Abstract
Background
It has been hypothesized that human cytomegalovirus (HCMV) may be associated with breast cancer progression. However, the role of HCMV infection in breast cancer remains controversial. We aimed to assess whether HCMV genes (UL122 and UL83) could be detected in breast carcinomas and reinvestigated their possible association with breast cancer progression. DNA from paraffin-embedded tissues was analyzed by real-time PCR. We investigated 20 fibroadenomas and 27 primary breast carcinomas (stages II, III, and IV).
Findings
Two carcinomas were positive for HCMV, one was positive for two TaqMan viral detection probes, and one was positive for a sole TaqMan viral detection probe (UL83), whereas the remainder of the samples was negative.
Conclusions
Samples studied showed no association between HCMV infection and breast cancer progression.
Keywords: Breast, Cancer, Progression, Virus, Human cytomegalovirus, Polymerase chain reaction, DNA
Introduction
The number of new cancer cases and deaths is expected to increase worldwide. In Mexico, breast cancer has an incidence of 21.2%, mortality of 13.2%, and 5-year prevalence of 30.8% [1]. Cases of breast cancer were recognized from the second decade of life and there was a peak incidence between 40 and 54 years of age. The majority of cases were at advanced stages (II, III, and IV). Metastasis is frequently a final and fatal step in the progression of solid malignancies. The molecular requirements for some of these steps may be specific tissues. Viruses have been central to modern cancer research and provide profound insight into both infectious and non-infectious cancer causes and players in disease progression. This diverse group of viruses reveals unexpected connections among innate immunity, immune sensors, and tumor suppressor signaling that control both viral infection and cancer. Infectious agents can be promoters of neoplastic transformation. Viruses associated with cancer (EBV, KSHV, HPV, HBV, HCV, HTLV-1, SV40, JCV, BKV, and MCV, HIV indirect carcinogen, HCMV oncomodulator, and controversial HMTV/MMTV) have been found to cause 16.1% of human cancers worldwide [2]. Human cytomegalovirus (HCMV) is a widespread opportunistic herpesvirus that causes severe and fatal diseases in immunocompromised individuals including those who are organ transplant recipients, HIV-infected and patients with cancer. In vitro, HCMV can transform cells and deregulate other pathways relevant to adenocarcinoma pathogenesis, especially those affecting the cell cycle, mutagenesis, apoptosis, and angiogenesis when it has an oncomodulator role [3]. The purpose of this study was to determine and investigate a possible association between progression of breast cancer and HCMV infection in our tumor collective of advanced breast carcinomas consisting of 27 primary breast carcinomas and 20 fibroadenomas by real-time PCR in order to compare malignant vs. non-malignant tumors.
Materials and methods
Samples of archived paraffin-embedded breast tissue (years: 2001−2008) were obtained from the Oncology Hospital of the XXI Century National Medical Center, Mexican Social Security Institute. The local ethics and scientific research committees approved the protocol.
The study group was comprised of 27 patients with breast carcinomas (stages II, III, and IV) who had recurrent disease and progression to anthracyclines and taxanes. All samples had a minimum of 70% cancer cells. The control group was comprised of 20 samples of fibroadenomas.
Genomic DNA extraction
To purify DNA from paraffin-embedded tissue, 10-μm-thick slices of carcinomas and fibroadenomas were subsequently treated with 900 μL of xylene to remove residual xylene. The tissue was washed twice with 1000 μL of 100% ethanol and was finally resuspended in 540 μL of RLT buffer (Qiagen). Dewaxed tissues were incubated three times with 60 μL of proteinase K at 56°C/24 h. DNA was extracted with AllPrep DNA/RNA/Protein Mini Kit (Qiagen) according to the manufacturer’s instructions. Briefly, lysated tissues were placed in a column and spun for 1 min/10000 rpm; later, the column was washed twice with AW1 and AW2 buffers. Finally, the DNA was eluted with 25 μL of EB buffer preheated to 70°C. DNA quantification was performed at 260 nm in Nanodrop (Thermo Scientific).
Multiplex PCR for glyceraldehyde-3-phosphate-dehydrogenase
DNA integrity was established by the Multiplex PCR kit (Qiagen) for glyceraldehyde-3-phosphate-dehydrogenase (GADPH) amplification with increase of 100 bp (range: 100–700 bp) [4]. PCR was performed with 5 ng genomic DNA in a 12.5 μL volume containing 1X Master Mix and 0.2 μM of each primer (Table 1). Amplification was performed in a Biometra Thermocycler (T Professional) as follows: 95°C/15 min for 40 cycles at 94°C/30 sec; at 57°C/90 sec; at 72°C/90 sec and, finally, at 72°C/10 min. PCR products were separated by electrophoresis on agarose gel 2.0%, stained with 1X Gel network (Biotium) and visualized under an ultraviolet light transilluminator and photodocumenter system (Syngene).
Table 1.
Sequence primers (multiplex PCR for glyceraldehyde-3-phosphate-dehydrogenase amplification)
| Forward primers | Reverse primers | Base pairs |
|---|---|---|
| GTT CCA ATA TGA TTC CAC CC |
CTC CTG GAA GAT GGT GAT GG |
100 |
| AGG TGG AGC GAG GCT AGC |
TTT TGC GGT GGA AAT GTC CT |
200 |
| AGG TGA GAC ATT CTT GCT GG |
TCC ACT AAC CAG TCA GCG TC |
300 |
| ACA GTC CAT GCC ATC ACT GC |
GCT TGA CAA AGT GGT CGT TG |
400 |
| AGC CCC TAA GGT CTT CAA GC |
CAT GCC TGT AGC TGG GAC TA |
500 |
| GGC TCC CTT GGG TAT ATG GT |
GGA GCC AGT CTT GGA TGA |
600 |
| CCC CAC ACA CAT GCA CTT AC | AAT GAA GGG GTC ATT GAT GG | 700 |
Detection of HCMV by real-time PCR
HCMV detection was performed by real-time PCR (qPCR), amplifying a 96-bp fragment, immediate early gene expression, Ie2 (UL122, forward primer: GGCTCACCTCGTCAATCTTGA; reverse primer: AGAAGGTGCGCAATATCATGAAAGA; TaqMan probe: FAM-CCCCCTTCTGCACACCC) and 84 bp, late gene expression, pp65 (UL83, forward primer: GGGACACAACACCGTAAAGC; reverse primer: GTGGAAGAGGACCTAACGATGAC; TaqMan probe: FAM-CCGCAACCCTTCATGC). qPCR was performed with 100 ng of genomic DNA at a final volume of 8 μL: 1X TaqMan Universal PCR Master Mix (Applied Biosystems), 0.54 μL UL122- or 0.54 μL UL83- TaqMan probe. Amplification was performed in an ABI 7500 thermal cycler as follows: 60°C/2 min and 95°C/10 min, and 40 cycles: 95°C/15 sec and 60°C/1 min, and 60°C/30 sec. Positive control was done with HCMV strain AD-169 (ATCC VR-583). Negative controls were DNA white blood cells from healthy individuals (blood donors) and reaction mix without DNA. All qPCR reactions were carried out in duplicate.
Statistical analysis
We used Fisher’s exact test to compare the number cytomegalovirus-positive samples between fibroadenomas and breast carcinomas.
Results
The study group included subjects with a mean age of 53.26 ± 11.99 years (range: 27−76 years). Mean overall survival was 44 months (range: 12−84 months). All had progression with active tumor sites affecting bone, soft tissue, liver, lung, and central nervous system. The control group had a mean age of 45 ± 9.27 years (range: 27−67 years). Females with fibroadenoma were younger than females with carcinoma.
Sample 19 was HCMV-positive with both TaqMan probes: one detected an immediate early gene (synonym of viral replication) and the other detected a late gene (presence of virus). The sample ´eight´ only tested positive for a late gene. The remaining samples were HCMV-negative.
Patients positive for HCMV were detected in undifferentiated infiltrating ductal carcinomas and Luminal A molecular subtype. A 54-year-old patient had 49 months of surveillance: T4, IIIC, ER+, PR+, Her2/Neu-negative, and undetermined p53, whereas another patient aged 68 years had 54 months of surveillance: T4, IIB, ER+, PR+, Her2/Neu-negative, p53-positive. Patient data are shown in Table 2. There was no association (p = 0.50) between the presence of HCMV and the clinicopathological parameters (survival, progression, tumoral activity sites, alive without disease, alive with stable disease, alive without disease progression, died of disease) analyzed. Molecular detection was qualitative and HCMV-positive samples had a high number (>35) of threshold of detection (Ct) cycles.
Table 2.
Clinicopathological features of breast cancer patients included in the study
| n = 27 | Cases (%) |
|---|---|
| Histological type |
|
| Infiltrating ductal carcinoma |
23 (86) |
| Lobular carcinoma |
2 (7) |
| Mixed |
2 (7) |
| Age (years) |
|
| 20–40 |
6 (22) |
| 41–69 |
13 (48) |
| >60 |
8 (30) |
| Tumor size |
|
| T0-T2 |
7 (26) |
| T3 |
9 (33) |
| T4 |
11 (41) |
| ER status |
|
| Positive |
16 (59) |
| Negative |
8 (30) |
| NA |
3 (11) |
| PR status |
|
| Positive |
13 (48) |
| Negative |
11 (41) |
| NA |
3 (11) |
| Her2/neu status |
|
| Positive |
8 (30) |
| Negative |
11 (40) |
| NA | 8 (30 |
T0-T2 tumor size <5 cm of diameter; T3 tumor size >5 cm of diameter; T4 tumor any size with extension to skin or chest.
ER, estrogen receptor; PR, progesterone receptor; NA, data not available.
Discussion
This study analyzed breast carcinoma ‘progression’ according to HCMV infection. By investigating progression of breast carcinoma resection specimens, we were able to study 27 primary tumors (stages II, III, and IV) and 20 fibroadenomas by qPCR. Several studies have been linked to HCMV infections with malignant phenotype, particularly with glioblastoma [5]. We are unable to support these latter findings with our collective tumors. Only two primary tumors (7.4%) from all 27 specimens (100%) were HCMV-positive qualitatively, with the remainder being negative. These results were coincident with colon cancer [6] but not coincident with other reports of breast cancer [7] where the target viral genes, molecular techniques and inclusion criteria of samples were different. The fact that we examined quality DNA from tumor specimens of breast carcinoma by Multiplex PCR for GAPDH [4] in all samples amplified a set of different-sized targets (Figure 1). PCR is able to detect acute as well as latent infections. PCR positivity in the two primary carcinomas of our tumor collective is due to replication and viral latency and the other to only a latent virus. Positivity was observed in the primary tumor. There was no positivity in the fibroadenomas. We were unable to support the finding that HCMV infection may be correlated with tumor progression. The only mechanism compatible with the observations reported is the “hit-and-run” hypothesis, which claims that a virus can mediate cellular transformation through an initial “hit”, whereas maintenance of the transformed state is compatible with viral molecule (“run”) loss. Although the concept of hit-and-run transformation has been controversial for many years, it remains the only plausible explanation for the observations of neoplastic transformation following in vitro transfection of herpesvirus and cytomegalovirus DNA, which have been presented by multiple laboratories for more than two decades [8].
Figure 1.
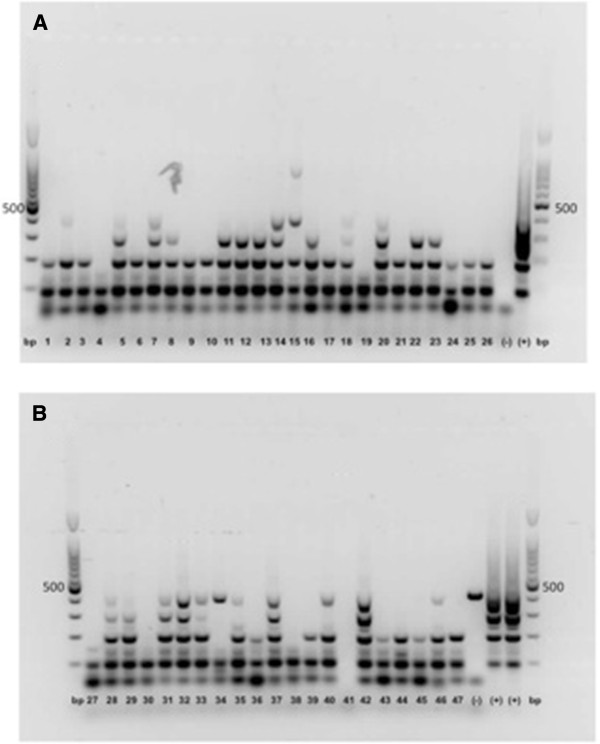
Quality control for breast carcinomas and fibroadenomas. Multiplex PCR for GADPH. (A) Lines 1−9 and 19−26 breast carcinomas; 10−18 fibroadenomas. (B) Lines 27, 37−45 breast carcinomas; 19−30, 46 and 47 fibroadenomas. Base pairs (bp); control positive (+), HCMV strain AD-169 (ATCC® VR-583); control negative (−), DNA white blood cells from healthy individual (blood donor) and reaction mix without DNA.
Conclusions
In the samples analyzed, there was no statistically significant association between malignant vs. non-malignant tumors (p = 0.50) and breast cancer progression vs. HCMV infection (p = 0.50), but we detected HCMV in two malignant tumors samples: one of which was positive for two detection probes, whereas the other sample was positive for one.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DUB conceived the study, coordinated and assisted in drafting the manuscript. HVS carried out molecular techniques and participated in the analysis of results. DGR was responsible for clinical aspects of the study and participated in the writing of the manuscript. IAC carried out the inclusion in paraffin of the tissues and immunohistochemistry assays for their classification. PA carried out molecular detection probes and helped to draft the manuscript. AGD performed the statistical analysis, critical review and editing of the manuscript. MERT conceived the study, participated in its design and coordination and was responsible for writing the manuscript and for acquiring financial support. All authors read and approved the final manuscript.
Contributor Information
Dolores Utrera-Barillas, Email: dutrera@prodigy.net.mx.
Hilda-Alicia Valdez-Salazar, Email: hildaavs@hotmail.com.
David Gómez-Rangel, Email: oncomed791105@hotmail.com.
Isabel Alvarado-Cabrero, Email: keme2.tijax12@gmail.com.
Penélope Aguilera, Email: penelope.aguilera@gmail.com.
Alejandro Gómez-Delgado, Email: agomez1992@att.net.mx.
Martha-Eugenia Ruiz-Tachiquin, Email: mertachiquin@yahoo.com.mx.
Acknowledgements
The authors acknowledge the assistance of Sharon Morey, Executive Editor, Scientific Communications for English editing. This work was partially supported by grant FIS/IMSS/PROT/G10/820 and FIS/IMSS/PROT/G09/761 from the Mexican Social Security Institute.
References
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. GLOBOCAN 2008 v2.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10 [Internet] Lyon, France: International agency for Research on Cancer; 2010. Available from: http://globocan.iarc.fr, accessed on day/month/year. [Google Scholar]
- de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- Cinatl J, Scholz M, Kotchetkov R, Vogel JU, Doerr HW. Molecular mechanisms of the modulatory effects of HCMV infection in tumor cell biology. Trends Mol Med. 2004;10:19–23. doi: 10.1016/j.molmed.2003.11.002. [DOI] [PubMed] [Google Scholar]
- van Beers EH, Joosse SA, Ligtenberg MJ, Fles R, Hogervorst FB, Verhoef S, Nederlof PM. A multiplex PCR predictor for a CGH success of FFPE samples. Br J Cancer. 2006;94:333–337. doi: 10.1038/sj.bjc.6602889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziurzynski K, Chang SM, Heimberger AB, Kalejta RF, McGregor Dallas SR, Smit M, Soroceanu L, Cobbs CS. the HCMV and Gliomas Symposium. Consensus on the role of human cytomegalovirus in glioblastomas. Neuro Oncol. 2012;14:246–255. doi: 10.1093/neuonc/nor227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knösel T, Schewe C, Dietel M, Petersen I. Cytomegalovirus is not associated with progression and metastasis of colorectal cancer. Cancer Lett. 2004;211:243–247. doi: 10.1016/j.canlet.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Harkins LE, Matlaf LA, Soroceanu L, Klemm K, Britt WJ, Wang W, Bland KI, Cobbs CS. Detection of human cytomegalovirus in normal and neoplastic breast epithelium. Herpesviridae. 2010;1:8. doi: 10.1186/2042-4280-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougall JK. “Hit and run” transformation leading to carcinogenesis. Dev Biol (Basel) 2001;106:77–82. discussion 82−83, 143−160. [PubMed] [Google Scholar]


