Abstract
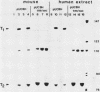
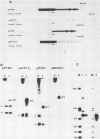
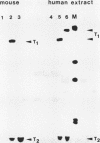
We have analyzed the sequences required for termination of human rDNA transcription. The human ribosomal transcription unit is shown to extend about 350 nucleotides into the 3'-terminal spacer and ends immediately upstream of a region with a distinct sequence heterogeneity. This heterogeneous region contains a cluster of conserved 10-base pair sequence elements which exert a striking homology to the proximal part of the 18-base pair murine rDNA transcription termination signal sequence, termed SalI box. Exonuclease III protection assays and in vitro transcription experiments with both homologous and heterologous human-mouse minigene constructs, and extracts from HeLa or Ehrlich ascites cells, reveal a functional analogy of the human sequence to the mouse SalI box. It mediates binding of a nuclear protein which functions as a transcription termination factor. The murine signal sequence is recognized by the human factor but not vice versa. The different sequence specificities and electrophoretic properties of the functionally equivalent protein factors suggest that a molecular coevolution has taken place between the termination signal sequences and the genes coding for the termination factors.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Braga E. A., Yussifov T. N., Nosikov V. V. Structural organization of rat ribosomal genes restriction endonuclease analysis of genomic and cloned ribosomal DNAs. Gene. 1982 Dec;20(2):145–156. doi: 10.1016/0378-1119(82)90033-6. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J. M., Schmickel R. D. A molecular basis for discrete size variation in human ribosomal DNA. Am J Hum Genet. 1985 Mar;37(2):311–325. [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Maier U., Ohrlein A., Hassouna N., Bachellerie J. P. Transcription of mouse rDNA terminates downstream of the 3' end of 28S RNA and involves interaction of factors with repeated sequences in the 3' spacer. Cell. 1985 Dec;43(3 Pt 2):801–810. doi: 10.1016/0092-8674(85)90253-3. [DOI] [PubMed] [Google Scholar]
- Grummt I., Rosenbauer H., Niedermeyer I., Maier U., Ohrlein A. A repeated 18 bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell. 1986 Jun 20;45(6):837–846. doi: 10.1016/0092-8674(86)90558-1. [DOI] [PubMed] [Google Scholar]
- Grummt I., Roth E., Paule M. R. Ribosomal RNA transcription in vitro is species specific. Nature. 1982 Mar 11;296(5853):173–174. doi: 10.1038/296173a0. [DOI] [PubMed] [Google Scholar]
- Grummt I., Sorbaz H., Hofmann A., Roth E. Spacer sequences downstream of the 28S RNA coding region are part of the mouse rDNA transcription unit. Nucleic Acids Res. 1985 Apr 11;13(7):2293–2304. doi: 10.1093/nar/13.7.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I. Specific transcription of mouse ribosomal DNA in a cell-free system that mimics control in vivo. Proc Natl Acad Sci U S A. 1981 Feb;78(2):727–731. doi: 10.1073/pnas.78.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., Oliemans J., Offenberg H., Dekker A. F., Piper P. W., Planta R. J., Klootwijk J. 3'-End formation of transcripts from the yeast rRNA operon. EMBO J. 1986 Oct;5(10):2703–2710. doi: 10.1002/j.1460-2075.1986.tb04554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal M., Arnheim N. Length heterogeneity in a region of the human ribosomal gene spacer is not accompanied by extensive population polymorphism. J Mol Biol. 1978 Nov 25;126(1):91–104. doi: 10.1016/0022-2836(78)90281-4. [DOI] [PubMed] [Google Scholar]
- La Volpe A., Simeone A., D'Esposito M., Scotto L., Fidanza V., de Falco A., Boncinelli E. Molecular analysis of the heterogeneity region of the human ribosomal spacer. J Mol Biol. 1985 May 25;183(2):213–223. doi: 10.1016/0022-2836(85)90214-1. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Characterization of three sites of RNA 3' end formation in the Xenopus ribosomal gene spacer. Cell. 1986 May 9;45(3):431–443. doi: 10.1016/0092-8674(86)90329-6. [DOI] [PubMed] [Google Scholar]
- Miesfeld R., Arnheim N. Species-specific rDNA transcription is due to promoter-specific binding factors. Mol Cell Biol. 1984 Feb;4(2):221–227. doi: 10.1128/mcb.4.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y., Financsek I., Kominami R., Muramatsu M. Fractionation and reconstitution of factors required for accurate transcription of mammalian ribosomal RNA genes: identification of a species-dependent initiation factor. Nucleic Acids Res. 1982 Nov 11;10(21):6659–6670. doi: 10.1093/nar/10.21.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmickel R. D., Waterson J. R., Knoller M., Szura L. L., Wilson G. N. HeLa cell identification by analysis of ribosomal DNA segment patterns generated by endonuclease restriction. Am J Hum Genet. 1980 Nov;32(6):890–897. [PMC free article] [PubMed] [Google Scholar]
- Simeone A., La Volpe A., Boncinelli E. Nucleotide sequence of a complete ribosomal spacer of D. melanogaster. Nucleic Acids Res. 1985 Feb 25;13(4):1089–1101. doi: 10.1093/nar/13.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss F., Varshavsky A. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984 Jul;37(3):889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Tautz D., Dover G. A. Transcription of the tandem array of ribosomal DNA in Drosophila melanogaster does not terminate at any fixed point. EMBO J. 1986 Jun;5(6):1267–1273. doi: 10.1002/j.1460-2075.1986.tb04356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil P. A., Luse D. S., Segall J., Roeder R. G. Selective and accurate initiation of transcription at the Ad2 major late promotor in a soluble system dependent on purified RNA polymerase II and DNA. Cell. 1979 Oct;18(2):469–484. doi: 10.1016/0092-8674(79)90065-5. [DOI] [PubMed] [Google Scholar]
- Yavachev L. P., Georgiev O. I., Braga E. A., Avdonina T. A., Bogomolova A. E., Zhurkin V. B., Nosikov V. V., Hadjiolov A. A. Nucleotide sequence analysis of the spacer regions flanking the rat rRNA transcription unit and identification of repetitive elements. Nucleic Acids Res. 1986 Mar 25;14(6):2799–2810. doi: 10.1093/nar/14.6.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]