Abstract
Objective
To determine the role of repressors in cell type and organ-specific activation of von Willebrand factor (VWF) promoter sequences −487 to +247 in vivo.
Methods and Results
Activation patterns of wild-type and mutant VWF promoters (sequences −487 to +247) containing mutations in repressors NFI- and NFY-binding sites were analyzed in transgenic mice. Mutation of the NFI-binding site activated the promoter in heart and lung endothelial cells, whereas mutation of the NFY-binding site activated the promoter in kidney vasculature. Immunofluorescence analyses showed that NFIB was predominant in heart and lung endothelial cells, whereas NFIX was predominantly detected in kidney endothelial cell nuclei. Using ChIP, we demonstrated that the distal lung-specific enhancer (containing a YY1 site) of the VWF gene is brought in proximity to the NFI binding site.
Conclusion
NFI and NFY repressors contribute differentially to organ-specific regulation of the VWF promoter, and the organ-specific action of NFI may reflect its organ-specific isoform distribution. Additionally, the lung-specific enhancer region of the endogenous VWF gene may inhibit NFI repressor function through chromatin looping that can approximate the two regions.
Introduction
Although sharing a common lineage, endothelial cells of different organs exhibit substantial tissue-to-tissue variation in vivo, reflected by their functions and patterns of gene expression1, 2. The activation patterns of endothelial-specific gene promoters in vivo are also manifested in the heterogeneity of endothelial cell phenotype3–5. Despite well-established endothelial-subset restricted activity of a number of endothelial-specific promoters, distinct regions that target expression to specific endothelial subsets, the mechanisms, and regulators of this differential expression are not known.
Previously, we demonstrated that nucleotides −487 to +247 of the VWF gene function as an endothelial-specific promoter in cultured cells and transgenic mice4, 6. The activity of this promoter fragment in adult transgenic mice, however, was restricted to a subset of brain vascular endothelial cells4, 7, suggesting that other sequences of the VWF gene in addition to the region −487 to +247 are required for the pattern of transcription observed with endogenous gene. Aird et. al. have demonstrated that an extended fragment of the VWF promoter (designated VWF2, spanning −2182 to the end of the first intron) directs expression in microvascular endothelial cells of heart, skeletal muscle, and brain8. Recently, we have demonstrated that a region in intron 51 of the VWF gene confers lung-specific activation to the VWF promoter9.
Analyses of the VWF promoter sequence −487 to +247 in vitro demonstrated that several transacting factors function as activators and repressors of the promoter. GATA6, HLP (Histone H1-like protein), NFY (interacting with a consensus CCAAT element) and Ets activate the VWF promoter, while NFI, Oct1 and NFY (interacting with a novel non-consensus DNA element) are repressors of the promoter6, 10–15. Thus, we hypothesized that along with a requirement for other VWF DNA sequences, some of these repressor elements may also participate in the organ-restricted activation pattern of the VWF promoter. Here, we explored the roles of NFI- and NFY-binding sites (novel non-consensus NFY-binding DNA element) in the regulation of the VWF promoter sequence −487 to +247 in vivo.
Material and Methods
Detailed description of Materials and Methods is provided in supplementary materials.
Generation and analyses of transgenic mice
LacZK plasmid containing the wild-type VWF promoter sequence −487 to +247 fused to LacZ gene and SV40 poly-A signal was generated as previously described4. Plasmids LacZK NFI, LacZK NFY and LacZKNFI-NFY contain sequences similar to LacZK except for 3 bp substitution mutations in NFI, NFY or both repressors’ binding sites. C57BL/6 strain of mice was used to generate transgenic mice by Ozgene (Bentley, WA, Australia). A minimum of two independent lines for each newly-generated transgenic mice were analyzed. Since the LacZK transgenic mouse was previously reported4, only one line was analyzed.
RNA preparation and real-time polymerase chain reaction (RT-PCR) analyses
RNA was prepared from heart, lung, liver, kidney and brain using the Qiagen RNeasy minikit (Valencia, CA) according to manufacturer’s instructions and used as template for RT-PCR analyses as previously described16. The primers used for detection of GAPDH and transgene LacZ are shown in Table 1.
Table 1.
List of primers used for Real-Time PCR
| Mouse GAPDH forward | TGTGTCCGTCGTGGATCTGA |
| Mouse GAPDH Reverse | CCTGCTTCACCACCTTCTTGA |
| LacZ forward | ATTAGGGCCGCAAGAAAACTATC |
| LacZ Reverse | GGTATACATGTCTGACAATGGCAG |
| Human VWF Intron 51-YY1 binding region (I51) Forward | CGCAGGGAAGAGAAGGGAAATAAACTGG |
| Human VWF Intron 51-YY1 binding region (I51) Reverse | GACTGATCTTCAAGAATTGTGGCCC |
| Human VWF NF1 binding region Forward | TAGGCTTGTGGCCAAGACCTTCAT |
| Human VWF NF1 binding region Reverse | CGGTCCTGGCCCTGACAAA |
Immunohistochemical detection of β-galactosidase
Tissue macro-arrays of heart, lung, liver, kidney and brain were prepared by Histobest Inc. (Edmonton, Alberta) and subjected to immunohistochemistry using primary anti-LacZ antibodies (Abcam ab116-100) as previously described9.
Immunofluorescent detection of NFI, PECAM and LacZ was performed as described (supplementary materials).
Cell culture and Chromatin Immunoprecipitation
HEK 293 cells were maintained as previously described10. Human microvascular lung endothelial cells were purchased from Lonza. Chromatin immunoprecipitation (ChIP) and RT-PCR were as previously described16 with primers detecting NFI- and YY1-binding sequences of the VWF gene (Table 1).
Statistical analysis
All data are presented as means with error bars representing standard deviations. Statistical analyses used the paired Student T-test.
Results
Mutation of NFI-binding sites results in VWF promoter activation specifically in lung and heart vasculature
To determine whether the repressors NFI and NFY participate in the organ-restricted activity of the VWF promoter in vivo, we generated transgenic mice harboring the LacZ gene under the regulation of the VWF promoter that was either wild-type (LacZK) or contained mutation of the NFI-binding site (LacZKNFI), NFY-binding site (LacZKNFY) or both (LacZKNFI-NFY). The VWF promoter in all transgenes constituted sequence −487 to +247 and the mutations contained three-base substitutions that were used previously to analyze NFI- and NFY-binding sites in vitro10, 12, 14 (Figure 1A). We analyzed two-three independent lines of mice carrying each transgene, except only one line for LacZK (previously analyzed extensively). Organs were harvested from adult transgenic mice LacZK (one line), LacZKNFI (three lines), LacZKNFY and LacZKNFI-NFY (two lines each) and processed for quantitative RT-PCR to detect LacZ mRNA, and immunohistochemistry to detect LacZ protein.
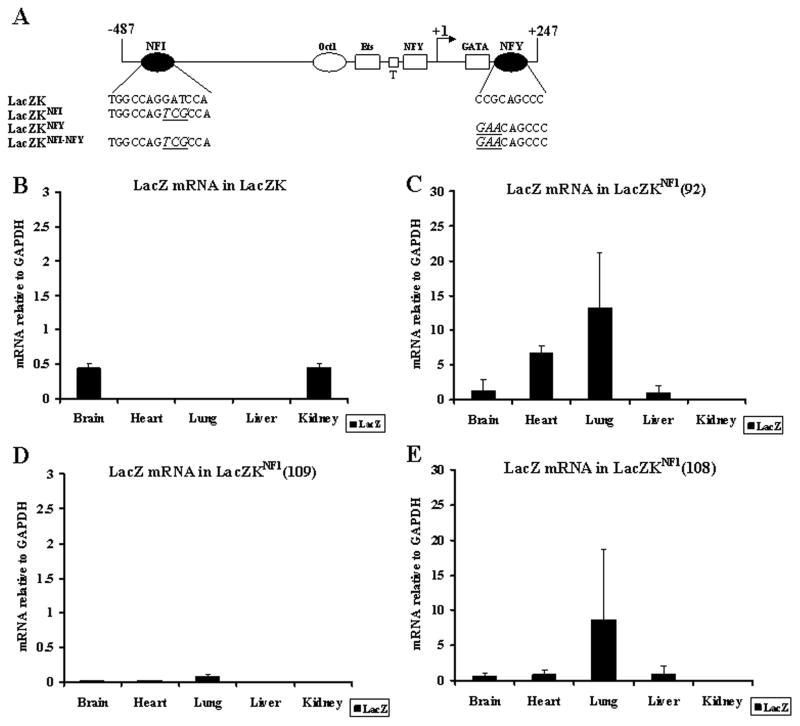
Figure 1. Real-time PCR (RT-PCR) analyses of LacZ transgene mRNA in LacZK and LacZKNFI transgenic mice.
(A) Schematic of the wild-type and mutant VWF promoter sequence −487 to +247 fused to the LacZ gene. Transacting factors functioning as repressors (NFI, Oct-1, and NFY: ovals) and activators (Ets, NFY and GATA6: rectangles) are shown. TATA element: T (small square). Arrow at +1 represents transcription start site. Binding sequence of the repressors NFI and NFY in the wild-type (LacZK) and mutant promoters (LacZKNFI, LacZKNFY and LacZKNFI-NFY) are shown with base substitution mutations underlined and bolded. (B–E) RNA (1 μg) from various organs of (B) LacZK and (C–E) three independent lines of LacZKNFI transgenic mice (F2 generations) were analyzed by RT-PCR to detect LacZ transgene and endogenous GAPDH mRNA for normalization. Results represent the averages of mRNA from 2 littermates of each line for each organ.
RT-PCR of the LacZK (the wild-type promoter) transgenic mouse detected LacZ mRNA only in the brain and kidney (Figure 1B). In contrast, in three lines of LacZKNFI, LacZ mRNA was detected most significantly in the lung, but also in the heart and brain, and to a lesser degree in the liver. Although the level of expression was variable among the three lines (potentially reflecting transgene variation in integration site and copy number), the pattern of expression was similar [Figure 1 C–E, note the scales on Y axis reflecting significantly higher levels in lines 92 and 108 (C and E) compared to line 109 (D)].
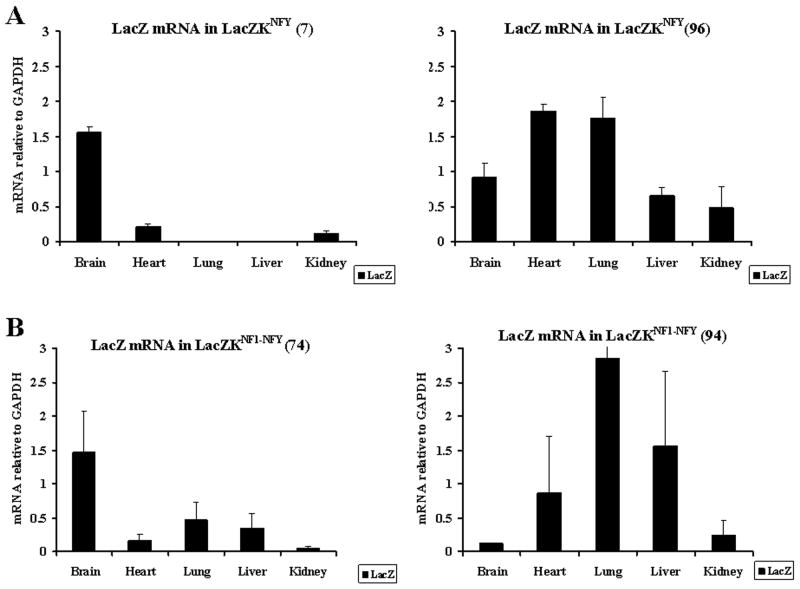
RT-PCR of the two lines of LacZKNFY detected LacZ mRNA in the brain, kidney and heart of one line, whereas in the other line expression was also detected in the lung and liver (Figure 2A). RT-PCR of organs from two lines of double mutant LacZKNFI-NFY detected LacZ mRNA in all five organs of both lines (Figure 2B).
Figure 2. RT-PCR analyses of LacZ transgene mRNA in LacZKNFY and LacZKNFI-NFY transgenic mice.
RT-PCR analyses of LacZ mRNA in 2 independent lines of LacZKNFY (A) and LacZKNFI-NFY (B) transgenic mice were done as in Figure 1.
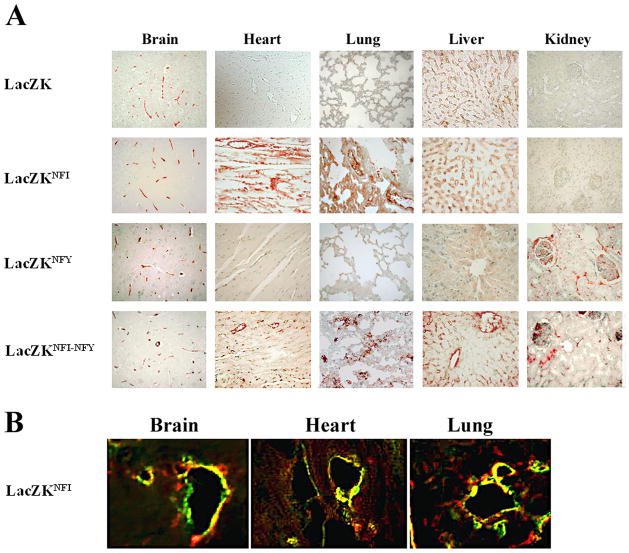
To determine LacZ protein expression, tissue arrays from formaldehyde-treated organs were processed for immunohistochemistry with specific anti-β-galactosidase antibody. In these analyses, sections from multiple tissues were processed on the same slide to minimizing inter-experimental staining variability. Consistent with previous reports4, 7, immunohistochemistry showed that LacZ protein was detected only in the brain vasculature of the transgenic mouse LacZK (Figure 3 panel LacZK).
Figure 3. Immunohistochemical and immunofluorescence analyses of LacZ expression in organs of transgenic mice.
(A) Formalin-embedded sections (5 μm) of heart, kidney, liver, lung and brain from four transgenic mice (adult F1 generations) LacZK, LacZKNFI, LacZKNFY and LacZKNFI-NFY on tissue arrays were immunostained with anti-β-galactosidase antibody as previously described9 (magnifications 400X). Two littermates from each line and two independent lines for each transgene (except LacZK only one line) were analyzed and results from one representative line of each transgene shown. No staining was detected in sections from organs of non-transgenic mice (data not shown). (B) Sections (5 μm) of OCT-frozen brain, heart and lung from a LacZKNFI transgenic mice (line 92) were treated with anti-β-galactosidase (green) and anti-PECAM (red) antibodies to detect LacZ and endothelial cells as described (Materials and methods). The results are representative of two independent experiments using two littermates of this line (magnifications 400X).
In contrast, analyses of LacZKNFI transgenic mice detected significant LacZ protein expression in the brain, lung and heart vasculature. Similar results were obtained for all three LacZKNFI lines. Results for one line are shown (Figure 3, LacZKNFI). In LacZKNFY transgenic mice, expression of LacZ was detected in the brain and kidney vasculature in both lines (results for one line shown in Figure 3, LacZKNFY). However, in one of the lines that demonstrated RNA expression in all organs, barely detectable expression in heart and lung vasculature was also observed (data not shown). In LacZKNFI-NFY transgenic mice, LacZ expression was detected in the vasculature of brain, heart, lung, kidney and liver in both lines (results for one line shown in Figure 3, LacZKNFI-NFY).
The above results demonstrate that mutation of the NFI-binding site (LacZKNFI ) activates the VWF promoter resulting in LacZ transgene expression detectable at both RNA and protein levels in heart and lung vasculature, in addition to brain. Double mutation in both NFI- and NFY-binding sites (LacZKNFI-NFY) activates the VWF promoter in the vasculature of all five major organs. The analyses of LacZKNFY transgenic mice demonstrated variation in the LacZ expression pattern between the two lines, most significantly at the RNA levels. However, consistent RNA and protein expression in the kidney vasculature of both lines of this transgenic mouse suggests that mutation of the NFY-binding site is necessary for activation of the promoter in kidney.
Based on the consistent results from three lines of LacZKNFI transgenic mice, attention was focused on these lines and confocal immunofluorescence was used to determine the localization of LacZ expression in OCT-frozen sections of brain, heart and lung of these mice. These analyses showed that LacZ [Alexa488-conjugated-secondary antibodies (green)] colocalizes specifically with PECAM, a marker of endothelial cells [Alexa594-conjugated-secondary antibodies (red)] in brain, lung and heart, thus confirming the endothelial-specific activation of the NFI mutant promoter in the vasculature of these organs (Figure 3B).
NFIB and NFIX are differentially expressed in vascular endothelial cells of the lung, heart and kidney
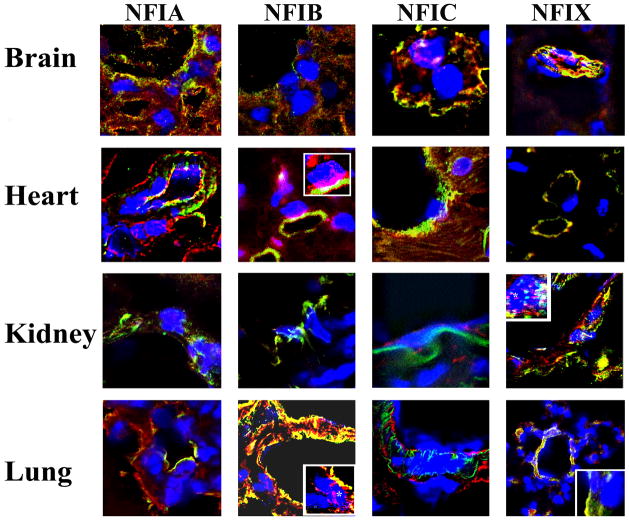
Based on the transgenic mice analyses, we hypothesized that NFI transacting factor specifically represses the VWF promoter in lung and heart, but not kidney vasculature. There are four known NFI isoforms designated as A, B, C and X 17. Thus, we explored whether endothelial cells of lung, heart, kidney and brain have different expression patterns of these isoforms. Since cultured endothelial cells may not accurately reflect organ-specific endothelial cell characteristics, we performed immunofluorescence/confocal microscopy analyses of mouse organ sections using specific antibodies to detect A, B, C and X isoforms of NFI [Alexa 594-conjugated-secondary antibodies (red)] and PECAM [Alexa488-conjugated-secondary antibody (green)] as a general marker for endothelial cells. Nuclei were stained with DAPI to determine nuclear localization of NFI. There were significant variations in the expression of NFI isoforms that were detected in the nuclei of endothelial cells of the organs studied (Figure 4). Expression of NFIC and NFIX were most significantly detected in brain endothelial cell nuclei, whereas barely detectable levels of NFIA and no NFIB were detected. NFIB followed by NFIA were most significantly detected in heart endothelial cell nuclei, with no detection of NFIX or NFIC. Predominantly, NFIX was detected in kidney endothelial cell nuclei with significantly lower levels of NFIC and no NFIA or NFIB. In lung endothelial cell nuclei, NFIB was detected most, whereas NFIA, C or X were not detectable.
Figure 4. Immunofluorescent analyses of NFI isoform expression in organs of mice.
Sections (5 μm) from OCT-frozen heart, kidney, lung and brain of a control C57Bl/6 mouse that were arranged on tissue arrays were treated with antibodies specific to NFI-A, NFI-B, NFI-C and NFI-X isoforms of NFI (red) and simultaneously with anti-PECAM antibodies (green) to detect endothelial cells as described (Materials and Methods). The results are representative of two independent experiments for each NFI antibody plus PECAM antibodies (magnifications 600X). The insets show magnified endothelial cell nuclei (Dapi).
These results demonstrate that distinct, non-overlapping isoforms of NFI are present specifically in lung, heart and kidney endothelial cell nuclei. NFIX is the major isoform detected in kidney with no detectable NFIB, whereas in the lung and heart, NFIB is the major isoform detected with no detectable NFIX.
Since mutation of the NFI-binding site activates the VWF promoter in lung and heart, but not kidney, we hypothesize that NFIB, but not NFIX, may function as a repressor of the VWF promoter. Our experiments do not exclude the possibility that NFIC and NFIA (when present) may also function as repressors of the VWF promoter.
DNA looping mediates proximity of the lung-specific enhancer located in the VWF intron 51 to the upstream NFI-binding site
Recently, we have reported that a region of intron 51 of the VWF gene that interacts with transacting factor YY1 functions as a lung-specific enhancer9. In vitro, this enhancer activity required YY1 interaction with this region. When the intron 51 region was inserted upstream or downstream of the VWF promoter, the promoter was activated specifically in the lung and brain endothelial cells of transgenic mice9. Since NFI functions as a repressor of the VWF promoter in lung (and heart) endothelium, we hypothesized that (without mutation of NFI-binding site) intron 51 sequences potentially overcome the inhibitory effect of NFI in lung endothelial cells through YY1. Since the YY1-binding site of intron 51 lies more than 100 kb downstream of the NFI-binding site, we also hypothesized that for this to occur, YY1 (interacting with a region of intron 51) may be brought in proximity to NFI, potentially through chromatin looping.
To test this hypothesis, we reasoned that if the intron 51-YY1-binding site and NFI-binding site are in close proximity, the cross-linking reagents in the ChIP assay may result in a complex containing: NFI-binding sequences-NFI factor-YY1 factor-YY1 binding sequences (Figure 5A). Thus, we would expect the detection of both NFI- and YY1-binding sequences using either NFI or YY1 antibodies. Analyses of intron 51 did not reveal an NFI-binding site and upstream VWF sequences (within 1Kb) did not contain a YY1-binding site. In addition, immunoprecipitation assays did not demonstrate co-precipitation of free (not bound to DNA) nuclear NFI and YY1 proteins (data not shown). Therefore, using NFI-specific antibodies for ChIP, if we detect both amplified YY1-binding intron 51 and NFI-binding sequences and vice versa, this strongly suggests the close proximity of these elements potentially through chromatin looping (Figure 5A).
Figure 5. Chromatin immunoprecipitation detects the association of NFI and YY1 with their cognate binding sites and chromatin looping.
ChIP and quantitative PCR analyses were performed on LMEC and HEK293 cells as described (Materials and Methods). (A) Schematic of VWF chromatin looping that could bring NFI in proximity to intron 51 YY1-binding site. Cross-linked chromatin from (B) LMEC and (C) HEK293 cells were immunoprecipitated with antibodies specific to NFI-A, B, C, X and YY1. Rabbit IgG antibody was used as negative control. Immunoprecipitates were subjected to quantitative RT-PCR using primers that specifically amplified an 89 bp upstream region of the VWF promoter containing the NFI binding site (NFI primers) or a 182 bp fragment of the VWF intron 51 sequence containing the YY1-binding site (I51 primers). Results for each antibody are presented as fold increase compared to control IgG antibody, and all ChIP represent analyses of 2–4 independent ChIP experiments for each antibody.
Human lung microvascular endothelial cells (LMEC) and non-endothelial HEK 293 cells were used for ChIP analyses. ChIP from LMEC usingYY1 antibody or all four isoform-specific NFI antibodies demonstrated that the NFI-binding sequences of the VWF promoter were amplified. Fold increases over results from ChIP with control IgG were indicative of specific immunoprecipitation with the target antibodies (Figure 5B, NFI primers). The reciprocal experiment also demonstrated that intron 51 sequences were amplified in ChIP with NFI antibodies as well as YY1 antibody (Figure 5B, I51 primers). These analyses also demonstrated that the amplification of NFI-binding sequences was highest when NFIB-specific antibody was used, suggesting that NFIB preferentially interacts with the VWF promoter in LMEC. This result is consistent with the organ-analyses demonstrating preferential expression of NFIB in lung endothelial cells (Figure 4, lung panel), although in cultured cells, other NFI isoforms can interact with the VWF promoter, suggesting the presence of multiple isoforms. In ChIP of LMEC, the intron 51 sequences were highly amplified when YY1-specific antibodies were used, demonstrating significant interaction of YY1 with intron 51 sequences. Together, these results show significant interaction of NFIB with the VWF promoter and YY1 with intron 51 sequences, and are consistent with the hypothesis that chromatin looping brings the YY1-binding intron 51 sequences in close proximity to the NFI-binding site in lung endothelial cells (LMEC). This arrangement may promote association of YY1 and NFI and/or allow YY1 to somehow influence the inhibitory function of NFI.
In HEK 293 however, ChIP showed that the NFI-binding site sequence was not amplified when YY1 antibodies were used (less than 1-fold compared to IgG) whereas it was amplified when NFIC- (almost 4-fold), or to a lesser extent when NFIA- (2-fold) specific antibodies were used. Furthermore, the YY1-binding site of intron 51 was not amplified (less than 1-fold) when either NFIC- or YY1-specific antibodies were used (Figure 5C). These results demonstrate that interaction of YY1 with VWF intron 51 in the context of chromatin does not occur in HEK293 cells. These results also demonstrate that in HEK293, NFIC predominantly interacts with the NFI-binding site, whereas NFIB or NFIX do not.
Discussion
Results of in vivo analyses of the VWF promoter with mutations in NFI- and NFY-binding sites support the hypothesis that the activity of the VWF promoter sequence −487 to +247 is repressed by at least these two repressors. RNA and immunohistochemical analyses demonstrated the expression of the LacZ transgene activated by mutant VWF promoters in distinct sets of organs, whereas, consistent with previous observations4, the wild-type promoter activated LacZ protein expression only in brain vasculature, although expression of mRNA in the kidney was also detected. The transgenic mice were generated using standard approaches and subject to transgene variation in integration sites and copy numbers, which could influence expression level, lead to aberrant transcription and/or cause ectopic expression. Therefore, the detection of both protein and mRNA LacZ transgene expression in any organ, and similarly, in two or more lines for each transgene, were used as the criteria for promoter activation. Accordingly, detection of LacZ mRNA in the kidney of LacZK transgenic mice without detectable LacZ protein suggests low level ectopic expression, and/or aberrant transcripts (untranslatable) in this line. Previously, multiple lines of transgenic mice harboring this VWF promoter sequence fused to LacZ or amyloid precursor protein (APP) genes demonstrated transgene activation in the brain, but not heart, lung, liver or kidney4, 7.
Based on analyses of three lines of LacZKNFI demonstrating LacZ expression at both mRNA and protein levels in heart and lung (and brain), we conclude that mutation of the NFI-binding site results in promoter activation specifically in heart and lung vasculature, but not in kidney or liver (where only mRNA was detected). On the other hand, mutation of the NFY-binding site (LacZKNFY) was required for activation of the VWF promoter in kidney vasculature. However, since in one of the two transgenic lines of LacZKNFY, barely detectable protein with significant mRNA expression were also observed in the heart and lung, analyses of additional transgenic lines are necessary to unambiguously determine LacZKNFY activation pattern. Nevertheless, our results unambiguously demonstrate that mutation of the NFY- but not NFI-binding site activates the VWF promoter in kidney vasculature. Analyses of transgenic mice with double mutations in both NFI- and NFY-binding sites (LacZKNFI-NFY), demonstrated promoter activity in heart, lung, kidney and liver vasculatures. This supports the results of individual mutations by demonstrating that double mutant transgenic mice exhibit a pattern of expression that overlaps the patterns observed with individual mutations. In addition, the results also suggest that the simultaneous mutation of both repressor elements has a synergistic effect on the VWF promoter activity, since it results in promoter activation in liver vasculature that was not detected at the protein level in either of single mutants. Together, the results of transgenic mice analyses demonstrate that repressors NFI and NFY function differentially to inhibit the VWF promoter in distinct organs. While NFI inhibits VWF promoter activity in heart and lung, but not kidney, NFY inhibits promoter activity primarily in kidney vasculature.
To date, a repressive function for NFY specifically in endothelial cells has been reported only for the VWF promoter10, 12. However, an organ-specific repressive function for NFI in the regulation of tPA gene (an endothelial-specific gene) promoter was previously reported18. It was hypothesized that NFI repression of tPA occurs differentially in endothelial cells of different organs, specifically in lung and brain, but not in kidney and liver. This hypothesis is consistent with our results demonstrating that NFI repression of the VWF promoter occurs in lung (and heart) but not in kidney and liver.
Several NFI isoforms (A, B, C and X) and various alternatively-spliced mRNAs of these isoforms have been reported17. An overlapping but distinct expression pattern for each isoform during embryonic development has been shown17. In addition, analyses in vitro have demonstrated differential activation/repression functions for various NFI isoforms depending on the target promoter17, 19. Consistent with these in vitro observations, our in vivo analyses of mouse organ sections showed that whereas NFIB was predominantly detected in lung and heart endothelial cell nuclei, NFIX was predominantly detected in kidney endothelial cell nuclei. Based on these observations and that the NFI mutant VWF promoter (LacZKNFI) is activated in heart and lung, but not kidney endothelial cells, we hypothesize that NFIB predominantly functions as repressor of VWF promoter activity. Analyses of knock-out mice specific for each NFI isoform have demonstrated that whereas all isoforms contribute to development of the central nervous system, NFIB is required specifically for lung development20, 21. Also, a report on the role of NFI in regulating the p21 promoter in cultured cells demonstrated that while NFIB functions as repressor, NFIX and A appear to have an opposing role19. Taken together, we hypothesize that the NFIB appears to have specific functions in the lung, including repression of the VWF promoter in lung endothelial cells.
We have previously reported that intron 51 sequences confer lung-specific activity on the VWF promoter9. We hypothesize that in regard to the endogenous gene, activators that interact with intron 51 sequences including YY1, overcome the repressive function of NFI in lung endothelial cells. We reasoned that a prerequisite for this function of intron 51 sequences may be a chromatin structure of the VWF gene that brings two distant regulatory elements (NFI-binding site and intron 51 sequences) into close proximity. We tested this hypothesis by ChIP and demonstrated that anti-NFI antibodies can amplify intron 51 YY1-binding sequences as well as NFI binding sequences (and vice versa) in human lung microvascular endothelial cells (LMEC), but not in HEK 293 cells. These results support the hypothesis that the NFI and intron 51 YY1-binding region are in close proximity in endothelial cells.
Using ChIP and in contrast to LMEC, YY1 in HEK293 cells does not interact with its cognate intron 51-binding site in the context of VWF chromatin. We cannot, however, eliminate the possibility that chromatin looping also occurs in HEK293 cells, but cannot be detected using ChIP, that is based on potential association of both YY1 and NFI with their cognate binding sites. Recently, we have reported that YY1 forms a specific complex with actin in HEK293 cell nuclei, which was not observed in human umbilical vein endothelial cells (HUVECs). This was demonstrated by gel mobility, DNA pull-down and immunoprecipitation/Western blot analyses. Based on ChIP failing to detect YY1 interaction with intron 51 sequences in HEK 293 cells, we hypothesize that nuclear actin association with YY1 may function as a regulatory switch to prevent YY1 interaction with intron 51 sequences, thus rendering it ineffective as an enhancer of the VWF gene in non-endothelial cells.
In summary, we propose the following hypothesis for lung-specific activation of the VWF promoter. NFI, predominantly NFIB, functions as a repressor in the lung, whereas YY1 binding to intron 51 that is in close proximity to the NFI-binding site overcomes the repression by NFI, thereby promoting transcription. In non-endothelial cells, even if chromatin looping is similar, interaction of nuclear actin with YY1 inhibits YY1 association with intron 51 sequences, thus rendering it ineffective as an activator.
Differential levels of VWF expression in endothelial cells of different organs and vessel types are reflective of the heterogeneity of endothelial cells22, 23. Furthermore, vasculatures of different organs were reported to modulate VWF expression differentially in response to stimuli23. Multiple repressors that participate in VWF promoter activity differentially in distinct organs, and different regulatory elements that overcome these repressors in an organ-specific manner may indicate transcriptional regulatory mechanisms that are susceptible to diverse and differential vascular-bed specific regulation.
Supplementary Material
Acknowledgments
This work was supported by research grants from Heart & Stroke foundation of Canada and Canadian Institute of Health Research (N.J).
Footnotes
Disclosure: None
Authorship
M.N. designed and performed experiments, analyzed data and wrote the manuscript. J.L. and S.K. designed and performed experiments and analyzed data. Richard Uwiera performed experiments and analyzed data. W.C., B.B. and N.J. designed research, analyzed data and wrote the manuscript.
All authors declare no competing financial interests.
References
- 1.Aird WC. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ Res. 2007;100:158–173. doi: 10.1161/01.RES.0000255691.76142.4a. [DOI] [PubMed] [Google Scholar]
- 2.Aird WC. Phenotypic heterogeneity of the endothelium: II. Representative vascular beds. Circ Res. 2007;100:174–190. doi: 10.1161/01.RES.0000255690.03436.ae. [DOI] [PubMed] [Google Scholar]
- 3.Fish JE, Marsden PA. Endothelial nitric oxide synthase: insight into cell-specific gene regulation in the vascular endothelium. Cell Mol Life Sci. 2006;63:144–162. doi: 10.1007/s00018-005-5421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aird WC, Jahroudi N, Weiler-Guettler H, Rayburn HB, Rosenberg RD. Human von Willebrand factor gene sequences target expression to a subpopulation of endothelial cells in transgenic mice. Proc Natl Acad Sci U S A. 1995;92:4567–4571. doi: 10.1073/pnas.92.10.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prandini MH, Dreher I, Bouillot S, Benkerri S, Moll T, Huber P. The human VE-cadherin promoter is subjected to organ-specific regulation and is activated in tumour angiogenesis. Oncogene. 2005;24:2992–3001. doi: 10.1038/sj.onc.1208483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jahroudi N, Lynch DC. Endothelial-cell-specific regulation of von Willebrand factor gene expression. Mol Cell Biol. 1994;14:999–1008. doi: 10.1128/mcb.14.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jahroudi N, Schmaier A, Srikanth S, Mahdi F, Lutka FA, Bowser R. Von Willebrand factor promoter targets the expression of amyloid beta protein precursor to brain vascular endothelial cells of transgenic mice. J Alzheimers Dis. 2003;5:149–158. doi: 10.3233/jad-2003-5209. [DOI] [PubMed] [Google Scholar]
- 8.Aird WC, Edelberg JM, Weiler-Guettler H, Simmons WW, Smith TW, Rosenberg RD. Vascular bed-specific expression of an endothelial cell gene is programmed by the tissue microenvironment. J Cell Biol. 1997;138:1117–1124. doi: 10.1083/jcb.138.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleinschmidt AM, Nassiri M, Stitt MS, Wasserloos K, Watkins SC, Pitt BR, Jahroudi N. Sequences in intron 51 of the von Willebrand factor gene target promoter activation to a subset of lung endothelial cells in transgenic mice. J Biol Chem. 2008;283:2741–2750. doi: 10.1074/jbc.M705466200. [DOI] [PubMed] [Google Scholar]
- 10.Peng Y, Jahroudi N. The NFY transcription factor inhibits von Willebrand factor promoter activation in non-endothelial cells through recruitment of histone deacetylases. J Biol Chem. 2003;278:8385–8394. doi: 10.1074/jbc.M213156200. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Peng Y, Ma Y, Jahroudi N. Histone H1-like protein participates in endothelial cell-specific activation of the von Willebrand factor promoter. Blood. 2004;104:1725–1732. doi: 10.1182/blood-2004-01-0082. [DOI] [PubMed] [Google Scholar]
- 12.Peng Y, Jahroudi N. The NFY transcription factor functions as a repressor and activator of the von Willebrand factor promoter. Blood. 2002;99:2408–2417. doi: 10.1182/blood.v99.7.2408. [DOI] [PubMed] [Google Scholar]
- 13.Schwachtgen JL, Janel N, Barek L, Duterque-Coquillaud M, Ghysdael J, Meyer D, Kerbiriou-Nabias D. Ets transcription factors bind and transactivate the core promoter of the von Willebrand factor gene. Oncogene. 1997;15:3091–3102. doi: 10.1038/sj.onc.1201502. [DOI] [PubMed] [Google Scholar]
- 14.Jahroudi N, Ardekani AM, Greenberger JS. An NF1-like protein functions as a repressor of the von Willebrand factor promoter. J Biol Chem. 1996;271:21413–21421. doi: 10.1074/jbc.271.35.21413. [DOI] [PubMed] [Google Scholar]
- 15.Schwachtgen JL, Remacle JE, Janel N, Brys R, Huylebroeck D, Meyer D, Kerbiriou-Nabias D. Oct-1 is involved in the transcriptional repression of the von willebrand factor gene promoter. Blood. 1998;92:1247–1258. [PubMed] [Google Scholar]
- 16.Peng Y, Stewart D, Li W, Hawkins M, Kulak S, Ballermann B, Jahroudi N. Irradiation modulates association of NF-Y with histone-modifying cofactors PCAF and HDAC. Oncogene. 2007;26:7576–7583. doi: 10.1038/sj.onc.1210565. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhry AZ, Lyons GE, Gronostajski RM. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev Dyn. 1997;208:313–325. doi: 10.1002/(SICI)1097-0177(199703)208:3<313::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 18.Pham NL, Franzen A, Levin EG. NF1 regulatory element functions as a repressor of tissue plasminogen activator expression. Arterioscler Thromb Vasc Biol. 2004;24:982–987. doi: 10.1161/01.ATV.0000126679.70877.d0. [DOI] [PubMed] [Google Scholar]
- 19.Ouellet S, Vigneault F, Lessard M, Leclerc S, Drouin R, Guerin SL. Transcriptional regulation of the cyclin-dependent kinase inhibitor 1A (p21) gene by NFI in proliferating human cells. Nucleic Acids Res. 2006;34:6472–6487. doi: 10.1093/nar/gkl861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mason S, Piper M, Gronostajski RM, Richards LJ. Nuclear factor one transcription factors in CNS development. Mol Neurobiol. 2009;39:10–23. doi: 10.1007/s12035-008-8048-6. [DOI] [PubMed] [Google Scholar]
- 21.Steele-Perkins G, Plachez C, Butz KG, Yang G, Bachurski CJ, Kinsman SL, Litwack ED, Richards LJ, Gronostajski RM. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol. 2005;25:685–698. doi: 10.1128/MCB.25.2.685-698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pusztaszeri MP, Seelentag W, Bosman FT. Immunohistochemical expression of endothelial markers CD31, CD34, von Willebrand factor, and Fli-1 in normal human tissues. J Histochem Cytochem. 2006;54:385–395. doi: 10.1369/jhc.4A6514.2005. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto K, de Waard V, Fearns C, Loskutoff DJ. Tissue distribution and regulation of murine von Willebrand factor gene expression in vivo. Blood. 1998;92:2791–2801. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.