurs under both aerobic and anaerobic conditions. J Biol Chem. 2011;286:3851–3862. doi: 10.1074/jbc.M110.190959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Zhang X, Sumanasekera C, Lester RL, Dickson RC. Signalling functions for sphingolipid long-chain bases in Saccharomyces cerevisiae. Biochem Soc Trans. 2005;33:1170–1173. doi: 10.1042/BST20051170. [DOI] [PubMed] [Google Scholar]
- Madeo F, Herker E, Maldener C, Wissing S, Lachelt S, Herlan M, Fehr M, Lauber K, Sigrist SJ, Wesselborg S, Frohlich KU. A caspase-related protease regulates apoptosis in yeast. Mol Cell. 2002;9:911–917. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- Philpott CC. Iron uptake in fungi: a system for every source. Biochim Biophys Acta. 2006;1763:636–645. doi: 10.1016/j.bbamcr.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Philpott CC, Rashford J, Yamaguchi-Iwai Y, Rouault TA, Dancis A, Klausner RD. Cell-cycle arrest and inhibition of G1 cyclin translation by iron in AFT1-1(up) yeast. EMBO J. 1998;17:5026–5036. doi: 10.1093/emboj/17.17.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants FM, Baltz AG, Trott AE, Fereres S, Thorner J. A protein kinase network regulates the function of aminophospholipid flippases. Proc Natl Acad Sci U S A. 2010;107:34–39. doi: 10.1073/pnas.0912497106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants FM, Breslow DK, Muir A, Weissman JS, Thorner J. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2011;108:19222–19227. doi: 10.1073/pnas.1116948108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants FM, Torrance PD, Bezman N, Thorner J. Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol Biol Cell. 2002;13:3005–3028. doi: 10.1091/mbc.E02-04-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakoury-Elizeh M, Protchenko O, Berger A, Cox J, Gable K, Dunn TM, Prinz WA, Bard M, Philpott CC. Metabolic response to iron deficiency in Saccharomyces cerevisiae. J Biol Chem. 2010;285:14823–14833. doi: 10.1074/jbc.M109.091710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Miao Y, Yamane Y, Zhang C, Shokat KM, Takematsu H, Kozutsumi Y, Drubin DG. Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Mol Biol Cell. 2012 doi: 10.1091/mbc.E12-03-0209. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touati D. Iron and oxidative stress in bacteria. Arch Biochem Biophys. 2000;373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- Valentine JS, Wertz DL, Lyons TJ, Liou LL, Goto JJ, Gralla EB. The dark side of dioxygen biochemistry. Curr Opin Chem Biol. 1998;2:253–262. doi: 10.1016/s1367-5931(98)80067-7. [DOI] [PubMed] [Google Scholar]
- Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- Van Ho A, Ward DM, Kaplan J. Transition metal transport in yeast. Annu Rev Microbiol. 2002;56:237–261. doi: 10.1146/annurev.micro.56.012302.160847. [DOI] [PubMed] [Google Scholar]
- Villa NY, Kupchak BR, Garitaonandia I, Smith JL, Alonso E, Alford C, Cowart LA, Hannun YA, Lyons TJ. Sphingolipids function as downstream effectors of a fungal PAQR. Mol Pharmacol. 2009;75:866–875. doi: 10.1124/mol.108.049809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.