SUMMARY
Eukaryotic transcription factors (TFs) perform complex and combinatorial functions within transcriptional networks. Here, we present a synthetic framework for systematically constructing eukaryotic transcription functions using artificial zinc fingers, modular DNA-binding domains found within many eukaryotic TFs. Utilizing this platform, we construct a library of orthogonal synthetic transcription factors (sTFs) and use these to wire synthetic transcriptional circuits in yeast. We engineer complex functions, such as tunable output strength and transcriptional cooperativity, by rationally adjusting a decomposed set of key component properties, e.g., DNA specificity, affinity, promoter design, protein-protein interactions. We show that subtle perturbations to these properties can transform an individual sTF between distinct roles (activator, cooperative factor, inhibitory factor) within a transcriptional complex, thus drastically altering the signal processing behavior of multi-input systems. This platform provides new genetic components for synthetic biology and enables bottom-up approaches to understanding the design principles of eukaryotic transcriptional complexes and networks.
INTRODUCTION
The genetic program of a living cell is governed by the faithful execution of a number of fundamental molecular functions by transcription factors (TFs). These include wiring specific connections to promoter regulatory elements, modulating the transcriptional output of a gene, tuning molecular noise, recruiting coactivator/repressor complexes and basal transcriptional machinery, cooperating with other TFs to regulate a gene, integrating an array of environmental signals, and even physically manipulating the geometrical configuration of chromosomes (Hahn and Young, 2011; Pedraza and van Oudenaarden, 2005; Ptashne, 1986, 1988; Rosenfeld et al., 2005). A tremendous amount of progress has been made toward understanding eukaryotic transcription regulation. Yet, there is still much to be learned about how the molecular properties of TFs give rise to the complex behavior of transcriptional networks. A synthetic approach, whereby minimal and insulated components and circuitry can be constructed to recapitulate eukaryotic transcription function, would be valuable for studying how transcriptional regulatory complexes are assembled and how TFs wire together transcriptional networks.
A framework for eukaryotic transcription regulation would also be broadly valuable to synthetic biology efforts, which seek to uncover the design principles of gene regulatory networks and program novel biological functions for a range of biotechnological and industrial applications (Andrianantoandro et al., 2006; Bashor et al., 2010; Khalil and Collins, 2010; Mukherji and van Oudenaarden, 2009; Nandagopal and Elowitz, 2011; Smolke and Silver, 2011). Engineering synthetic transcriptional networks has been a major focus of the field, and a variety of circuit behaviors have been implemented, including memory, oscillations, logic operations, filtering, and noise propagation (Basu et al., 2005; Becskei and Serrano, 2000; Elowitz and Leibler, 2000; Friedland et al., 2009; Gardner et al., 2000; Guet et al., 2002; Pedraza and van Oudenaarden, 2005; Rosenfeld et al., 2005). In these and virtually all other studies of synthetic transcriptional networks, circuitry has been constructed using a handful of well-studied prokaryotic TFs; these “off-the-shelf parts” represent the extent of well-understood and reliable transcriptional components. Indeed, the synthetic construction of transcriptional networks in eukaryotes has relied heavily upon importing these same bacterial TF-promoter pairs (Lu et al., 2009; Weber and Fussenegger, 2009). This approach has advantages, as bacterial TFs are largely orthogonal to eukaryotic transcriptional machinery. Additionally, because bacterial TFs perform relatively simple molecular tasks (as compared with eukaryotic TFs), assembling and programming simple circuitry with them can be straightforward. Yet, for this reason, and because they regulate transcription in fundamentally different ways than their eukaryotic counterparts, bacterial TFs are a poor starting point for engineering many of the complex transcriptional functions enumerated above. Furthermore, bacterial TFs are severely limiting with respect to extensibility—they bind to specific target sequences and often oligomerize cooperatively when bound. Typically, these functions are integrated and coupled, making the tuning of any one property difficult. Laborious re-engineering schemes, such as directed evolution, may be required to generate an expanded set of components. As a result, the use of bacterial TFs is unlikely to scale to the more sophisticated circuitry needed for engineering transcriptional regulatory function in eukaryotic systems.
Here, we present an alternative approach to engineering transcriptional regulation in eukaryotes using synthetic transcription factors (sTFs) constructed from Cys2-His2 zinc finger (ZF) domains (Figure 1). The sTFs feature a modular design in which separate protein domains carry out individual molecular functions: ZF domains enable binding to DNA at user-specified sequences embedded within an engineered promoter, the transcriptional output for that promoter is driven by an activation domain that recruits basal transcription machinery, and a protein-protein interaction domain allows cooperative interactions with adjacent TFs. This decomposed design permits the facile tuning of individual sTF component properties. ZF domains were selected to carry out sTF DNA binding function because of their potential for engineered sequence specificity. ZFs are small (~30 amino acid) domains that bind to ~3 bps of DNA (Elrod-Erickson et al., 1998; Pavletich and Pabo, 1991). ZFs are utilized in natural transcriptional networks in virtually all eukaryotic taxa to solve the combinatorial problem of DNA recognition by binding to promoter sequences in tandem arrays (Pabo et al., 2001). Recent advances have made it possible to purposefully re-engineer the DNA-binding specificity of individual ZFs to bind to a wide variety of 3 bp sequences, and then covalently link them together into artificial, multifinger arrays capable of recognizing longer DNA sequences with a high degree of specificity (Beerli and Barbas, 2002; Maeder et al., 2008, 2009; Pabo et al., 2001; Sander et al., 2011). Notably, with oligomerized pool engineering (OPEN) (Maeder et al., 2008) and other “context-dependent” engineering methods (Sander et al., 2011), multifinger arrays with defined specificities have been generated to design ZF nucleases (ZFNs) for targeted gene and genome modification (Foley et al., 2009; Maeder et al., 2008; Sebastiano et al., 2011; Townsend et al., 2009; Zou et al., 2009).
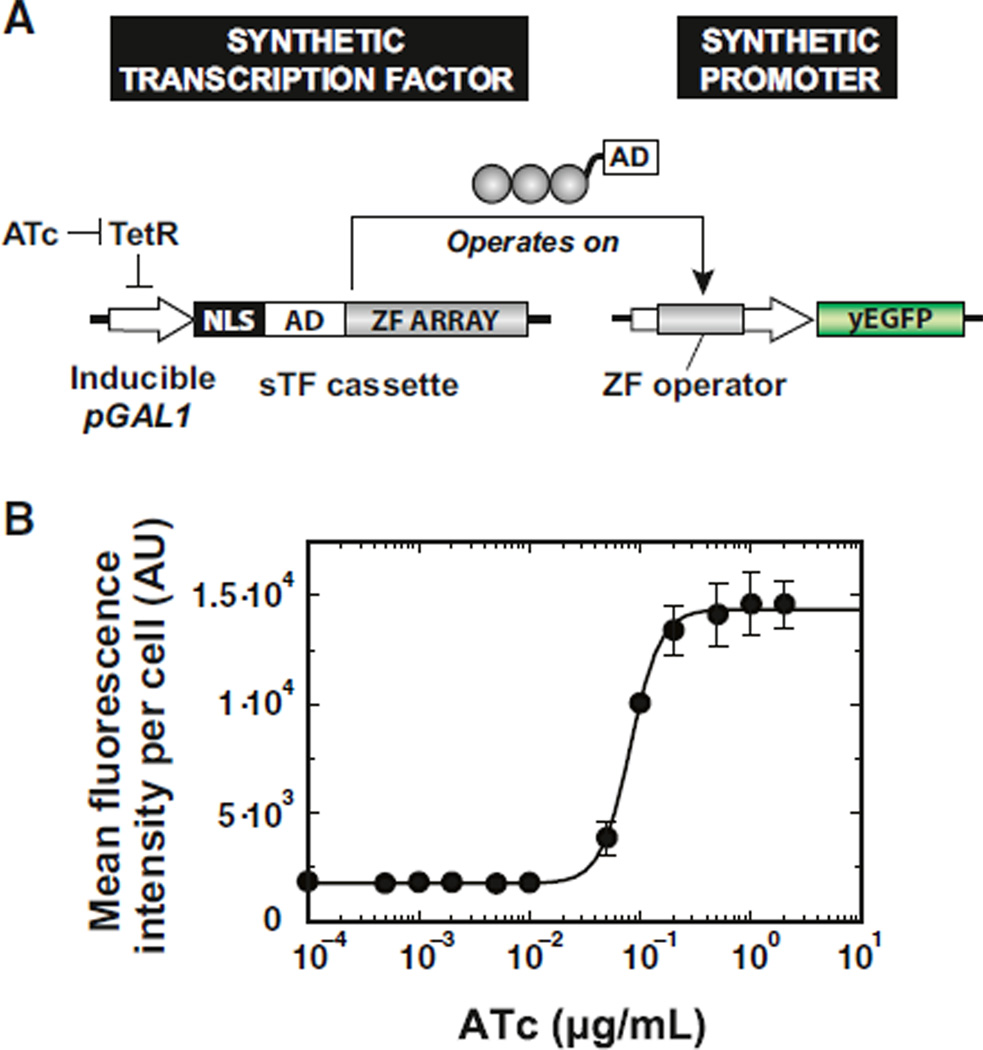
Figure 1. Synthetic Construction of Eukaryotic Transcription Functions.
Eukaryotic transcription factors (TFs) perform a variety of molecular functions to control promoters and facilitate the operation of genetic networks (top panel). Zinc fingers (ZFs) are modular domains found in many eukaryotic TFs that make sequence-specific contacts with DNA. Artificial ZF arrays were used as core building blocks for constructing synthetic TFs (sTFs) and gene circuitry in S. cerevisiae (bottom panel). The use of artificial ZF domains permits a fully decomposed design of a sTF, for which the molecular component properties are accessible, modular, and tunable (red italicized). The independent control of these component properties enables the systematic construction and modulation of transcriptional behavior. AD, transcriptional activation domain; GOI, gene of interest; REs, regulatory elements.
Using the OPEN platform, we construct a library of specific and orthogonal sTF-promoter pairs, and demonstrate that these pairs can be used to wire synthetic transcriptional cascades in Saccharomyces cerevisiae. We then use these circuits as a testbed system for exploring the relationship between circuit output and sTF function. We find that a few, key properties, e.g., DNA specificity, DNA affinity, promoter-operator design, and protein interactions, can be rationally and independently adjusted to tune transcriptional behavior. For example, we demonstrate the tuning of transcriptional output through the perturbation of ZF binding affinity and operator number. Additionally, we engineer cooperative transcriptional systems by multimerizing weakly-activating sTF monomers using modular protein-protein interaction domains. Finally, in order to synthetically explore transcriptional signal integration, we construct a set of simple two-input promoters that recruit two individual sTFs. By systematically altering the architecture of the complex through subtle changes to the component properties of the sTFs, we can assign entirely different transcriptional roles to an individual sTF and thus dramatically alter the signal processing of the system.
RESULTS
Wiring Specific and Orthogonal Transcriptional Connections with a Library of Synthetic TF-Promoter Pairs
Transcriptional networks, natural and synthetic, are wired together with sequence-specific protein-DNA interactions. We sought to program DNA-binding specificity, via artificial ZF proteins, in order to wire specific and orthogonal transcriptional connections in the eukaryote, S. cerevisiae. To do so, we first devised a platform by which ZF-based sTFs could be readily constructed and customized. The platform consists of a cassette, into which artificial three-finger arrays with engineered specificities are inserted to generate sTF species. The sTF cassette is paired with a synthetic promoter bearing ZF binding sequences that act as operators for the sTFs (Figure 2A).
Figure 2. Artificial ZFs Can Be Used to Construct Synthetic Transcriptional Activators.
(A) Circuit design for synthetic transcriptional cascade. Synthetic transcription factors (sTFs) are expressed from an ATc-inducible GAL1 promoter (pGAL1). sTF activators are composed of artificial ZF arrays fused to a herpes simplex VP16 activation domain (AD) and a nuclear localization sequence (NLS). Upon induction, the sTF operates on a cognate synthetic promoter—minimal CYC1 promoter engineered with ZF binding sequences directly upstream of the TATA box—to direct the expression of a yeast-enhanced green fluorescent protein (yEGFP) reporter. Circuits were chromosomally integrated into S. cerevisiae.
(B) sTF activator circuits built from artificial ZF arrays activate transcription from cognate synthetic promoters in a dose-dependent fashion (ZF 37-12 shown here). Points represent mean values for three experiments ± SD. See also Figure S1.
Transcriptional activation is one of the most common mechanisms for the control of gene regulation and appears to be a universally conserved process in all eukaryotes, from fungi to metazoans (Fischer et al., 1988; Ma et al., 1988; Webster et al., 1988). We utilized the principle of activation by recruitment (Ptashne, 1988; Ptashne and Gann, 1997) to test our sTFs as minimal transcriptional activators. In our design, the engineered ZF array recapitulates the TF function of binding to a specific DNA site, in this case, to its cognate 9 bp operator in a synthetic promoter. The ZF protein is fused to a VP16 minimal activation domain (AD), which autonomously facilitates recruitment of the RNA polymerase II machinery for mRNA initiation (Ptashne, 1988). This scheme provides a decoupled, modular approach to transcriptional activation, whereby TFs and the initiation machinery can be synthetically recruited in combinatorial fashion. From these components, we constructed a synthetic transcriptional cascade and used it as a test bed for rationally customizing the properties of our transcriptional components to program in vivo behaviors (Figure 2A). Within the circuit, sTF activators are first transcribed from a previously described TetR-controlled GAL1 promoter (Ellis et al., 2009; Murphy et al., 2007), which is induced by anhydrotetracycline (ATc). Addition of ATc activates flux through the circuit to produce sTF activators, which in turn activate downstream transcription from cognate synthetic promoters to produce yEGFP expression (Figure 2B; Figure S1 available online). The resulting gene regulatory transfer function, which combines the effects of the TetR expression system and the operation of sTFs on their synthetic promoters, exhibits monotonic, dose-dependent production of yEGFP (Figure 2B). These results suggest that desired synthetic transcriptional connections can be made based on the specificity of engineered ZF proteins to their target sites.
With the OPEN selection system, we have the ability to rapidly alter the ZF-DNA interaction specificity to create a large library of interaction partners (i.e., engineered ZF proteins and corresponding target sites). We used artificial ZF arrays constructed by OPEN to generate a library of sTF-promoter pairs. In particular, we identified 19 three-finger arrays with binding specificities predicted to be orthogonal to one another (we predominately chose OPEN ZF arrays that had been engineered to bind sequences in orthologous genes found in plants, insects, and metazoans) (Figure 3A). The artificial arrays and cognate binding sequences were inserted into our framework, and the resulting library of sTF-promoter pairs were tested for activation by triggering our synthetic circuits. We found that the sTFs activated yEGFP expression from cognate promoters by factors of 1.3–6.6 (compared to uninduced cells) (Figure 3B), showing that we could indeed make sequence-specific transcriptional connections with artificially designed ZF arrays. Notably, yEGFP expression levels in uninduced cells were mostly found to be similar to the basal expression levels of cells harboring only synthetic promoters (Figure S2). Thus, in general, a signal was produced only when we induced expression of an sTF in the presence of a cognate promoter.
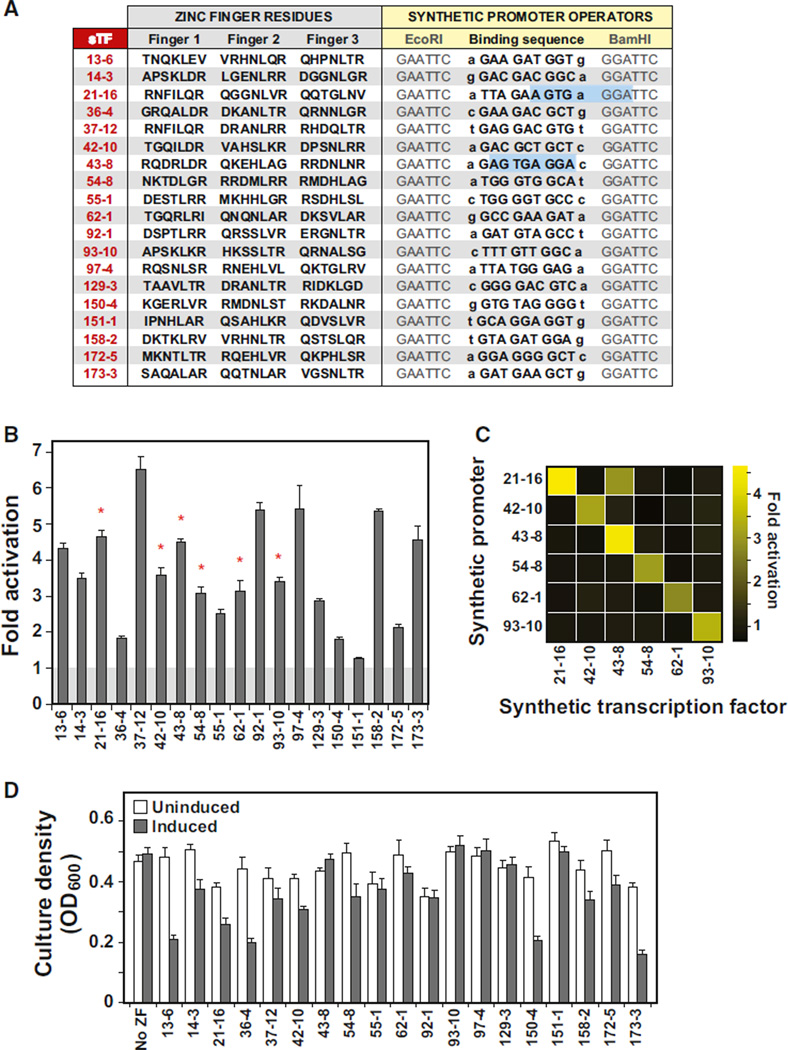
Figure 3. Wiring a Library of Specific and Orthogonal Transcriptional Connections with Engineered ZF Arrays.
(A) sTF-promoter pair library sequences. Amino acid residues of the recognition helices for 19 OPEN-engineered three-finger arrays, and corresponding DNA binding sequences (ZF binding sequences were inserted between EcoRI and BamHI sites within synthetic promoters).
(B) sTFs activate transcription from cognate synthetic promoters. “Fold activation” values were calculated as the ratio of fluorescence values from induced cells (500 ng/ml ATc) to those from uninduced cells. Red stars denote the six sTF-promoter pairs chosen to test for orthogonality.
(C) sTFs constructed from OPEN-engineered ZFs are orthogonal to one another. sTF43-8 activated noncognate Promoter21-16 due to the fortuitous creation of a sequence that is significantly similar to the binding sequence of 43-8, when the downstream BamHI restriction site is considered (A, blue boxes).
(D) Fitness cost of sTF expression on host cell growth at 30 hr after circuit induction (“no ZF” = strain with synthetic promoter and sTF cassette lacking a ZF array). Error bars represent SD of three experiments.
See also Figures S2 and S3.
We next investigated whether the transcriptional connections made within our library of sTFs were indeed specific only to their cognate synthetic promoters. We selected a subset of six sTFs from our library that exhibited robust activation (>2.5-fold) (Figure 3B, red stars), and crossed them with each of the other non-cognate promoters. Upon triggering the circuit, we observed no cross-activation in the subset of tested sTFs (Figure 3C) with one notable exception: the effect of sTF43-8on Promoter21–16. Examination of the sequence just downstream of the ZF operator for Promoter21–16 revealed the fortuitous creation of a sequence possessing significant similarity to the binding sequence of 43-8 (at 8 out of 9 bps) (Figure 3A, blue boxes). Thus, we attribute the observed cross-activation to the presence of this binding sequence within the noncognate promoter. Overall, these results show that synthetic transcriptional connections can be designed to be orthogonal to one another by using the OPEN method to engineer the DNA-binding specificities of ZF arrays.
In the design of synthetic elements and gene circuitry, a further “orthogonality” criterion is the degree to which the synthetic system interacts with pathways and machinery native to the cellular host. Ideally, insulated networks would interact with host pathways only at desired nodes and otherwise function independently. Using our synthetic yeast platform, we investigated one potential and rapid method for assessing sTF-host interactions. Specifically, we measured the growth of cells with and without the induction of sTFs, under the assumption that unwanted interactions with the host genome would impose a fitness cost on the cells. We observed no adverse or modest effects on growth in the great majority of sTFs from our library (Figure 3D and Figure S3). Our scheme may thus represent a starting point for designing and screening sTFs with optimal functionality and orthogonality within a desired host.
These results show that engineered ZF arrays are effective building blocks for minimal sTF activators, and that DNA interaction specificity is a component property that can be programmed to mediate the construction of specific and orthogonal synthetic transcriptional connections in yeast. Moreover, largely through this ability to engineer DNA specificity for many interaction partners, our platform is able to make meaningful predictions about orthogonality (among synthetic components and with host machinery), which remains a major unaddressed issue in synthetic biology.
Tuning Transcriptional Output
ZFs are well-studied structural motifs with crystallographic information providing blueprints for harnessing their structure-function relationship to program more complex transcriptional behaviors. We investigated how we could rationally engineer various component properties pertaining to the ZF-DNA interaction to tune transcriptional outputs in our synthetic eukaryotic system. For these studies, we focused on the sTF pair 42-10 and 43-8 (sTF42-10 and sTF43-8) because they activate transcription robustly to similar levels but show orthogonal activities to one another. In addition, these two activators show some distinct properties, e.g., 42-10 seemed to impose a fitness cost on the yeast host, whereas 43-8 did not (Figure 3D).
To tune up the level of transcriptional activation, we focused on alterations to the promoter architecture. We multimerized ZF binding sequences to create promoters with repeat operators that would correspondingly recruit greater numbers of sTF interactions and thus ADs. With promoters harboring one, two, and eight tandem operators, we observed a corresponding increase in the transcriptional output of the system, confirming that we could tune up the level of activation (Figure 4A). Importantly, no cross-activation was observed between these sTFs and any of the tandem synthetic promoters (Figure S4).
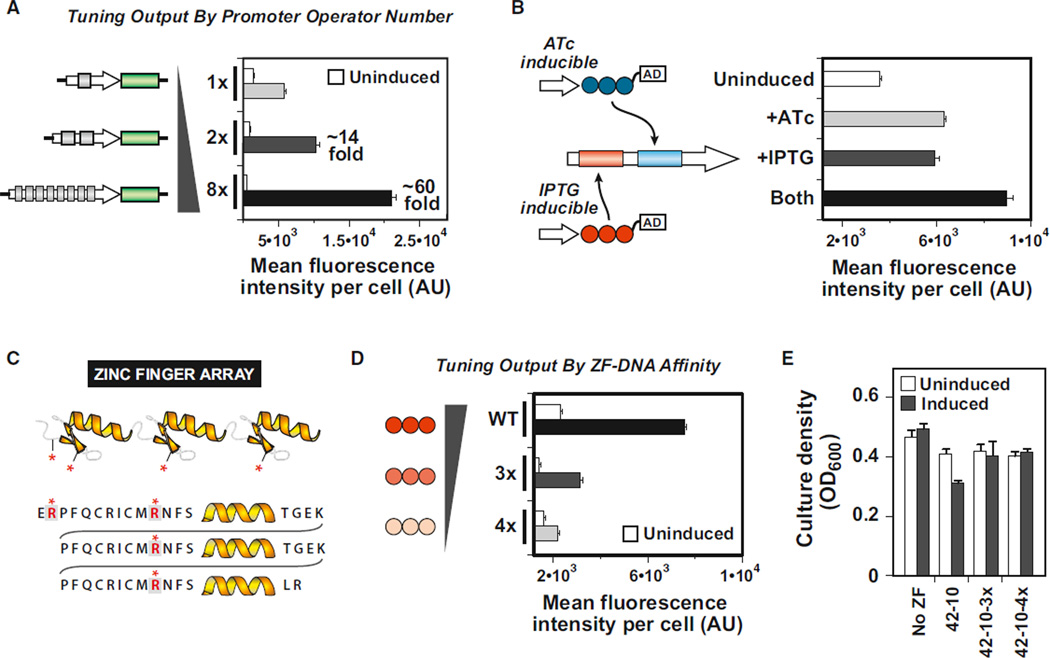
Figure 4. Tuning Transcriptional Outputs by Rationally Adjusting Multiple Component Properties.
(A) Tuning up output strength by increasing ZF operator number in synthetic promoter (sTF43-8).
(B) Integrating two distinct sTFs at a single synthetic promoter. sTF43-8 and sTF42-10 were expressed independently from ATc- and IPTG-inducible GAL1 promoters.
(C) Schematic representation of the canonical Cys2-His2 ZF protein (top). Each finger is composed of two β strands and a recognition helix, which makes sequence-specific contacts to three DNA bps. Four arginine residues in the ZF framework that mediate nonspecific interactions with the phosphate backbone were targeted for mutation to alanine residues (gray boxes and highlighted in red) in order to alter the affinity of the ZF for its cognate binding sequence.
(D) Tuning down activation output by engineering ZF affinity variants in sTF42-10 (3x: R2A/R39A/R67A, 4x: R2A/R11A/R39A/R67A). Horizontal axis begins at basal (promoter-only) fluorescence level (B and D).
(E) Phosphate backbone mutants of 42-10 rescue the fitness cost of sTF42-10 on host cell growth. Error bars represent SD of three experiments.
See also Figures S4 and S5.
Eukaryotic promoters are known to integrate multiple inputs by binding to distinct TFs. In fact, transcriptional networks may even act as logic gates through such regulation schemes. Our synthetic promoters can similarly be designed to recruit distinct sTFs through architectures that include distinct operators. We constructed a two-input promoter with operators specific for sTF43-8 and sTF42-10. We then directed the expression of sTF43-8 and sTF42-10 under the independent and respective control of TetR- and LacI-controlled GAL1 promoters, which could in turn be induced by the chemical inputs, ATc and isopropyl-β-D-1-thiogalactopyranoside (IPTG). Upon induction of either or both of the sTF species, we observed transcriptional activation over the uninduced case (Figure 4B), confirming that our promoter design can indeed integrate distinct transcriptional signals in Boolean OR-like fashion.
Promoter architecture can be designed to alter the number of sTFs recruited and thus tune transcriptional output strength. An alternative approach is to regulate the ZF-DNA interaction through structure-guided mutation of the ZF backbone to alter nonspecific DNA affinity. Along these lines, we targeted four arginine residues outside of the DNA recognition helices that are known, based on structural studies, to mediate nonspecific interactions of a three-finger array with the DNA phosphate backbone (Elrod-Erickson et al., 1996; Pavletich and Pabo, 1991). The first arginine residue (position 2) is located upstream of the first β strand of the amino-terminal finger, whereas the remaining three (positions 11, 39, 67) are found within the β sheets of each of the three fingers, immediately upstream of each recognition helix (Figure 4C). The arginine residues mediate nonspecific interactions, in part, through their positive charge; thus, we altered each of these to alanine residues.
We screened the DNA-binding activities of ZF arrays possessing various combinations of these four phosphate backbone mutations using a previously described bacterial-two-hybrid (B2H) system (Maeder et al., 2008; Wright et al., 2006). Single residue alanine substitution mutations yielded modest effects, whereas sets of mutations revealed a step-wise decrease in DNA-binding activity (Figure S5). We next incorporated the phosphate backbone mutations into sTF42-10, and tested the transcriptional activity of the resulting variants in our synthetic eukaryotic system. We found that transcriptional output could be analogously tuned down as the number of mutations was increased from zero (sTF42-10) to three (sTF42-10-3x) to four (sTF42-10-4x), in effect creating weaker-activating sTF variants from the lower-affinity variants (Figure 4D). We additionally investigated the effects the weaker-activating sTFs have on the yeast host. In the fitness growth assay, we found that the phosphate backbone mutations were able to systematically rescue the growth inhibition observed with sTF42-10 (Figure 4D and Figure S3). We presume that this effect occurs because the mutant ZF proteins are less able to mediate off-target DNA interactions that may be at the root of the fitness cost.
Taken together, these results demonstrate that the rational engineering of ZF binding sites in the promoter architecture and the ZF-DNA binding interaction—two component properties of our synthetic system—provide effective strategies for tuning transcriptional output.
Engineering Cooperative Transcriptional Systems from Weakly-Interacting Components
The assembly of TFs into multimeric complexes is a mechanism for achieving cooperativity and shaping input-output responses to regulate transcription. Inspired by natural cooperative systems, we next sought to assemble sTFs into multimeric complexes that could achieve synergistic transcriptional behaviors (Ptashne and Gann, 2002). To do so, we harnessed PDZ interaction domains from metazoan cells. These domains are naturally responsible for organizing intracellular signaling complexes, so we explored whether they could be utilized to assemble and organize our synthetic factors in transcriptional applications. Because these domains are modular, they provide an additional tunable component property to our framework and allow for generalizable designs for multimerization. Furthermore, canonical PDZ domains are extremely rare in nonmetazoans (Harris and Lim, 2001), and are therefore unlikely to interact with endogenous yeast machinery.
The weakly-activating sTFs represent ideal components with which to demonstrate multimerization and the synthetic construction of transcriptional cooperativity. These sTFs were built from ZF mutants with lower nonspecific DNA binding activity. We therefore investigated whether their assembly could stabilize a protein-DNA complex to better initiate transcription, presumably by slowing the off-rate of each component from the bound promoter. The well-studied PDZ domain from the mammalian protein α1-syntrophin (Craven and Bredt, 1998; Harris et al., 2001) was fused to the C terminus of 43-8-4x, and its cognate peptide ligand (GSGS-VKESLV), to which it binds with low micromolar affinity, was fused to the C terminus of 42-10-4x. For these studies, only sTF43-8-4x carried an AD, making it a single locus for the recruitment of transcription initiation machinery. We did not attach an AD to the dimeric partner (sTF42-10-4x) in order to test whether this additional (nonactivating) factor could stabilize the complex through dimerization and thus aid in the cooperative activation of transcription by sTF43-8-4x. In addition, we generated a nonbinding partner by fusing a mutated form of the cognate ligand (GSGS-VKEAAA) to 42-10-4x. Expression of each “monomeric” sTF was driven by independently inducible GAL1 promoters. Upon induction, the sTFs operate on a synthetic “dimeric” promoter to activate downstream transcription (Figure 5).
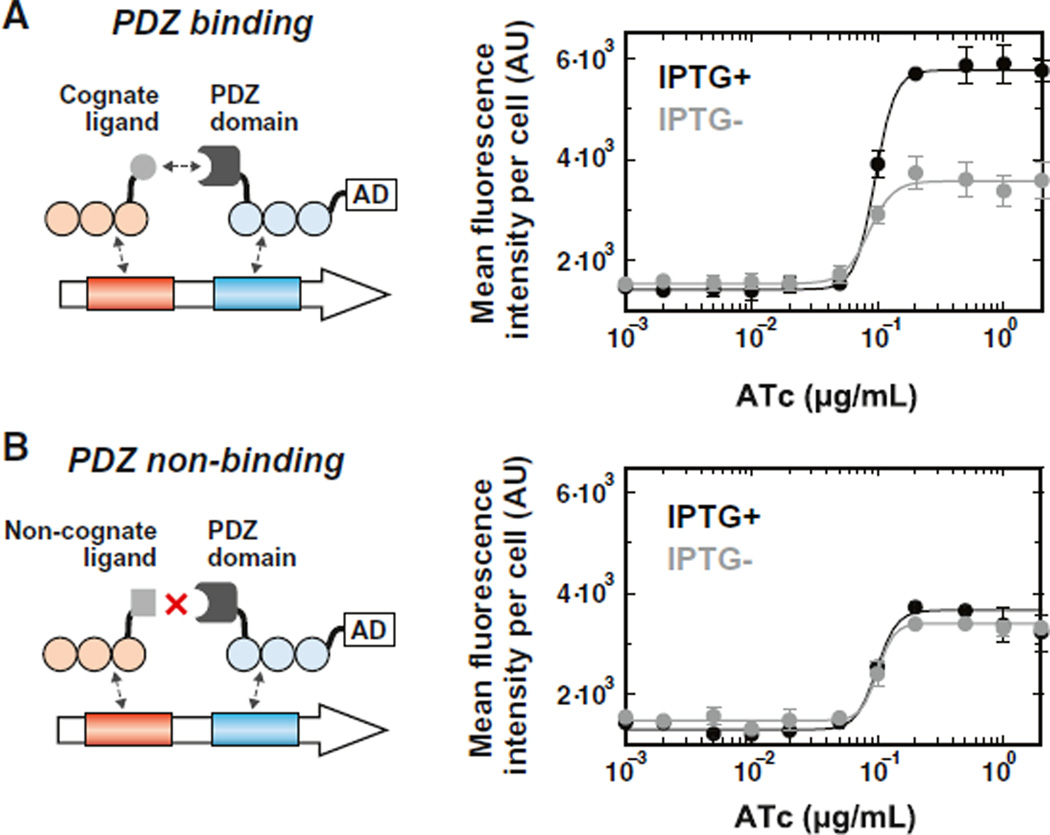
Figure 5. Transcriptional Cooperative Systems Can Be Engineered from Weakly-Activating sTF “monomers” that Are Dimerized with a PDZ Interaction Domain.
(A) The dimerization interaction promotes cooperative behavior in transcriptional activation. Syntrophin PDZ domain (dark gray) was fused to the C-terminal of ZF affinity mutant 43-8-4x, and the resulting AD-carrying sTF “monomer” was expressed from ATc-inducible pGAL1. The heterologous ligand (light gray) was fused to the C-terminal of 42-10-4x, and the resulting AD-less factor was expressed from IPTG-inducible pGAL1. The factors assemble at a synthetic “dimeric” promoter to cooperatively activate downstream transcription (“IPTG+” = full induction with 20 mM IPTG).
(B) Disruption of the dimerization interaction abolishes cooperative behavior in transcriptional activation. A nonbinding ligand variant (GSGS-VKEAAA) was instead fused to 42-10-4x. Points represent mean values for three experiments ± SD.
We titrated expression of the PDZ-harboring sTF43-8-4x, both in the presence and absence of its ligand-carrying partner. In the resulting dose responses, we observed a significant synergistic effect on the transcriptional output of sTF43-8-4x when the partner factor was present, as compared to when it was not present (Figure 5A). Critically, we observed no cooperative effect on the transcriptional output of the system in the analogous dose response with the nonbinding ligand partner (Figure 5B). These results suggest that the monomeric sTFs, aided by engineered protein-protein interactions, cooperate to recruit and stabilize one another at the synthetic promoter, thereby increasing the promoter occupancy and the resulting transcriptional activity of the complex.
Engineering Diverse Two-Input Signal Processing Behavior
With this synthetic framework, we can construct and study cooperative transcriptional assemblies. Additionally, we can explore cooperativity and other complex transcriptional behaviors within the context of signal integration, as a result of having synthetic control over combinatorial inputs and components. We thus harnessed the various control “knobs” of our framework to engineer and explore signal processing behavior for synthetic two-input promoters.
These studies were enabled by the design of dimeric promoters harboring distinct operators for ZFs 43-8 and 42-10, and the independently-controlled expression of two customizable sTF cassettes by the chemical inputs ATc and IPTG (Figure 6). Induction of a single sTF (PDZ-carrying sTF43-8) by the addition of either the input controlling its expression or both inputs resulted in robust and equal levels of transcriptional output from the two-input promoter (Figure 6A). We utilized this system to engineer a variety of transcriptional input combinations. Our previous cooperativity results (Figure 5) established an interesting starting point for investigating how a pair of transcriptional signals can be integrated. So, we first used the dimerizing sTFs, constructed from ZF mutants 43-8-4x and 42-10-4x. The sTF43-8-4x activator was directed to the operator closest to the downstream gene’s start codon (proximal position), and the AD-less partner monomer to the distal position (Figure 6, “proximal activator” architecture). When the distal monomer was engineered to carry the heterologous PDZ ligand, we observed cooperative-like amplification in transcriptional output in the presence of both inputs (Figure 6B). In this case, the distal monomer participates in binding to and stabilizing the proximal sTF activator at the promoter to enhance transcription. Interestingly, the two-component complex achieved transcriptional output levels similar to those of the single WT activator (Figure 6A), but only through the addition of both inputs and a total DNA operating specificity of 18 bps rather than 9 bps. Furthermore, we found that we could boost this effect by adding an AD onto the distal monomer, thus engineering a two-activator system and providing another source for transcriptional machinery recruitment at the promoter (Figure 6C).
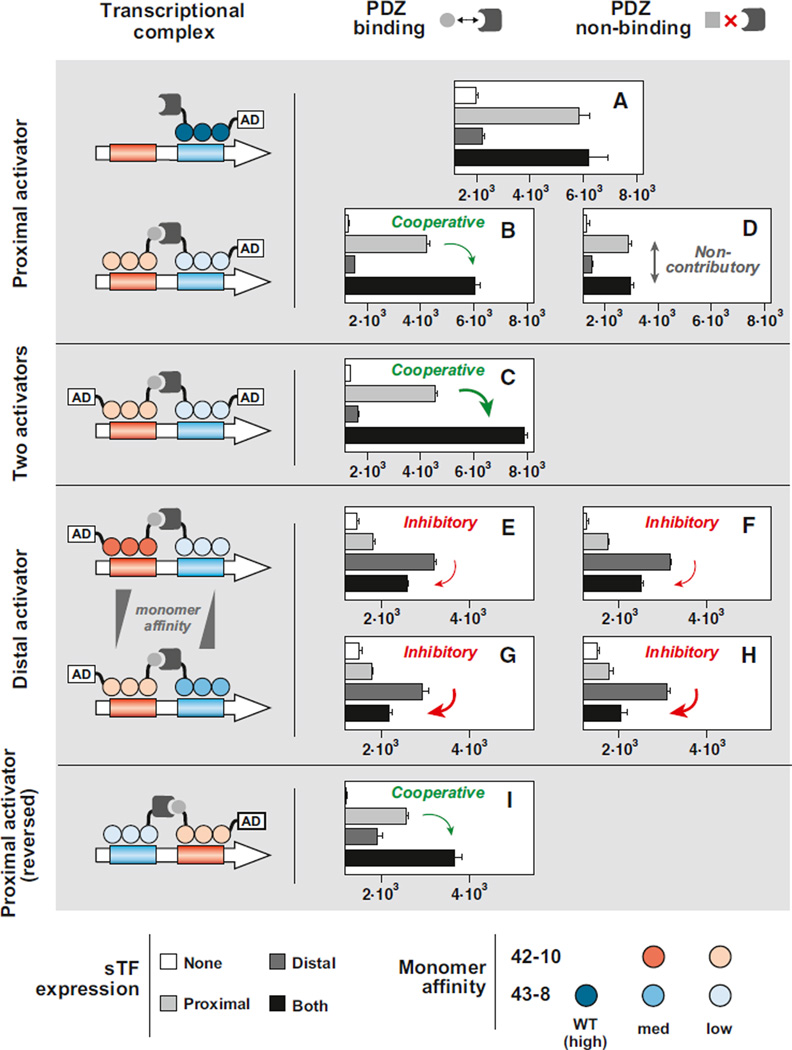
Figure 6. Synthetic ZF-Based Transcription Framework Can Be Used to Engineer Diverse Two-Input Behaviors.
(A) The transcriptional operation of a single sTF43-8 (carrying a PDZ domain) at the proximal position of a two-input promoter.
(B) Cooperative two-input synergy engineered with PDZ-carrying sTF43-8-4x as the proximal activator and cognate ligand-carrying sTF42-10-4x as the distal partner.
(C) Cooperative two-input synergy further enhanced by the addition of an AD onto the distal partner to create a two-activator system.
(D) A “null” two-input system engineered by abolishing the dimerization interaction with a PDZ nonbinding ligand on the distal partner, thus rendering it noncontributory.
(E and F) Inhibitory two-input behavior engineered by reversing the activator location (from proximal to distal) and using either PDZ binding (E) or nonbinding ligands (F).
(G and H) Inhibition by the proximal monomer can be further increased by increasing the proximal ZF affinity to DNA (43-8-4x to 43-8-3x) and decreasing the distal ZF affinity to DNA (42-10-3x to 42-10-4x) in both PDZ binding (G) and nonbinding cases (H).
(I) By reversing the orientation of the operators, sTF43-8-4x is converted from an inhibitor to a cooperative factor to, once again, obtain cooperative transcriptional synergy in the two-input behavior. All sTFs were expressed from either ATc- or IPTG-inducible pGAL1 (500 ng/ml ATc and/or 20 mM IPTG). Horizontal axes correspond to “mean fluorescence intensity per cell (AU)” and begin at basal (promoter-only) fluorescence level. Error bars represent SD of three experiments.
The PDZ-mediated sTF dimerization therefore serves as a key component property for enabling this type of synergistic two-input behavior. By simply modifying the ligand to abolish the binding interaction (i.e., mutating it to the noncognate GSGSVKEAAA), we rendered the distal monomer transcriptionally noncontributory in the proximal activator scenario, and subsequently engineered a different two-input behavior: one that shows equal output levels in the presence of either both inputs or the input directing the proximal activator (Figure 6D). In other words, we created a two-input system with a “null” effect when both inputs are present.
We next sought to reverse the monomer roles by simply switching the placement of the AD. We loaded the AD onto ligand-carrying sTF42-10-3x and removed it from PDZ-carrying sTF43-8-4x, while directing the two monomers at the same dimeric promoter (Figure 6, “distal activator” architecture). Strikingly, we did not observe transcriptional output synergy. Instead, we observed an inhibition of the output signal in the presence of both inputs (Figure 6E). Furthermore, the inhibitory behavior was conserved in both PDZ-binding and nonbinding cases (Figure 6F). These results suggest that, with this particular combination of components in the distal activator scenario, the proximal monomers take on an inhibitory as opposed to a cooperative role. If this were the case, then we reasoned that we should be able to further strengthen the inhibition by increasing the ZF affinity of the inhibitory monomer (43-8-4x) to its operator and decreasing the ZF affinity of the distal activator (42-10-3x). Indeed, when we replaced ZF mutants 43-8-4x and 42-10-3x with 43-8-3x and 42-10-4x, respectively, we observed a commensurate decrease in transcriptional output in the presence of both inputs down to near baseline levels (Figures 6G and 6H). These results further suggest that, through this slight change to the complex architecture (proximal activator to distal activator scenarios), the AD-less partner monomer has shifted its transcriptional function from cooperative to inhibitory, in effect completely transforming the logical behavior of the system.
Finally, we expected that flipping the orientation of the operators, such that the 42-10 operator was placed in the proximal position, could “rescue” the cooperative behavior (Figure 6, “proximal activator (reversed)” architecture). Indeed, with a reversed dimeric promoter, we once again observed a cooperative enhancement in the system’s output in the presence of both inputs as compared to that of the single inputs (Figure 6I). In effect, this change served to transform the transcriptional role of the 43-8-based species from inhibitor to a cooperative factor.
Taken together, our results demonstrate that sTF monomers can be customized to different roles (e.g., activating, cooperative, nonparticipatory, inhibitory) within a simple two-input system through the rational perturbation of component properties made accessible by our synthetic framework. These distinct roles can differentially shape the signal-processing behavior at a promoter. The results also highlight the importance in how a promoter’s geometry couples TF recruitment and binding to a downstream transcriptional behavior (Ptashne, 1986).
DISCUSSION
Synthetic approaches to understand, rewire, and construct complex transcriptional networks, particularly in eukaryotes, have been severely hindered by a lack of reliable components and a framework for designing and assembling them. We have developed an extensible synthetic biology framework for regulating eukaryotic transcription, whereby artificial ZF proteins are used as core building blocks from which to construct complex transcription functions and circuitry. The use of a context-dependent ZF selection scheme allows us to rapidly alter and program the ZF-DNA interaction specificity, and identify orthologous pairs of ZF arrays-DNA sites that can be engineered into sTFs for wiring networks within yeast. This work brings additional forms and levels of connectivity to synthetic transcriptional circuits, beyond that which is achievable with the few, classical prokaryotic TF-promoter pairs. Using our methodology, one should be able to create a virtually unlimited number of sTF-promoter pairs, with which to make transcriptional circuit connections. In this regard, we note that three-finger arrays have been engineered for more than 500 different nine-bp sites using the OPEN (Maeder et al., 2008) and context-dependent assembly (CoDA) (Sander et al., 2011) methods (J. Sander, M. Maeder, C. Khayter, E. Dahlborg, and J.K.J, unpublished data).
Engineered transcription activator-like (TAL) effectors have recently emerged as an important alternative to customized ZFs for programming DNA specificity. Naturally occurring TAL effectors encoded by Xanthamonas bacteria bind to target DNA sequences using arrays of highly conserved 33–35 amino acid repeat domains. A single TAL effector repeat binds to one nucleotide of DNA with specificity of binding associated with the identities of amino acids at two positions known as repeat variable di-residues (RVDs). TAL effector repeats bearing different RVDs have been described for specific binding to each of the four possible DNA nucleotides, and these repeats can be simply joined together to create arrays capable of binding to extended target DNA sequences (Bogdanove and Voytas, 2011). The simplicity and modularity of the TAL effector repeats suggests that nearly any DNA sequence should be targetable, and recent work has demonstrated that engineered TAL effector nuclease (TALEN) fusions can be robustly generated for a wide variety of different sequences (Reyon et al., 2012). Nonetheless, engineered ZFs have several important advantages, including their considerably smaller size, their less repetitive coding sequence (potentially important for packaging into viral vectors), and a greater understanding of their biochemical properties, structure, and function, which is important for creating variations in affinity, specificity, and tunability.
We constructed a synthetic transcriptional cascade in yeast that was used as a test bed for systematically tuning sTF component properties. For instance, we tuned the strength of transcriptional outputs through modifications of ZF binding sites in the promoter architecture as well as through structure-guided modifications to the ZF protein to alter ZF-DNA interactions. Additionally, our framework provides the ability to engineer and tune transcriptional cooperativity. To date, there exists no simple way of building cooperative transcriptional systems, even though their importance is well-documented in both natural and synthetic gene regulation. As a result, in most synthetic studies, researchers have used TFs with integrated, cooperative properties. In contrast, our approach establishes a modular framework for constructing cooperative transcriptional activation schemes de novo, through the multimerization of weakly-activating ZF-TFs using low-affinity protein interaction domains (i.e., PDZ domains). This has important consequences for constructing higher-order complexes that more accurately mimic eukaryotic transcription regulation schemes, lead to sharper switch-like responses, and modulate cooperativity within circuits. Indeed, multimerization and cooperativity are ubiquitous molecular regulation schemes that underlie complex gating and decision-making in cells. For example, the yeast GAL1 promoter is able to integrate coactivator proteins in specific temporal order by utilizing the cooperativity of certain interactions to gate subsequent recruitment events (Bryant and Ptashne, 2003). It is of great interest to understand how activators function cooperatively to assemble specific initiation complexes and regulate transcription. Our bottom-up and modular approach to transcriptional cooperativity could be used to synthetically recapitulate such phenomena so as to study these fundamental mechanisms of regulation. This type of approach has been used to understand transcriptional synergy in prokaryotes (Joung et al., 1993, 1994), and our platform should now enable this strategy to be used to model more combinatorially complex eukaryotic promoters. Furthermore, our extensible and modular framework for multimeric and cooperative transcriptional systems may allow for the implementation of expanded computational operations in eukaryotes, such as logic devices with more input possibilities.
Previous reports have described various frameworks for creating dimeric ZF proteins. In all of these studies, elements derived from naturally occurring TFs (Pomerantz et al., 1998; Wolfe et al., 2000) or ones selected from combinatorial peptide libraries (Wang et al., 2001; Wang and Pabo, 1999) were used to dimerize two-finger units. A disadvantage of this strategy is that a dimerized two-finger complex would have a maximum specificity of 12 bps (assuming that each of the four fingers in the dimer specifies 3 bps). Our approach differs by utilizing three-finger monomers that have had their binding activities reduced by mutagenesis of nonspecific phosphate-contacting residues. This strategy creates dimeric proteins that can have specificities as high as 18 bps, a sequence long enough to be potentially unique in a mammalian genome. Furthermore, the use of modular protein-protein interaction domains for multimerization is advantageous for various reasons. For example, the interaction is orthogonal and tunable, allowing us to “match” affinities of all the component interactions making up the sTF complex, and it provides extensibility for further expanding the complex architecture and dynamically increasing or decreasing the DNA binding specificity of the complexes.
We also showed that complex signal processing behaviors can arise when control of TF cooperativity is combined with the ability to engineer promoters of multimerized ZF binding sites. Cellular signal processing is a mechanism by which environmental and other signals are integrated to modulate transcription and thus critical cellular processes, such as growth and stress responses. We constructed a simple, synthetic two-input transcriptional system that allowed us to decompose contributions from the sTF component properties to the system’s processing behavior. We showed that, with the same two core TFs and promoter operator sites, a cell could process and integrate signals in a variety of ways. For example, subtle changes, such as reversing promoter operators and disrupting protein-protein interactions, can have striking effects on the output of the system. This led to the construction of not just varied digital logic behavior, but a range of analog tunability. In an inhibitory system (Figures 6E–6H), we arrived at an interesting Boolean logic gate that produced a positive signal only in the presence of a single input: A > B (A does not imply B). A broad observation from our studies was that specific perturbations to an sTF’s component properties (DNA affinity, multimerization with other species, location of operator, etc.) could allow it to interconvert between different transcriptional roles within the complex, such as activator, cooperative factor, noncontributory, and inhibitor. This synthetic approach could be utilized to explore the diversity of behaviors that can be programmed by even just a few transcriptional components; furthermore, our findings provide simple strategies for reprogramming the signal processing behavior of a cell. Similar strategies are undoubtedly employed naturally, where there are many examples of individual proteins that can take on either activating or repressing roles depending on the cellular and environmental states (Ma and Ptashne, 1988; Maxon and Herskowitz, 2001; Rubin-Bejerano et al., 1996).
Given that TFs containing ZFs play a central role in eukaryotic promoter regulation (Pabo et al., 2001), our system represents a promising means for synthetic recapitulation of eukaryotic promoter function, and thus will significantly enhance our ability to construct synthetic gene networks in mammalian cells, an area of tremendous potential in synthetic biology (Weber and Fussenegger, 2009). Indeed, yeast may serve as a well-characterized testbed for the design and validation of synthetic gene circuits that can be subsequently ported to higher organisms. Synthetic transcriptional regulators based on the sTFs described here can be used to create classifier circuits to identify cell state (Nissim and Bar-Ziv, 2010; Xie et al., 2011), memory devices to record cellular events, and logic gates for cellular signal processing (Kramer et al., 2004), which can aid in the study and control of stem cell differentiation, therapeutics, and complex human diseases. Additionally, ZF-based proteins have been shown to be powerful targeting elements of endogenous genomic loci in many mammalian cells, including cancer, immune, and stem cells. These proteins include ZF nucleases (Zou et al., 2009), which are being tested in therapeutic applications for modifying/ disrupting disease-causing alleles and genes, and artificial TFs (Blancafort et al., 2005), which can be used to modify the expression of native genes for reprogramming purposes. Together with these ZF technologies, our work may lead to the construction of integrated gene circuits—artificial circuitry that seamlessly and specifically integrates into endogenous gene networks and/or leads to the modification of endogenous genes—for more dynamical and sophisticated genetic control in cell-based therapeutic applications.
Synthetic biology is helping us to understand how organisms behave and develop through the forward engineering of molecular circuitry with well-understood genetic components (Elowitz and Lim, 2010). The present work expands the synthetic biology toolkit with new genetic components, beyond re-purposed bacterial transcriptional components, to program eukaryotic cells. Additionally, it provides a bottom-up framework for exploring the complexity of eukaryotic promoters and their combinatorial regulation by TF complexes and circuitry. This framework can be a starting point for determining the transcriptional components, modules, and circuitry needed to implement the sophisticated behaviors that control the development and function of eukaryotic cells.
EXPERIMENTAL PROCEDURES
Strains and Media
S. cerevisiae YPH500 (α, ura3-52, lys2-801, ade2-101, trp1Δ63, his3Δ200, leu2Δ1) (Stratagene) was used as the host strain in all yeast experiments. Culturing, genetic transformation, and verification of transformation were done as previously described (Murphy et al., 2007), using either the URA3, TRP1, or LEU2 genes as selectable markers.
Plasmid Construction
Synthetic promoter plasmids were constructed from integrative plasmid pRS406 (Stratagene) by cloning ZF binding sequences (BS) directly upstream of the CYC1 minimal promoter TATA box. The corresponding ZF-activated promoter drives the expression of a yeast enhanced green fluorescent protein (yEGFP) (Cormack et al., 1997), which is preceded by a Kozak consensus sequence.
sTF circuit plasmids were constructed from the previously described yeast integrative plasmids pTPG1 (TX: TetR-regulated control promoter) and pLOG1 (LX: LacI-regulated control promoter) (Ellis et al., 2009). Briefly, these plasmids consist of a GAL1 upstream activation signal (UAS) followed by either a TetR- or LacI-regulated wild-type GAL1 promoter, which drives the expression of our sTF cassettes. The strong constitutive TEF1 promoter also directs the expression of yeast codon-optimized versions of TetR (Tn10.B tetracycline repressor) and the Escherichia coli Lac inhibitor (LacI). Constitutive expression of the repressors ensures low basal levels of expression of our sTF cassettes from the engineered GAL1 promoter, which can be relieved by the respective addition of the chemical inputs, ATc and IPTG, to the medium.
The sTF cassette, which was synthesized (DNA2.0) and cloned as a KpnI/XhoI fragment into pTPG1 and pLOG1, consists of an open cloning site for engineered ZF arrays. Upon insertion of a ZF gene, the resulting minimal sTF becomes (N- to C-terminal): 3xFLAG – NLS – VP16 AD – ZF array. All ZF genes were codon-optimized, individually synthesized (IDT), and cloned as XbaI/BamHI fragments into the cassette. Protein-protein interaction domains, namely, syntrophin PDZ domain and peptide ligands, were added as C-terminal fusions to the sTF, separated by a GSGS linker, and cloned from synthesized, codon-optimized gene fragments (DNA2.0).
Induction Experiments
Single yeast colonies for each strain were picked and used to inoculate 500 ml of SD-Glu (synthetic drop-out media containing 2% glucose with selectable amino acid mixtures) in Costar 96-well assay blocks (V-bottom; 2 ml max volume; Fisher Scientific). The cultures were grown at 30°C with 900 rpm shaking for 24–48 hr. A triplicate set of 500 µl YEP-Gal (yeast extract peptone media containing 2% galactose) cultures, with and without inducers, were inoculated to an OD600 of ~0.08–0.1 and grown at 30°C with 900 rpm shaking for ~14–16 hr. Cells were then treated with cycloheximide to inhibit protein synthesis, and then assayed for yEGFP expression by flow cytometry.
Flow Cytometry and Data Analysis
For all data, we acquired 5,000–10,000 events using a BD LSRFortessa equipped with a High Throughput Sampler (BD Biosciences). Events were gated by forward and side scatter. The geometric means of the fluorescence distributions were calculated. The autofluorescence value of S. cerevisiae YPH500 cells harboring no genomic integrations was subtracted from these values to give the fluorescence values reported in this study. “fold activation” values were calculated as the ratio of fluorescence values from induced cells to those from uninduced cells.
Growth Assays
Growth assays were performed similarly to induction experiments, except that experimental cultures were inoculated to an OD600 of ~0.03–0.05 and grown at 30°C with 900 rpm shaking for 30 hr. OD600 measurements were taken using a SpectraMax M5 fluorescence microplate reader (Molecular Devices) using culture volumes of 100 µL. A “no ZF” control—a strain engineered with synthetic promoter and sTF cassette lacking a ZF array—was assayed in parallel.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Collins Lab for helpful discussions, Carl O. Pabo for helpful suggestions on reducing nonspecific affinity of zinc fingers, and Katie M. Flynn for help with artwork. This work was supported by the Howard Hughes Medical Institute (J.J.C.), NIH Director’s Pioneer Awards DP1 OD003644 (J.J.C.) and DP1 OD006862 (J.K.J.), an Office of Naval Research Multidisciplinary University Research Initiative (MURI) grant (T.K.L.), the National Science Foundation CCF-1124247 (T.K.L.), a Defense Advanced Research Projects Agency grant (DARPA-BAA-11-23), and a National Science Foundation Graduate Research Fellowship (C.L.R.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures and five figures and can be found with this article online at http://dx.doi.org/ 10.1016/j.cell.2012.05.045.
REFERENCES
- Andrianantoandro E, Basu S, Karig DK, Weiss R. Synthetic biology: new engineering rules for an emerging discipline. Mol. Syst. Biol. 2006;2:2006.0028. doi: 10.1038/msb4100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashor CJ, Horwitz AA, Peisajovich SG, Lim WA. Rewiring cells: synthetic biology as a tool to interrogate the organizational principles of living systems. Annu. Rev. Biophys. 2010;39:515–537. doi: 10.1146/annurev.biophys.050708.133652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- Beerli RR, Barbas CF., III Engineering polydactyl zinc-finger transcription factors. Nat. Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- Blancafort P, Chen EI, Gonzalez B, Bergquist S, Zijlstra A, Guthy D, Brachat A, Brakenhoff RH, Quigley JP, Erdmann D, Barbas CF., III Genetic reprogramming of tumor cells by zinc finger transcription factors. Proc. Natl. Acad. Sci. USA. 2005;102:11716–11721. doi: 10.1073/pnas.0501162102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Bryant GO, Ptashne M. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell. 2003;11:1301–1309. doi: 10.1016/s1097-2765(03)00144-8. [DOI] [PubMed] [Google Scholar]
- Cormack BP, Bertram G, Egerton M, Gow NA, Falkow S, Brown AJ. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- Craven SE, Bredt DS. PDZ proteins organize synaptic signaling pathways. Cell. 1998;93:495–498. doi: 10.1016/s0092-8674(00)81179-4. [DOI] [PubMed] [Google Scholar]
- Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechnol. 2009;27:465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz M, Lim WA. Build life to understand it. Nature. 2010;468:889–890. doi: 10.1038/468889a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- Elrod-Erickson M, Benson TE, Pabo CO. High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition. Structure. 1998;6:451–464. doi: 10.1016/s0969-2126(98)00047-1. [DOI] [PubMed] [Google Scholar]
- Fischer JA, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988;332:853–856. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- Foley JE, Yeh JR, Maeder ML, Reyon D, Sander JD, Peterson RT, Joung JK. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PLoS ONE. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, Lu TK, Wang X, Shi D, Church GM, Collins JJ. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Guet CC, Elowitz MB, Hsing W, Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- Hahn S, Young ET. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics. 2011;189:705–736. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- Harris BZ, Hillier BJ, Lim WA. Energetic determinants of internal motif recognition by PDZ domains. Biochemistry. 2001;40:5921–5930. doi: 10.1021/bi010142l. [DOI] [PubMed] [Google Scholar]
- Joung JK, Le LU, Hochschild A. Synergistic activation of transcription by Escherichia coli cAMP receptor protein. Proc. Natl. Acad. Sci. USA. 1993;90:3083–3087. doi: 10.1073/pnas.90.7.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Koepp DM, Hochschild A. Synergistic activation of transcription by bacteriophage lambda cI protein and E. coli cAMP receptor protein. Science. 1994;265:1863–1866. doi: 10.1126/science.8091212. [DOI] [PubMed] [Google Scholar]
- Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat. Rev. Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer BP, Fischer C, Fussenegger M. BioLogic gates enable logical transcription control in mammalian cells. Biotechnol. Bioeng. 2004;87:478–484. doi: 10.1002/bit.20142. [DOI] [PubMed] [Google Scholar]
- Lu TK, Khalil AS, Collins JJ. Next-generation synthetic gene networks. Nat. Biotechnol. 2009;27:1139–1150. doi: 10.1038/nbt.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Ptashne M. Converting a eukaryotic transcriptional inhibitor into an activator. Cell. 1988;55:443–446. doi: 10.1016/0092-8674(88)90030-x. [DOI] [PubMed] [Google Scholar]
- Ma J, Przibilla E, Hu J, Bogorad L, Ptashne M. Yeast activators stimulate plant gene expression. Nature. 1988;334:631–633. doi: 10.1038/334631a0. [DOI] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Sander JD, Voytas DF, Joung JK. Oligomerized pool engineering (OPEN): an ‘open-source’ protocol for making customized zinc-finger arrays. Nat. Protoc. 2009;4:1471–1501. doi: 10.1038/nprot.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxon ME, Herskowitz I. Ash1p is a site-specific DNA-binding protein that actively represses transcription. Proc. Natl. Acad. Sci. USA. 2001;98:1495–1500. doi: 10.1073/pnas.98.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji S, van Oudenaarden A. Synthetic biology: understanding biological design from synthetic circuits. Nat. Rev. Genet. 2009;10:859–871. doi: 10.1038/nrg2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KF, Balázsi G, Collins JJ. Combinatorial promoter design for engineering noisy gene expression. Proc. Natl. Acad. Sci. USA. 2007;104:12726–12731. doi: 10.1073/pnas.0608451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandagopal N, Elowitz MB. Synthetic biology: integrated gene circuits. Science. 2011;333:1244–1248. doi: 10.1126/science.1207084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim L, Bar-Ziv RH. A tunable dual-promoter integrator for targeting of cancer cells. Mol. Syst. Biol. 2010;6:444. doi: 10.1038/msb.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Pedraza JM, van Oudenaarden A. Noise propagation in gene networks. Science. 2005;307:1965–1969. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- Pomerantz JL, Wolfe SA, Pabo CO. Structure-based design of a dimeric zinc finger protein. Biochemistry. 1998;37:965–970. doi: 10.1021/bi972464o. [DOI] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986;322:697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Genes & signals. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional repressor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol. Cell. Biol. 1996;16:2518–2526. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiano V, Maeder ML, Angstman JF, Haddad B, Khayter C, Yeo DT, Goodwin MJ, Hawkins JS, Ramirez CL, Batista LF, et al. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells. 2011;29:1717–1726. doi: 10.1002/stem.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolke CD, Silver PA. Informing biological design by integration of systems and synthetic biology. Cell. 2011;144:855–859. doi: 10.1016/j.cell.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BS, Pabo CO. Dimerization of zinc fingers mediated by peptides evolved in vitro from random sequences. Proc. Natl. Acad. Sci. USA. 1999;96:9568–9573. doi: 10.1073/pnas.96.17.9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BS, Grant RA, Pabo CO. Selected peptide extension contacts hydrophobic patch on neighboring zinc finger and mediates dimerization on DNA. Nat. Struct. Biol. 2001;8:589–593. doi: 10.1038/89617. [DOI] [PubMed] [Google Scholar]
- Weber W, Fussenegger M. Engineering of synthetic mammalian gene networks. Chem. Biol. 2009;16:287–297. doi: 10.1016/j.chembiol.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Webster N, Jin JR, Green S, Hollis M, Chambon P. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL4 trans-activator. Cell. 1988;52:169–178. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]
- Wolfe SA, Ramm EI, Pabo CO. Combining structure-based design with phage display to create new Cys(2)His(2) zinc finger dimers. Structure. 2000;8:739–750. doi: 10.1016/s0969-2126(00)00161-1. [DOI] [PubMed] [Google Scholar]
- Wright DA, Thibodeau-Beganny S, Sander JD, Winfrey RJ, Hirsh AS, Eichtinger M, Fu F, Porteus MH, Dobbs D, Voytas DF, Joung JK. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat. Protoc. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.