Abstract
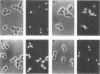
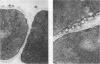
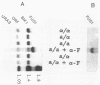
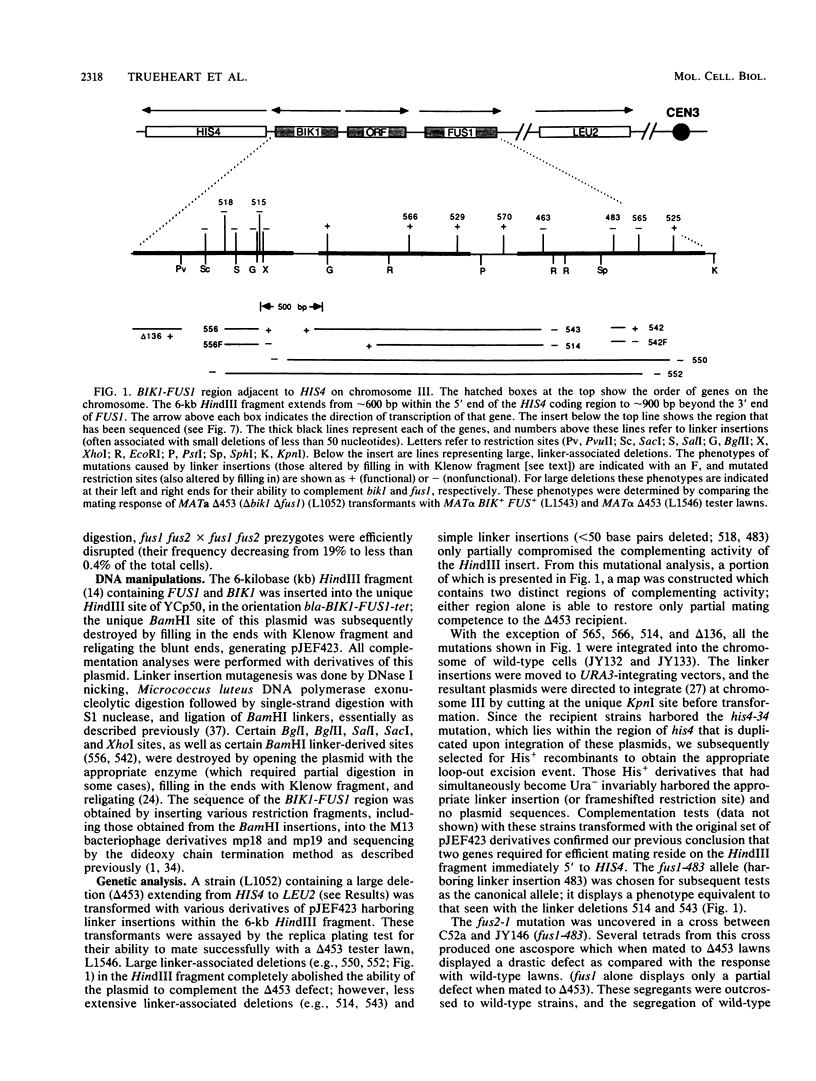
We characterized two genes, FUS1 and FUS2, which are required for fusion of Saccharomyces cerevisiae cells during conjugation. Mutations in these genes lead to an interruption of the mating process at a point just before cytoplasmic fusion; the partition dividing the mating pair remains undissolved several hours after the cells have initially formed a stable "prezygote." Fusion is only moderately impaired when the two parents together harbor one or two mutant fus genes, and it is severely compromised only when three or all four fus genes are inactivated. Cloning of FUS1 and FUS2 revealed that they share some functional homology; FUS1 on a high-copy number plasmid can partially suppress a fus2 mutant, and vice versa. FUS1 remains essentially unexpressed in vegetative cells, but is strongly induced by incubation of haploid cells with the appropriate mating pheromone. Immunofluorescence microscopy of alpha factor-induced a cells harboring a fus1-LACZ fusion showed the fusion protein to be localized at the cell surface, concentrated at one end of the cell (the shmoo tip). FUS1 maps near HIS4, and the intervening region (including BIK1, a gene required for nuclear fusion) was sequenced along with FUS1. The sequence of FUS1 revealed the presence of three copies of a hexamer (TGAAAC) conserved in the 5' noncoding regions of other pheromone-inducible genes. The deduced FUS1 protein sequence exhibits a striking concentration of serines and threonines at the amino terminus (46%; 33 of 71), followed by a 25-amino acid hydrophobic stretch and a predominantly hydrophilic carboxy terminus, which contains several potential N-glycosylation sites (Asn-X-Ser/Thr). This sequence suggests that FUS1 encodes a membrane-anchored glycoprotein with both N- and O-linked sugars.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresch C., Müller G., Egel R. Genes involved in meiosis and sporulation of a yeast. Mol Gen Genet. 1968;102(4):301–306. doi: 10.1007/BF00433721. [DOI] [PubMed] [Google Scholar]
- Bulawa C. E., Slater M., Cabib E., Au-Young J., Sburlati A., Adair W. L., Jr, Robbins P. W. The S. cerevisiae structural gene for chitin synthase is not required for chitin synthesis in vivo. Cell. 1986 Jul 18;46(2):213–225. doi: 10.1016/0092-8674(86)90738-5. [DOI] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975 Oct;124(1):511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Conde J., Fink G. R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran B. P., Carter B. L. Alpha-factor enhancement of hybrid formation by protoplast fusion in Saccharomyces cerevisiae II. Curr Genet. 1986;10(12):943–945. doi: 10.1007/BF00398292. [DOI] [PubMed] [Google Scholar]
- Davis C. G., Elhammer A., Russell D. W., Schneider W. J., Kornfeld S., Brown M. S., Goldstein J. L. Deletion of clustered O-linked carbohydrates does not impair function of low density lipoprotein receptor in transfected fibroblasts. J Biol Chem. 1986 Feb 25;261(6):2828–2838. [PubMed] [Google Scholar]
- DeFeo-Jones D., Scolnick E. M., Koller R., Dhar R. ras-Related gene sequences identified and isolated from Saccharomyces cerevisiae. Nature. 1983 Dec 15;306(5944):707–709. doi: 10.1038/306707a0. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. The nucleotide sequence of the HIS4 region of yeast. Gene. 1982 Apr;18(1):47–59. doi: 10.1016/0378-1119(82)90055-5. [DOI] [PubMed] [Google Scholar]
- Duntze W., MacKay V., Manney T. R. Saccharomyces cerevisiae: a diffusible sex factor. Science. 1970 Jun 19;168(3938):1472–1473. doi: 10.1126/science.168.3938.1472. [DOI] [PubMed] [Google Scholar]
- Duntze W., Stötzler D., Bücking-Throm E., Kalbitzer S. Purification and partial characterization of -factor, a mating-type specific inhibitor of cell reproduction from Saccharomyces cerevisiae. Eur J Biochem. 1973 Jun;35(2):357–365. doi: 10.1111/j.1432-1033.1973.tb02847.x. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Fink G. R. Insertion of the eukaryotic transposable element Ty1 creates a 5-base pair duplication. Nature. 1980 Jul 24;286(5771):352–356. doi: 10.1038/286352a0. [DOI] [PubMed] [Google Scholar]
- Fink G. R., Styles C. A. Curing of a killer factor in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1972 Oct;69(10):2846–2849. doi: 10.1073/pnas.69.10.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring E. S., Grossman L. I., Krupnick D., Cryer D. R., Marmur J. The petite mutation in yeast. Loss of mitochondrial deoxyribonucleic acid during induction of petites with ethidium bromide. J Mol Biol. 1970 Sep 14;52(2):323–335. doi: 10.1016/0022-2836(70)90033-1. [DOI] [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984 Feb;36(2):503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Hereford L., Fahrner K., Woolford J., Jr, Rosbash M., Kaback D. B. Isolation of yeast histone genes H2A and H2B. Cell. 1979 Dec;18(4):1261–1271. doi: 10.1016/0092-8674(79)90237-x. [DOI] [PubMed] [Google Scholar]
- Holland J. P., Holland M. J. Structural comparison of two nontandemly repeated yeast glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1980 Mar 25;255(6):2596–2605. [PubMed] [Google Scholar]
- Holland M. J., Holland J. P., Thill G. P., Jackson K. A. The primary structures of two yeast enolase genes. Homology between the 5' noncoding flanking regions of yeast enolase and glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1981 Feb 10;256(3):1385–1395. [PubMed] [Google Scholar]
- Julius D., Brake A., Blair L., Kunisawa R., Thorner J. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell. 1984 Jul;37(3):1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- Kilmartin J. V., Adams A. E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984 Mar;98(3):922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laz T. M., Pietras D. F., Sherman F. Differential regulation of the duplicated isocytochrome c genes in yeast. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4475–4479. doi: 10.1073/pnas.81.14.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlean P., Ammer H., Watzele M., Tanner W. Synthesis of an O-glycosylated cell surface protein induced in yeast by alpha factor. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6263–6266. doi: 10.1073/pnas.83.17.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi M., Shimoda C., Yanagishima N. Mating reaction in Saccharomyces cerevisiae. V. Changes in the fine structure during the mating reaction. Arch Mikrobiol. 1974 Apr 10;97(1):27–38. [PubMed] [Google Scholar]
- Roeder G. S., Fink G. R. DNA rearrangements associated with a transposable element in yeast. Cell. 1980 Aug;21(1):239–249. doi: 10.1016/0092-8674(80)90131-2. [DOI] [PubMed] [Google Scholar]
- Rogers D., Bussey H. Fidelity of conjugation in Saccharomyces cerevisiae. Mol Gen Genet. 1978 Jun 14;162(2):173–182. doi: 10.1007/BF00267874. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Price B. R., Fink G. R. Saccharomyces cerevisiae nuclear fusion requires prior activation by alpha factor. Mol Cell Biol. 1986 Oct;6(10):3490–3497. doi: 10.1128/mcb.6.10.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sakai K., Yanagishima N. Mating reaction in Saccharomyces cerevisiae. II. Hormonal regulation of agglutinability of a type cells. Arch Mikrobiol. 1972;84(3):191–198. doi: 10.1007/BF00425197. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos T., del Rey F., Conde J., Villanueva J. R., Nombela C. Saccharomyces cerevisiae mutant defective in exo-1,3-beta-glucanase production. J Bacteriol. 1979 Aug;139(2):333–338. doi: 10.1128/jb.139.2.333-338.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz P. J., Pillus L., Grisafi P., Solomon F., Botstein D. Two functional alpha-tubulin genes of the yeast Saccharomyces cerevisiae encode divergent proteins. Mol Cell Biol. 1986 Nov;6(11):3711–3721. doi: 10.1128/mcb.6.11.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D. A genetic system for analysis of staphylococcal nuclease. Gene. 1983 May-Jun;22(2-3):181–189. doi: 10.1016/0378-1119(83)90102-6. [DOI] [PubMed] [Google Scholar]
- Singh A., Chen E. Y., Lugovoy J. M., Chang C. N., Hitzeman R. A., Seeburg P. H. Saccharomyces cerevisiae contains two discrete genes coding for the alpha-factor pheromone. Nucleic Acids Res. 1983 Jun 25;11(12):4049–4063. doi: 10.1093/nar/11.12.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatchell K., Chaleff D. T., DeFeo-Jones D., Scolnick E. M. Requirement of either of a pair of ras-related genes of Saccharomyces cerevisiae for spore viability. Nature. 1984 Jun 7;309(5968):523–527. doi: 10.1038/309523a0. [DOI] [PubMed] [Google Scholar]
- Terrance K., Lipke P. N. Sexual agglutination in Saccharomyces cerevisiae. J Bacteriol. 1981 Dec;148(3):889–896. doi: 10.1128/jb.148.3.889-896.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkacz J. S., MacKay V. L. Sexual conjugation in yeast. Cell surface changes in response to the action of mating hormones. J Cell Biol. 1979 Feb;80(2):326–333. doi: 10.1083/jcb.80.2.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson L. E., Pringle J. R. Transient G1 arrest of S. cerevisiae cells of mating type alpha by a factor produced by cells of mating type a. Exp Cell Res. 1974 Nov;89(1):175–187. doi: 10.1016/0014-4827(74)90200-6. [DOI] [PubMed] [Google Scholar]