Abstract
The identification of autism susceptibility genes has been hampered by phenotypic heterogeneity of autism, among other factors. However, the use of endophenotypes has shown preliminary success in reducing heterogeneity and identifying potential autism-related susceptibility regions. To further explore the utility of using language related endophenotypes, we performed linkage analysis on multiplex autism families stratified according to delayed expressive speech and also assessed the extent to which parental phenotype information would aid in identifying regions of linkage. A whole genome scan using a multipoint nonparametric linkage approach was performed in 133 families, stratifying the sample by phrase speech delay and word delay. None of the regions reached suggested genome-wide or replication significance thresholds. However, several loci on chromosomes 1, 2, 4, 6, 7, 8, 9, 10, 12, 15, and 19 yielded nominally higher linkage signals in the delayed groups. The results did not support reported linkage findings for loci on chromosomes 7 or 13 that were a result of stratification based on the language delay endophenotype. In addition, inclusion of information on parental history of language delay did not appreciably affect the linkage results. The nominal increase in NPL scores across several regions using language delay endophenotypes for stratification suggests that this strategy may be useful in attenuating heterogeneity. However, the inconsistencies in regions identified across studies highlight the importance of increasing sample sizes to provide adequate power to test replications in independent samples.
Keywords: Autism, linkage, endophenotypes, language, AGRE
INTRODUCTION
Autism and the related autism spectrum disorders (ASD) are complex neurodevelopmental disorders characterized by core deficits in three major domains: social interaction and social relatedness, verbal and non-verbal communication, and restricted interests and/or repetitive or stereotyped behaviors and resistance to change. The expression of the deficits encompasses a wide continuum extending from mild peculiarities to severe developmental disabilities. There is strong evidence from two major lines of investigation that the genetic contribution to ASD is substantial (Cook 2001; Folstein and Rosen-Sheidley 2001). First, indirect evidence comes from the high incidence of neurogenetic disorders and chromosomal anomalies occurring in 5–9% of autism patients (Lewis et al. 1995; Fombonne et al. 1997; Cook 2001; Wassink et al. 2001). Second, twin and family studies provide direct evidence of a genetic etiology in idiopathic autism (Folstein and Rutter 1977; Ritvo et al. 1985; Steffenburg et al. 1989; Ritvo et al. 1991; Bailey et al. 1995; Le Couteur et al. 1996).
Although heritability estimates for ASD range from 60 to 90% (Folstein and Rutter 1977; Ritvo et al. 1985) placing it among the most heritable of complex neuropsychiatric conditions; the identification of candidate loci for the disorder has been complicated by genetic and phenotypic heterogeneity. Results from the nine published whole genome scans using autism as a qualitative phenotype (IMGSAC 1998; Barrett et al. 1999; Philippe et al. 1999; Risch et al. 1999; IMGSAC 2001; Liu et al. 2001; Auranen et al. 2002; Shao et al. 2002b; Yonan et al. 2003) have been variable with the most consistent findings on chromosome 7 (Badner and Gershon 2002). The other regions of interest that have shown strong linkage signals and/or have support from multiple studies include: 2q (Philippe et al. 1999; IMGSAC 2001; Buxbaum et al. 2002; Shao et al. 2002b), 4 (IMGSAC 1998; Barrett et al. 1999; Yonan et al. 2003), 13 (Barrett et al. 1999), 17p (Risch et al. 1999; IMGSAC 2001; Liu et al. 2001; Yonan et al. 2003; Stone et al. 2004; Cantor et al. 2005) and X (Auranen et al. 2002; Shao et al. 2002b).
Investigators have begun to use endophenotypes related to autism in an attempt to reduce heterogeneity and identify factors that may relate more closely to genetic etiologies than the current broad diagnostic categories. Endophenotypes are components of a more complex phenotype, such as behavioral, cognitive, morphologic or biochemical features that may be more directly related to the underlying genetic etiologies (Gottesman and Gould 2003). Using behavioral endophenotypes such as insistence on sameness, obsessive-compulsive behavior or savant skills to stratify ASD families in linkage analysis has shown promise (Nurmi et al. 2003; Shao et al. 2003; Buxbaum et al. 2004; McCauley et al. 2004). The most significant linkage result reported by our group was based on stratifying families by the sex of the autistic proband; this revealed a locus with genome-wide significance on chromosome 17 for the families with only male autistic probands (Stone et al. 2004) and this locus was recently replicated, with genome-wide significance, in an independent sample (Cantor et al. 2005). Stratification of families by the proband’s language delay has also proved a useful linkage strategy, producing strengthened signals on chromosomes 2q (Buxbaum et al. 2001; Shao et al. 2002a), 7q (Bradford et al. 2001) and 13q (Bradford et al. 2001). In addition to stratifying families based on proband phrase speech delay, Bradford et al. (2001) also considered parents with language delay as affected and reported strengthened linkage signals to autism on chromosomes 7 and 13. In an alternative approach, use of a language endophenotype in a quantitative linkage analysis highlighted a region on chromosome 7q that was not present in a previous qualitative scan of the same data from the Autism Genetic Resource Exchange (AGRE; Alarcón et al. 2002; Liu et al. 2001).
Given these varied findings, replication is critical to assess the validity of reported linkage peaks in complex neuropsychiatric disorders. Based on the success of previous reports (Bradford et al. 2001; Buxbaum et al. 2001; Shao et al. 2002a) we stratified families from the AGRE sample by the presence of expressive speech delay and included parental information regarding a history of language delays or deficits in order to categorize parents as affected in the linkage analysis. Rather than limit our investigation to previously reported chromosomal regions with only modest evidence for linkage, we performed a whole genome scan.
MATERIALS AND METHODS
Participants
Families were obtained from the Autism Genetics Resource Exchange (AGRE) program. AGRE is a large shared database containing phenotype and genotype information of ASD families available to approved researchers (Geschwind et al. 2001). Families were ascertained based on the criteria that there were at least two siblings with a reported ASD diagnosis. Diagnoses are confirmed by the Autism Diagnosis Interview-Revised (ADI-R) (Lord et al. 1994). The present analysis used the broad-spectrum diagnosis for probands which included those with autistic disorder and those with similar but lesser deficits in communication, social relatedness and/or repetitive behaviors/restricted interests. Details on diagnostic algorithms are available on the AGRE website (www.agre.org).
Phenotypic assessment included the diagnostic testing using the ADI-R (Lord et al. 1994) and the Autism Diagnostic Observation Schedule (ADOS-G) ((Lord et al. 2000) and cognitive testing using the Raven Progressive Matrices – Coloured Version (Raven 1956) and the Peabody Picture Vocabulary Test (PPVT-III) (Dunn and Dunn 1981) which can be used as surrogate non-verbal and verbal IQ measures, respectively.
Families for this analysis were selected from the 345 genotyped families based on the availability of more extensive parental language data. MZ twins and individuals with known chromosomal abnormalities were excluded. This sample included 133 nuclear families with a total of 634 individuals, 131 fathers, 133 mothers, 89 unaffected siblings (40 males, 49 females), and 280 probands (221 males, 59 females).
Stratification
Delay in spoken language acquisition is frequently seen in autism but not required for the DSM-IV diagnosis of an autism spectrum disorder. We stratified the families based on proband phrase and word speech delay as reported on the ADI-R items: A12 or “age at first word” and A13 or “age at first phrase”. Phrase Delay (PD) families included two or more autistic or broad-spectrum probands with phrase speech acquired after 36 months of age (n=69). Word Delay (WD) families included two or more autistic or broad-spectrum probands with single words acquired after 18 months of age (n=60). We defined word delay as the acquisition of single spoken words past 18 months of age because this is the standard cut-off used in the clinical practice of neurology and is, thus, widely regarded as clinically meaningful. Probands who had not yet acquired the single words or phrases were considered delayed for purposes of this analysis.
Parental classification
Parental affectation status was based on a history of language development questionnaire adapted from those used for family studies of specific language impairment (Tomblin 1989; Tomblin et al. 1992) and dyslexia (Lefly and Pennington 2000). If the parent had a self-reported history of delayed speech and language or had difficulty learning to read, the individual was considered affected similar to the approach used by Bradford and colleagues (Bradford et al. 2001).
Genotypes
Laboratory and genotyping procedures have been previously detailed (Liu et al. 2001; Yonan et al. 2003). Briefly, 10–20 microliters of blood were collected from all available family members (parents, affected children, unaffected children) in the family home. Samples were shipped to Rutgers University Repository where immortalized lymphoblast cell lines were created and DNA was extracted for storage and distribution to approved AGRE researchers. DNA was genotyped at the Columbia University Genome Center using 365 DNA microsatellite markers. These included 335 markers used in the original genome scan (Liu et al. 2001) at an average density of 10cM, as well as an additional 30 fine map microsatellites on chromosome 7q with an average density of 2 cM. The markers had an average heterozygosity of 0.77.
Analysis
The SAS analysis software package (SAS/STAT 1990) was used to calculate descriptive statistics of the sample and to prepare input files for genetic analysis. Mendelian genotype errors were queried with PedCheck (O’Connell and Weeks 1998) and genotype error was detected in <0.01%.
A multipoint nonparametric linkage analysis was performed using the Genehunter program (Kruglyak et al. 1996) using the broad-spectrum diagnosis. Four analyses were performed: 1) all families (ALL; n=133); 2) phrase delayed (PD) families (n=69); 3) word delayed (WD) families (n=60); and 4) all families including parental information (ALL with PI; n=133). Given that none of the linkage peaks reached criteria for genome-wide significance (i.e. p = 2 × 10−5 based on Lander and Kruglyak, 1995), we reported only loci with nominal p values ≤ 0.05, as well as those corresponding results from the ALL group for comparison purposes.
RESULTS
Linkage with and without stratification
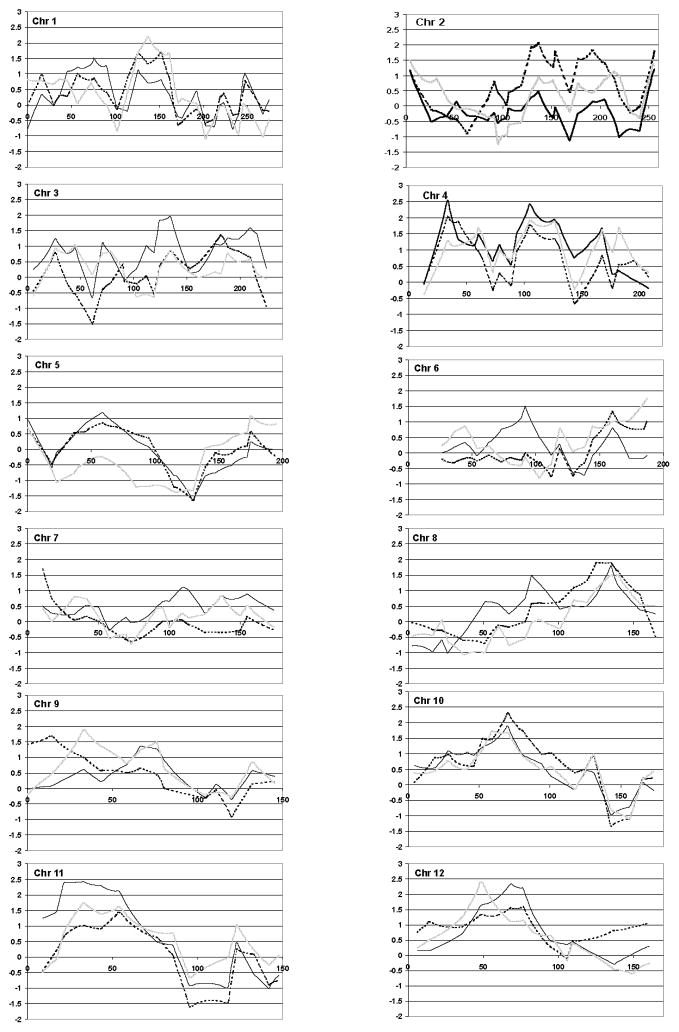
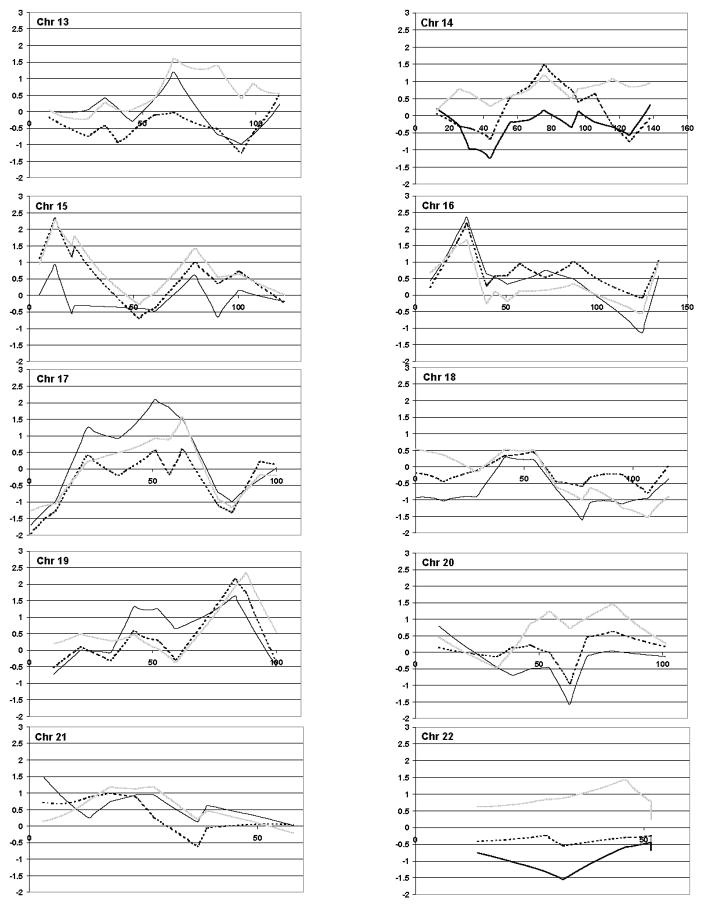
We stratified the families based on proband phrase and word speech delay to attempt to replicate previous findings from other smaller cohorts of multiplex autism pedigrees. We also performed the linkage scan in the entire cohort to use as a comparison to determine whether stratification actually increased the NPL score. None of the linkage results of the non-parametric multipoint scan (Methods) were significant at a genome-wide level for any of the cohorts (ALL, PD or WD). Moreover, the results on chr 2, 7 and 13 did not meet the suggested threshold for replication (Lander and Kruglyak, 1995). However, as shown in Table I, several peaks reached nominal significance (p<.05). Loci on chromosomes 1, 2, 4, 6, 7, 8, 9, 10, 12, 15 and 19 showed a modest increase in linkage scores either when stratified by PD or WD, and several of these (chr 1, 10, 12, 15, 19) had NPL scores greater than 2.2 yielding p values ≤.01 (Table I). Loci on chromosome 1, 15 and 19 had NPL scores with p values ≤.05 using either language-related phenotypes, PD or WD.
Table I.
Summary of peaks in stratification analysis in ALL and DELAYED families.
| chr | Region | All Families | Stratified by PD | Stratified by WD |
|---|---|---|---|---|
| cM | NPL | NPL | NPL | |
| 1 | 125.51–136.88 | 1.12 | 1.70 * | 2.2 ** |
| 2 | 136.34 | 0.48 | 2.07 * | ni |
| 2 | 153.65 | −0.039 | 1.78 * | ni |
| 2 | 191.87 1 | 0.14 | 1.82 * | ni |
| 2 | 255.7 | 1.20 | 1.81 * | ni |
| 4 | 33.42 | 2.53 ** | ni | ni |
| 4 | 104.94 | 2.42 ** | ni | ni |
| 4 | 167.55 | 1.67 * | ni | ni |
| 4 | 181.93 | 0.35 | ni | 1.69 * |
| 6 | 187.23 | −0.08 | ni | 1.76 * |
| 7 | 10.68 | 0.50 | 1.68 * | ni |
| 8 | 127.23 | 1.22 | 1.90 * | ni |
| 9 | 14.23 | 0.08 | 1.69 * | ni |
| 9 | 33.09 | 0.61 | ni | 1.89 * |
| 10 | 59.03–70.23 | 1.89 * | 2.32 ** | ni |
| 11 | 32.95 | 2.43 ** | ni | ni |
| 11 | 54.02 | 2.11 * | ni | ni |
| 12 | 48.7 | 1.64 * | ni | 2.41 ** |
| 12 | 68.16 | 2.34 ** | ni | ni |
| 15 | 12.22 | 0.94 | 2.35 ** | 2.28 ** |
| 16 | 28.3 | 2.36 ** | ni | ni |
| 17 | 50.74 | 2.10 * | ni | ni |
| 19 | 83.19–87.66 | 1.64 * | 2.16 ** | 2.34 ** |
p≤.05,
p≤.01, ni = no increase
Locus in region previously linked to language delay endophenotype in autism (Buxbaum et al. 2001; Shao et al. 2002a)
Locus in region previously linked to autism and endophenotypes regarding insistence on sameness (Shao et al. 2003) and savant skills (Nurmi et al. 2003)
Similar to previous studies, stratification by Phrase Delay did appear to strengthen the linkage signals on chromosome 2 (Buxbaum et al. 2001; Shao et al. 2002a). In our sample this was achieved at four loci, one of which is in the region (peak= 188 cM) identified by both the Buxbaum (peak=186 cM) and Shao groups (peak=198 cM). Although our results could be considered in support of Buxbaum’s chromosome 2 peak, after removal of the AGRE families that overlapped both studies (n=36 total, 16 in PD group) the peak at this locus disappeared in the Phrase Delayed group (NPL=1.03, p>.05). In contrast to a previous report (Bradford et al. 2001) stratification by phrase or word delay did not strengthen the signal compared to the entire sample on chromosomes 7 or 13. In fact, our data did not show a peak in the same regions on either of those chromosomes in any group.
Parental language information
Inclusion of parental language development information had a negligible effect on the linkage scores and, thus, these results are not presented. However, we examined the transmission of this trait in the nuclear families and observed that language-deficit information obtained retrospectively from the parents did not co-segregate with proband language delay as measured by the ADI in this cohort. The percentage of parents with language problems tended to be higher in the sets of families without proband language delays, although the differences were not significant. Specifically 13.5% of the 74 PD families were parents affected with language problems whereas 20.5% of the 214 NPD families were affected (X2=1.79 ns). A similar pattern was observed when stratifying families with word delay: 17% of the 123 WD families were affected parents versus 20% of the 165 NWD families (X2=.4 ns). Thus, it is not surprising that the inclusion of parental report information did not strengthen linkage signals.
Association with other cognitive or behavioral variables
To better characterize the subgroups and determine if there was some other feature co-segregating with the delayed groups that could contribute to the modestly strengthened linkage signals, the delayed and non delayed groups were compared on mean scores from the cognitive, language and behavioral testing (see Table IIa and b). The multiple statistical comparisons performed required an adjusted significance level of p<.0007; the resulting between-group significance values did not exceed this threshold.
Table IIa.
Test scores for PHRASE DELAYED vs NON-DELAYED groups show no significant differences when corrected for multiple comparisons.
| Mean Scores (sd) | T-score | df | p-value | ||
|---|---|---|---|---|---|
| Phrase delay | No Delay | ||||
| Raven | 93.4 (27.5) | 94.3 (25) | −.14 | 104 | .89 |
| PPVT | 75.2 (25.4) | 77.2 (30) | −.29 | 100 | .77 |
| ADI social | 20.4 (6.8) | 19.8 (7) | .67 | 282 | .5 |
| ADI comm. Non-Verbal | 14.7 (5.1) | 14.9 (4.8) | −.31 | 200 | .76 |
| ADI comm. Verbal | 11.2 (3.1) | 11.6 (3.2) | −.57 | 80 | .57 |
| ADI behav | 5.1 (2.4) | 6 (2.8) | −2.47 | 282 | .01 |
Table IIb.
Test scores for WORD DELAYED vs NON-DELAYED groups show no significant differences when corrected for multiple comparisons.
| Mean Scores (sd) | T-score | df | p-value | ||
|---|---|---|---|---|---|
| Word delay | No Delay | ||||
| Raven | 93.9 (24.2) | 94.2 (26.6) | −.050 | 104 | .96 |
| PPVT | 74 (27.2) | 78.8 (30.1) | −.831 | 100 | .41 |
| ADI social | 20.6 (6.7) | 19.4 (7.1) | 1.474 | 282 | .14 |
| ADI comm. Non-Verbal | 15.1 (5.4) | 14.7 (4.3) | .659 | 200 | .51 |
| ADI comm. Verbal | 10.8 (3.1) | 11.9 (3) | −1.597 | 80 | .11 |
| ADI behav | 5.9 (2.8) | 5.6 (2.7) | .891 | 282 | .37 |
DISCUSSION
We performed a nonparametric genome scan on a subset of 133 families from the AGRE sample with extended phenotypic information to test the approach used in previous studies to reduce heterogeneity and strengthen linkage signals by stratification based on proband language delay endophenotypes (Buxbaum et al. 2001; Shao et al. 2002a). We also incorporated information from parents of probands with reported language difficulties to determine whether this would similarly strengthen linkage signals as shown by Bradford et al. (Bradford et al. 2001).
Consistent with results from previous genome scans in independent samples, the present linkage results from the 133 AGRE families with language information identified regions on chromosomes 4 (Barrett et al., 1999), 10 (IMGSAC, 1998); 11 (Barrett et al., 1999), 16 (IMGSAC, 1998 & 2001), and 17 (IMGSAC, 2001) yielding NPL scores with p values of ≤.05.
As shown previously, stratification of families based on proband phrase or word delay did appear to strengthen linkage signals in a number of regions. However, none met suggested criteria for genome-wide significance or for replication (Lander and Kruglyak, 1995). A region on chromosome 2q was of particular interest as an autism susceptibility region for two reasons: 1) it had been previously reported by two groups (Buxbaum et al. 2001; Shao et al. 2002a) to have increased evidence for linkage in language-delayed families; and 2) there is an association between autism and a mitochondrial gene in this region (Ramoz et al. 2004). However, when the AGRE families included in the Buxbaum et al. analysis were removed from the current study, the peak in that region was diminished. This suggested that the same group of families contributed to the linkage signal on 2q in both studies. Thus, the present study does not provide additional support for linkage in the 2q region despite having a larger number of families than the Buxbaum report.
Our results from the stratified families also showed a nominal peak on chromosome 15q at 12.3cM which is within a region containing the GABRB3 gene (15q11-13, 9-23cM) that has been variably associated with autism (Cook et al. 1998; Maestrini et al. 1999; Buxbaum et al. 2002), with endophenotypes related to insistence on sameness (Shao et al. 2003) and savant skills (Nurmi et al. 2003), but not with language delay.
We did not observe any strengthening of the signal in the region of chromosome 7q identified by Bradford et al. (Bradford et al. 2001). Our group has demonstrated suggestive linkage to this region on chromosome 7q in AGRE using a quantitative trait locus (QTL) approach based on age at first word (Alarcón et al. 2002). Further analysis of this region has demonstrated that the linkage peak may not be related to the magnitude of language delay per se, but rather to a more general language-related susceptibility trait (Alarcón et al. 2005). Thus, not finding an effect of stratification based on delay in the AGRE sample would not be surprising. However, we did find a region at the telomere on chromosome 7 in the phrase delayed families as well as very modest evidence (p<.05) for linkage on chromosomes 1, 2, 4, 6, 8, 9, 10, 12, and 19 in areas that have not previously been reported as linked to autism and will need to be further studied in an independent sample.
We also examined the utility of incorporating historical information regarding parental language difficulties into the linkage analysis. Bradford et al. (Bradford et al. 2001) hypothesized that extending the specific endophenotype of language delay to other family members should increase the signal at any locus related to that phenotype. Expecting that the majority of the increased signal would come from families who also had probands with speech delay, they also stratified on phrase speech delay in a manner similar to our analysis. Their results on chromosome 7 and 13 showed a modest effect of inclusion of parent information, made stronger by combining with stratification based on the presence of proband phrase speech delay. Results of the present study did not support those described in Bradford’s report.
The lack of support observed in this study of previous linkage peaks strengthened by stratification and/or inclusion of parental information could be explained in a number of ways. First, none of the linkage results in the previous stratification studies reached genome-wide significance and therefore may have represented spurious results based on sub-setting of the cohort into smaller groups. Similarly, although the results for the Bradford et al. study (2001) that explored the utility of including parental information in the linkage analysis was based on 50 families, they still did not reach significance for genome-wide scans (Lander and Kruglyak 1995) and may represent spurious findings. Second, the present results may reflect a lack of power due to the small sample size that was a consequence of including only the subset of the complete AGRE sample with available parental language information. Third, in this cohort, it is also possible that the language deficits in parents are not related to the specific delay endophenotype in their children, and, thus, inclusion of parental information in the present linkage analysis increased heterogeneity rather than reducing it. This is supported by the lack of parent-child clustering of language delay within families. Finally, although the questions and methods used to obtain these data were similar to those used in other studies, it is possible that the self-report language history information we obtained from parents is not reliable.
This study provides an example of the difficulty phenotypic heterogeneity poses for the identification of autism susceptibility genes. Furthermore, the fact that different studies all using similar language-related endophenotypes yield linkage to different loci implies that even the language delay component of ASD could be genetically heterogeneous. For instance, a child could present with speech delay secondary to a true expressive language deficit (which may also be accompanied by a receptive language deficit), or due to a more specific motor speech disorder (e.g. speech apraxia). These different underlying pathologies could have distinct underlying genetic etiologies leading to the identification of a variety of loci in linkage analyses. Alternatively, this trait could co-segregate with another trait (so far undetected) that truly underlies linkage to the chromosomal regions identified. Better definition of the language endophenotype and perhaps further subgrouping of much larger samples into more homogeneous groups based on related aspects of the phenotype (e.g. receptive and expressive speech delay or speech apraxia) will be necessary to further explore this concept.
Figure 1.
NPL plots of each chromosome for ALL families (solid lines) and those stratified by phrase delay (dark dotted lines) and word delay (light dotted lines).
Acknowledgments
This work was supported by grants from the M.I.N.D. Institute to SJS and MA, and from the NARSAD to MA. We gratefully acknowledge the resources provided by the Autism Genetic Resource Exchange Consortium* and the participating AGRE families. The Autism Genetic Resource Exchange is a program of Cure Autism Now and is supported, in part, by grant MH64547 from the National Institute of Mental Health to Daniel H. Geschwind (PI).
*The AGRE Consortium:
Dan Geschwind, M.D., Ph.D., UCLA, Los Angeles, CA; Maja Bucan, Ph.D., University of Pennsylvania, Philadelphia, PA; W. Ted Brown, M.D., Ph.D., F.A.C.M.G., N.Y.S. Institute for Basic Research in Developmental Disabilities, Long Island, NY; Joseph Buxbaum, Ph.D., Mt. Sinai School of Medicine, NY, NY; Rita M. Cantor, Ph.D., UCLA School of Medicine, Los Angeles, CA; John N. Constantino, M.D., Washington University School of Medicine, St. Louis, MO; T. Conrad Gilliam, Ph.D., University of Chicago, Chicago, IL; Clara Lajonchere, Ph.D, Cure Autism Now, Los Angeles, CA; David H. Ledbetter, Ph.D., Emory University, Atlanta, GA; Christa Lese-Martin, Ph.D., Emory University, Atlanta, GA; Janet Miller, J.D., Ph.D., Cure Autism Now, Los Angeles, CA; Stanley F. Nelson, M.D., UCLA School of Medicine, Los Angeles, CA; Gerard D. Schellenberg, Ph.D., University of Washington, Seattle, WA; Carol A. Samango-Sprouse, Ed.D., George Washington University, Washington, D.C.; Sarah J. Spence, M.D., Ph.D., UCLA, Los Angeles, CA; Matthew State, M.D., Ph.D., Yale University, New Haven, CT.; Rudolph E. Tanzi, Ph.D., Massachusetts General Hospital, Boston, MA.
References
- Alarcón M, Cantor RM, Liu J, Gilliam TC, Geschwind DH. Evidence for a language quantitative trait locus on chromosome 7q in multiplex autism families. Am J Hum Genet. 2002;70:60–71. doi: 10.1086/338241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón M, Yonan AL, Gilliam TC, Cantor RM, Geschwind DH. Quantitative genome scan and Ordered-Subsets Analysis of autism endophenotypes support language QTLs. Mol Psychiatry. 2005 doi: 10.1038/sj.mp.4001666. [DOI] [PubMed] [Google Scholar]
- Auranen M, Vanhala R, Varilo T, Ayers K, Kempas E, Ylisaukko-Oja T, Sinsheimer JS, Peltonen L, Jarvela I. A genomewide screen for autism-spectrum disorders: evidence for a major susceptibility locus on chromosome 3q25-27. Am J Hum Genet. 2002;71:777–790. doi: 10.1086/342720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badner JA, Gershon ES. Regional meta-analysis of published data supports linkage of autism with markers on chromosome 7. Mol Psychiatry. 2002;7:56–66. doi: 10.1038/sj.mp.4000922. [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- Barrett S, Beck JC, Bernier R, Bisson E, Braun TA, Casavant TL, Childress D, et al. An autosomal genomic screen for autism. Collaborative linkage study of autism. Am J Med Genet. 1999;88:609–615. doi: 10.1002/(sici)1096-8628(19991215)88:6<609::aid-ajmg7>3.3.co;2-c. [DOI] [PubMed] [Google Scholar]
- Bradford Y, Haines J, Hutcheson H, Gardiner M, Braun T, Sheffield V, Cassavant T, Huang W, Wang K, Vieland V, Folstein S, Santangelo S, Piven J. Incorporating language phenotypes strengthens evidence of linkage to autism. Am J Med Genet. 2001;105:539–547. [PubMed] [Google Scholar]
- Buxbaum JD, Silverman J, Keddache M, Smith CJ, Hollander E, Ramoz N, Reichert JG. Linkage analysis for autism in a subset families with obsessive-compulsive behaviors: evidence for an autism susceptibility gene on chromosome 1 and further support for susceptibility genes on chromosome 6 and 19. Mol Psychiatry. 2004;9:144–150. doi: 10.1038/sj.mp.4001465. [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Silverman JM, Smith CJ, Greenberg DA, Kilifarski M, Reichert J, Cook EH, Jr, Fang Y, Song CY, Vitale R. Association between a GABRB3 polymorphism and autism. Mol Psychiatry. 2002;7:311–316. doi: 10.1038/sj.mp.4001011. [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Silverman JM, Smith CJ, Kilifarski M, Reichert J, Hollander E, Lawlor BA, Fitzgerald M, Greenberg DA, Davis KL. Evidence for a susceptibility gene for autism on chromosome 2 and for genetic heterogeneity. Am J Hum Genet. 2001;68:1514–1520. doi: 10.1086/320588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor RM, Kono N, Duvall JA, Alvarez-Retuerto A, Stone JL, Alarcón M, Nelson SF, Geschwind DH. Replication of Autism Linkage: Fine-Mapping Peak at 17q21. Am J Hum Genet. 2005:76. doi: 10.1086/430278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EH., Jr Genetics of autism. Child Adolesc Psychiatr Clin N Am. 2001;10:333–350. [PubMed] [Google Scholar]
- Cook EH, Jr, Courchesne RY, Cox NJ, Lord C, Gonen D, Guter SJ, Lincoln A, Nix K, Haas R, Leventhal BL, Courchesne E. Linkage-disequilibrium mapping of autistic disorder, with 15q11-13 markers. Am J Hum Genet. 1998;62:1077–1083. doi: 10.1086/301832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn LM, Dunn LM. Peabody Picture Vocabulary Test - Revised Manual. American Guidance Service; CIrcle Pines, MN: 1981. [Google Scholar]
- Folstein S, Rutter M. Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry. 1977;18:297–321. doi: 10.1111/j.1469-7610.1977.tb00443.x. [DOI] [PubMed] [Google Scholar]
- Folstein SE, Rosen-Sheidley B. Genetics of autism: complex aetiology for a heterogeneous disorder. Nat Rev Genet. 2001;2:943–955. doi: 10.1038/35103559. [DOI] [PubMed] [Google Scholar]
- Fombonne E, Du Mazaubrun C, Cans C, Grandjean H. Autism and associated medical disorders in a French epidemiological survey. J Am Acad Child Adolesc Psychiatry. 1997;36:1561–1569. doi: 10.1016/S0890-8567(09)66566-7. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Sowinski J, Lord C, Iversen P, Shestack J, Jones P, Ducat L, Spence SJ. The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet. 2001;69:463–466. doi: 10.1086/321292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- IMGSAC . A full genome screen for autism with evidence for linkage to a region on chromosome 7q. International Molecular Genetic Study of Autism Consortium. Hum Mol Genet. 1998;7:571–578. doi: 10.1093/hmg/7.3.571. [DOI] [PubMed] [Google Scholar]
- IMGSAC . Further characterization of the autism susceptibility locus AUTS1 on chromosome 7q. Hum Mol Genet. 2001;10:973–982. doi: 10.1093/hmg/10.9.973. [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results [see comments] Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Le Couteur A, Bailey A, Goode S, Pickles A, Robertson S, Gottesman I, Rutter M. A broader phenotype of autism: the clinical spectrum in twins. J Child Psychol Psychiatry. 1996;37:785–801. doi: 10.1111/j.1469-7610.1996.tb01475.x. [DOI] [PubMed] [Google Scholar]
- Lefly DL, Pennington BF. Reliability and validity of the Adult Reading History Questionnaire. Journal of Learning Disabilities. 2000;33:286–296. doi: 10.1177/002221940003300306. [DOI] [PubMed] [Google Scholar]
- Lewis KE, Lubetsky MJ, Wenger SL, Steele MW. Chromosomal abnormalities in a psychiatric population. Am J Med Genet. 1995;60:53–54. doi: 10.1002/ajmg.1320600110. [DOI] [PubMed] [Google Scholar]
- Liu J, Nyholt DR, Magnussen P, Parano E, Pavone P, Geschwind D, Lord C, Iversen P, Hoh J, Ott J, Gilliam TC. A genomewide screen for autism susceptibility loci. Am J Hum Genet. 2001;69:327–340. doi: 10.1086/321980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Risi S, Lambrecht L, Cook EH, Jr, Leventhal BL, DiLavore PC, Pickles A, Rutter M. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord. 2000;30:205–223. [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- Maestrini E, Lai C, Marlow A, Matthews N, Wallace S, Bailey A, Cook EH, Weeks DE, Monaco AP. Serotonin transporter (5-HTT) and gamma-aminobutyric acid receptor subunit beta3 (GABRB3) gene polymorphisms are not associated with autism in the IMGSA families. The International Molecular Genetic Study of Autism Consortium. Am J Med Genet. 1999;88:492–496. doi: 10.1002/(sici)1096-8628(19991015)88:5<492::aid-ajmg11>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- McCauley JL, Olson LM, Dowd M, Amin T, Steele A, Blakely RD, Folstein SE, Haines JL, Sutcliffe JS. Linkage and association analysis at the serotonin transporter (SLC6A4) locus in a rigid-compulsive subset of autism. Am J Med Genet. 2004;127B:104–112. doi: 10.1002/ajmg.b.20151. [DOI] [PubMed] [Google Scholar]
- Nurmi EL, Dowd M, Tadevosyan-Leyfer O, Haines JL, Folstein SE, Sutcliffe JS. Exploratory subsetting of autism families based on savant skills improves evidence of genetic linkage to 15q11-q13. J Am Acad Child Adolesc Psychiatry. 2003;42:856–863. doi: 10.1097/01.CHI.0000046868.56865.0F. [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, Van Maldergem L, Penet C, Feingold J, Brice A, Leboyer M, van Malldergerme L. Genome-wide scan for autism susceptibility genes. Paris Autism Research International Sibpair Study [published erratum appears in Hum Mol Genet 1999 Jul;8(7):1353] Hum Mol Genet. 1999;8:805–812. doi: 10.1093/hmg/8.5.805. [DOI] [PubMed] [Google Scholar]
- Ramoz N, Reichert JG, Smith CJ, Silverman JM, Bespalova IN, Davis KL, Buxbaum JD. Linkage and association of the mitochondrial aspartate/glutamate carrier SLC25A12 gene with autism. Am J Psychiatry. 2004;161:662–669. doi: 10.1176/appi.ajp.161.4.662. [DOI] [PubMed] [Google Scholar]
- Raven JC. Coloured progressive matrices. Western Psychological Services; Los Angeles, CA: 1956. [Google Scholar]
- Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, et al. A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet. 1999;65:493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritvo ER, Freeman BJ, Mason-Brothers A, Mo A, Ritvo AM. Concordance for the syndrome of autism in 40 pairs of afflicted twins. Am J Psychiatry. 1985;142:74–77. doi: 10.1176/ajp.142.1.74. [DOI] [PubMed] [Google Scholar]
- Ritvo ER, Ritvo R, Freeman BJ. Debate and argument. Concordance for the syndrome of autism [letter; comment] J Child Psychol Psychiatry. 1991;32:1031–1034. doi: 10.1111/j.1469-7610.1991.tb01928.x. [DOI] [PubMed] [Google Scholar]
- SAS/STAT UsG. SAS Institute; Cary, NC: 1990. [Google Scholar]
- Shao Y, Cuccaro ML, Hauser ER, Raiford KL, Menold MM, Wolpert CM, Ravan SA, Elston L, Decena K, Donnelly SL, Abramson RK, Wright HH, DeLong GR, Gilbert JR, Pericak-Vance MA. Fine mapping of autistic disorder to chromosome 15q11-q13 by use of phenotypic subtypes. Am J Hum Genet. 2003;72:539–548. doi: 10.1086/367846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Raiford KL, Wolpert CM, Cope HA, Ravan SA, Ashley-Koch AA, Abramson RK, Wright HH, DeLong RG, Gilbert JR, Cuccaro ML, Pericak-Vance MA. Phenotypic homogeneity provides increased support for linkage on chromosome 2 in autistic disorder. Am J Hum Genet. 2002a;70:1058–1061. doi: 10.1086/339765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Wolpert CM, Raiford KL, Menold MM, Donnelly SL, Ravan SA, Bass MP, McClain C, von Wendt L, Vance JM, Abramson RH, Wright HH, Ashley-Koch A, Gilbert JR, DeLong RG, Cuccaro ML, Pericak-Vance MA. Genomic screen and follow-up analysis for autistic disorder. Am J Med Genet. 2002b;114:99–105. doi: 10.1002/ajmg.10153. [DOI] [PubMed] [Google Scholar]
- Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, Jakobsson G, Bohman M. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry. 1989;30:405–416. doi: 10.1111/j.1469-7610.1989.tb00254.x. [DOI] [PubMed] [Google Scholar]
- Stone JL, Merriman B, Cantor RM, Yonan AL, Gilliam TC, Geschwind DH, Nelson SF. Evidence for sex-specific risk alleles in autism spectrum disorder. Am J Hum Genet. 2004;75:1117–1123. doi: 10.1086/426034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomblin JB. Familial concentration of developmental language impairment. J Speech Hear Disord. 1989;54:287–295. doi: 10.1044/jshd.5402.287. [DOI] [PubMed] [Google Scholar]
- Tomblin JB, Freese PR, Records NL. Diagnosing specific language impairment in adults for the purpose of pedigree analysis. J Speech Hear Res. 1992;35:832–843. doi: 10.1044/jshr.3504.832. [DOI] [PubMed] [Google Scholar]
- Wassink TH, Piven J, Patil SR. Chromosomal abnormalities in a clinic sample of individuals with autistic disorder. Psychiatr Genet. 2001;11:57–63. doi: 10.1097/00041444-200106000-00001. [DOI] [PubMed] [Google Scholar]
- Yonan AL, Alarcón M, Cheng R, Magnusson PK, Spence SJ, Palmer AA, Grunn A, Juo SH, Terwilliger JD, Liu J, Cantor RM, Geschwind DH, Gilliam TC. A genomewide screen of 345 families for autism-susceptibility loci. Am J Hum Genet. 2003;73:886–897. doi: 10.1086/378778. [DOI] [PMC free article] [PubMed] [Google Scholar]