Abstract
The role of the Salmonella Pathogenicity Islands (SPIs) in pathogenesis of Salmonella enterica Typhimurium infection in the chicken is poorly studied, while many studies have been completed in murine models. The Type VI Secretion System (T6SS) is a recently described protein secretion system in Gram-negative bacteria. The genus Salmonella contains five phylogenetically distinct T6SS encoded in differentially distributed genomic islands. S. Typhimurium harbors a T6SS encoded in SPI-6 (T6SSSPI-6), which contributes to the ability of Salmonella to colonize mice. On the other hand, serotype Gallinarum harbors a T6SS encoded in SPI-19 (T6SSSPI-19) that is required for colonization of chicks. In this work, we investigated the role of T6SSSPI-6 in infection of chicks by S. Typhimurium. Oral infection of White Leghorn chicks showed that a ΔT6SSSPI-6 mutant had reduced colonization of the gut and internal organs, compared with the wild-type strain. Transfer of the intact T6SSSPI-6 gene cluster into the T6SS mutant restored bacterial colonization. In addition, our results showed that transfer of T6SSSPI-19 from S. Gallinarum to the ΔT6SSSPI-6 mutant of S. Typhimurium not only complemented the colonization defect but also resulted in a transient increase in the colonization of the cecum and ileum of chicks at days 1 and 3 post-infection. Our data indicates that T6SSSPI-6 contributes to chicken colonization and suggests that both T6SSSPI-6 and T6SSSPI-19 perform similar functions in vivo despite belonging to different phylogenetic families.
Introduction
Nontyphoidal Salmonella gastroenteritis has an estimated global burden of 93.8 million cases per year, of which 80.3 million cases are likely to be food-borne [1]. The most prevalent serovars responsible for food-borne salmonellosis are S. enterica serovar Enteritidis and S. enterica serovar Typhimurium [2]. Salmonella enterica serovar Typhimurium (S. Typhimurium) is a broad host-range pathogen able to infect humans, mice and birds. In mice, this serovar causes a systemic infection similar to human typhoid fever that results from infection with serovar Typhi (as well as Paratyphi A, B, and C) [3], [4]; for this reason the murine model has been widely used to study the pathogenesis of Salmonella infection. In humans however, S. Typhimurium causes self-limiting gastroenteritis characterized by abdominal pain, vomiting and inflammatory diarrhea [5]. In contrast, this pathogen is able to colonize the chicken without clinical symptoms, and is thus a major vehicle for transmission of salmonellosis to humans.
Studies conducted using murine models of infection and in vitro cell culture systems have identified numerous genes required to establish a successful infection by S. Typhimurium. Most genes are clustered in genomic islands known as Salmonella Pathogenicity Islands (SPIs) [6]–[10]. Of the five SPIs (SPI-1 to SPI-5) common to all serovars of Salmonella enterica, the SPI-1 and SPI-2 are the two major virulence determinants of Salmonella. Each of these SPIs encodes two different type III secretion systems (T3SS) that deliver effector proteins directly into the cytoplasm of eukaryotic cells [11], [12]. The T3SSSPI-1 is mainly involved in invasion of intestinal epithelial cells [13], [14] but it is also required for intracellular proliferation and for the biogenesis of the Salmonella containing vacuole inside infected cells [15], [16]. The T3SSSPI-2 is essential for survival within phagocytic cells and systemic infection [17].
Studies on the role of the SPIs in the pathogenesis of S. Typhimurium infection in the chicken are few and are sometimes contradictory. While some authors reported that both T3SSSPI-1 and T3SSSPI-2 are required for the infection process [18]–[21], one study showed that neither T3SSSPI-1 nor T3SSSPI-2 is critical for colonization of chickens [9]. One report directly compared the intestinal and systemic colonization of Salmonella-resistant mice and one-week-old chickens by S. Typhimurium [22]. Infected chicks had very few organisms in internal organs and no symptoms of systemic effects, while in mice, spleen and liver were colonized by bacteria and showed significant enlargement. Furthermore, colonization of the intestine had a different dynamic in the chicken versus the mice models of infection, as SPI-1 was important for association to the intestinal epithelium of the chicken rather than for invasion, as is the case in mice [22]. From these studies, it is evident that the murine model has a limited applicability to Salmonella infection of the chicken, and that genes in addition to the highly conserved SPIs are required for chicken colonization and systemic spread.
Type VI secretion systems participate in a variety of different processes, ranging from inter-bacterial relationships to pathogenesis [23]–[27]. Gram-negative bacteria carrying T6SS clusters include human, animal and plant pathogens [28]–[34]. The genus Salmonella contains five phylogenetically distinct T6SS loci; four of them are differentially distributed among serovars of S. enterica, while the fifth T6SS is present in S. bongori [35], [36]. Two of these clusters, T6SSSPI-6 and T6SSSPI-19, have been linked to Salmonella pathogenesis. T6SSSPI-6 is required for intracellular replication in macrophages and systemic dissemination in mice by S. Typhimurium [37]–[41] and S. Typhi [29], while T6SSSPI-19 contributes to colonization of the gastrointestinal tract and internal organs of chickens by S. Gallinarum strain 287/1 [42].
In this study we have investigated the contribution of T6SSSPI-6 to S. Typhimurium ability to colonize the gastrointestinal tract and internal organs of White Leghorn chicks. We have also addressed whether T6SSSPI-19 of S. Gallinarum can rescue the colonization defect of a S. Typhimurium mutant lacking T6SSSPI-6. Through competitive index experiments we demonstrate that T6SSSPI-6 is crucial to gastrointestinal colonization and systemic spread of S. Typhimurium in chicks. In addition, we show that transfer of T6SSSPI-19 restores the colonization defect of a mutant lacking T6SSSPI-6, indicating that both T6SS perform similar functions in vivo despite belonging to different phylogenetic families.
Materials and Methods
Bacteria and Growth Conditions
The bacterial strains used in this work are listed in Table 1 . Bacteria were routinely cultivated in LB broth (10 g/l tryptone, 5 g/l yeast extract, 5 g/l NaCl) at 37°C with aeration or on LB plates (15 g/l agar) supplemented with the appropriate antibiotic at the following concentrations: Ampicillin (Amp), 100 µg/ml; Kanamycin (Kan), 50 µg/ml; Chloramphenicol (Cam), 20 µg/ml; Trimethoprim (Tm), 100 µg/ml; Spectinomycin (Sp), 250 µg/ml.
Table 1. Strains and plasmids used in this study.
| Strains | Features | Source of reference |
| Escherichia coli | ||
| DH5α | F-Φ80lacZΔM15Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk -, mk +) phoA supE44 thi-1 gyrA96 relA1 λ- | Laboratory collection |
| EC100D pir-116 | F-mcrAΔ(mrr-hsdRMS-mcrBC)Φ 80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ- rpsL (StrR) nupG pir-116(DHFR) | Laboratory collection |
| EC100D pir-116/R995+SPI-6 | Strain with T6SSSPI-6 from S. Typhimurium cloned in plasmid R995 | This study |
| EC100D pir-116/R995+SPI-19 | Strain with T6SSSPI-19 from S. Gallinarum cloned in plasmid R995 | [42] |
| DH5α/R995 | Strain harboring an empty R995 vector | This study |
| DH5α/R995-VC6 | Strain containing a derivative of plasmid R995 with a 1,209 bp DNAfragment of T6SSSPI-6 cloned from S. Typhimurium | This study |
| Salmonella Typhimurium | ||
| 14028s | Wild-type strain | Laboratory collection |
| MTM753 | 14028s ΔphoN | This study |
| MTM35 | 14028s ΔSPI-6 T6SS | This study |
| MTM2640 | 14028s ΔclpV | This study |
| WT/R995 | 14028s containing an empty R995 vector | This study |
| MTM35R | MTM35 harboring R995 plasmid | This study |
| MTM35R6 | MTM35 complemented with plasmid R995+SPI-6 | This study |
| MTM35R19 | MTM35 complemented with plasmid R995+SPI-19 | This study |
| Plasmids | ||
| pKD46 | bla PBAD bet exo pSC101 oriTs, AmpR | [43] |
| pEKA30 | IncQ plasmid that constitutively express Cre recombinase, AmpR | [44] |
| pCLF2 | Red-swap redesigned vector, CamR | [50] |
| pCLF4 | Red-swap redesigned vector, KanR | [50] |
| pVEX1212 | Suicide vector harboring a loxP site followed by a SpR cassette | [44] |
| pVEX2212 | Suicide vector harboring a loxP site followed by a CamR cassette | [44] |
| R995 | Self-transmissible broad-host range IncP vector | [44] |
| R995-VC6 | A derivative of plasmid R995 with a 1,209 bp DNA fragment of T6SSSPI-6cloned from S. Typhimurium | This study |
| R995+SPI-6 | T6SSSPI-6 cluster from S. Typhimurium 14028s cloned in vector R995 | This study |
| R995+SPI-19 | T6SSSPI-19 cluster from S. Gallinarum 287/91 cloned in vector R995 | [42] |
DNA Methods
DNA manipulations were performed using standard protocols. Plasmid DNA was isolated from overnight cultures using the QIAprep Spin Miniprep Kit (QIAGEN), according to the manufactureŕs instructions. Genomic DNA was isolated from overnight cultures utilizing the GenElute Bacterial Genomic DNA kit (Sigma) according to the manufactureŕs instructions. PCR products were purified using the QIAquick PCR Purification Kit (QIAGEN). XbaI restriction enzyme (Fermentas) and T4 DNA ligase (New England Biolabs) were used as per manufacturer instructions. DNA samples were routinely analyzed by electrophoresis in 1% agarose gels (1X Tris-acetate-EDTA buffer) and visualized under UV light after ethidium bromide staining.
PCR Amplifications
Primers were designed using the Vector NTI Advance 10.0 software (Invitrogen) and are listed in Table 2 . PCR amplifications were performed in a MultiGene TC9600-G thermal cycler (LabNet), using GoTaq Flexi DNA Polymerase (Promega). Conditions for tiling-PCR amplification were as follows: 3 min at 94°C followed by 30 cycles of incubations at 94°C for 30 s, 58°C for 30 s, and 72°C for 4 min, followed by a final extension step at 72°C for 7 min. Conditions for standard PCR amplification were as follows: 3 min at 94°C followed by 30 cycles of incubations at 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min, followed by a final extension step at 72°C for 5 min. When required, PCR products were purified by using the QIAquick PCR purification kit (Qiagen).
Table 2. Primers used in this study.
| Primer | Sequencea |
| Mutagenesis | |
| SPI-6_T6SS_(H1+P1) | AGGGTGTTTTTATACATCCTGTGAAGTAAAAAAAACCGTAGTGTAGGCTGGAGCTGCTTC |
| SPI-6_T6SS_(H2+P2) | GTGAACATGGCACATTAATTTGAAGCAGCTCTCATCCGGTCATATGAATATCCTCCTTAG |
| SPI-6_OUT5 | CCGAAGTGTATCTGGCGATGA |
| STM0272_(H1+P1) | GGCATAACACATGGAAACTCCTGTTTCACGCAGTGCGTTGGTGTAGGCTGGAGC TGCTTC |
| STM0272_(H2+P2) | ACGGCCGGTTTCAGCAAACGATCTCAAAAACAATCTGCTCCATATGAATATCCTCCTTAG |
| STM0272_OUT5 | GGCGGCAGTAAATACGATGT |
| STM_ΔphoN_(H1+P1) | GTGAGTCTTTATGAAAAGTCGTTATTTAGTATTTTTTCTAGTGTAGGCTGGAGCTGCTTC |
| STM_ΔphoN_(H2+P2) | ACTTTCACCTTCAGTAATTAAGTTCGGGGTGATCTTCTTTCATATGAATATCCTCCTTAG |
| STM_ΔphoN_OUT5 | TTGCCTGATCCGGAGTGA |
| K1 | CAGTCATAGCCGAATAGCCT |
| C3 | CAGCTGAACGGTCTGGTTATAGG |
| VEX Capture | |
| STM0266_VEX_H1_U1 | GGCCACGTGGGCCGTGCACCTTAAGCTT |
| STM0266_VEX_H2_U2 | GAGGTTATTCATGTCAACAGGATTACGTTTCACACTGGAGGTGCAGGCTGGAGCTGCTTC |
| STM0298_VEX_H1_D1 | GGGGAGGTTGTGCGACGTTTGCATAATCCAGCAAGAACTGGGTTTAACGGTTGTGGACAACAAGCCAGGG |
| STM0298_VEX_H2_D2 | ACACAGGCCAGACTGATTATACAGGCATGAAAAAGCTCTCCAGGTCGACGTCCCATGGCCATTCGAATTC |
| STM_VC_OUT5 | GCTCTAGACCGGAGGGGTTATCTTTTCC |
| STM_VC_OUT3 | GCTCTAGATTGAAGCAGCTCTCATCCGG |
| 5trfA | ACGTCCTTGTTGACGTGGAAAATGACCTTG |
| 3trfA | CCGGAAGGCATACAGGCAAGAACTGATCG |
| SPI-6_OUT_DOWN | AAACGGGTCTATTTACAGGGGCAC |
| Tiling-PCR | |
| 1_T6SS_SPI-6_FOR | TTCAAGAAGTTCCACCGTCTATCG |
| 1_T6SS_SPI-6_REV | ACCTGTTTGAGCTGCTACATACCAG |
| 2_T6SS_SPI-6_FOR | CATTCAGTTCGCCGTCAAAGTG |
| 2_T6SS_SPI-6_REV | CCGCTGCGAATTTTGTTATCG |
| 3_T6SS_SPI-6_FOR | CCACGTTCTTCGGCATTACCAG |
| 3_T6SS_SPI-6_REV | CGGTGTTGTAAACCAGATGCTCC |
| 4_T6SS_SPI-6_FOR | AGACGCTGGCGAACACGATC |
| 4_T6SS_SPI-6_REV | TAAGCACTGGCCGTAGCTCTGG |
| 5_T6SS_SPI-6_FOR | GCAGCCATCCTTTGCACAAG |
| 5_T6SS_SPI-6_REV | GGTTGTGTTATTGGCGGCTTC |
| 6_T6SS_SPI-6_FOR | TATGCGATCAGGCGAACCTG |
| 6_T6SS_SPI-6_REV | TCTTCCTGTAACCGGGTATCCAG |
| 7_T6SS_SPI-6_FOR | GGTTGGATCAGGGACTGGATACC |
| 7_T6SS_SPI-6_REV | CGTAACCCTCAACATCCTGCG |
| 8_T6SS_SPI-6_FOR | AAAGCACCGGTGAATGTGGCTG |
| 8_T6SS_SPI-6_REV | TCGGTGTGGTCATCCTTACGGG |
| 9_T6SS_SPI-6_FOR | TGTCAGCACCAACAGTCGCC |
| 9_T6SS_SPI-6_REV | CGCCCTTCGATAGAATCTGGC |
| 10_T6SS_SPI-6_FOR | TAGTAGGGCCAGATTCTATCGAAGG |
| 10_T6SS_SPI-6_REV | CCCTCCGGCTTTTACACATTATTC |
Italics indicate the region that anneals to the 5′ or 3′ end of the antibiotic resistance cassette used for the mutagenesis. Underline indicates XbaI restriction sites used for cloning an internal region of homology to T6SS of SPI-6 into R995 plasmid.
Construction of S. Typhimurium Mutant Strains
Mutants of S. Typhimurium carrying deletions of the T6SSSPI-6 gene cluster and the clpV (STM0272) or phoN genes were constructed using the Lambda-Red System [43]. The oligonucleotides used for the mutagenesis are shown in Table 2 and the sequences of plasmids pCLF2 and pCLF4 used as templates are available in GenBank (accession numbers HM047089 and EU629214.1, respectively). The correct insertion of the resistance cassettes was checked by PCR, and confirmed mutations were moved to a clean genetic background by generalized transduction using the high-frequency transducing phage P22 HT105/1 int-201. To be able to identify wild type versus mutant colonies in the mixed competition experiments, the S. Typhimurium ΔphoN mutant was used as the wild type strain. phoN+ and phoN- strains can be distinguished by blue-white selection on 5-bromo-4-chloro-3-indolyl phosphate (XP) containing media, phoN- strains form white colonies while phoN+ strains appear blue. Mutations in phoN do not affect the ability of S. Typhimurium to colonize and persist in the chick [22].
Cloning of S. Typhimurium SPI-6 by VEX-Capture
Cloning of a ∼39 Kb fragment containing the T6SSSPI-6 gene cluster from S. Typhimurium 14028s onto plasmid R995 was performed by the VEX-Capture technique for the targeted excision and cloning of large DNA fragments [44]. First, loxP sites were inserted at each side of the targeted genomic region by homologous recombination of PCR products by the Lambda-Red system, using as templates the plasmids pVEX1212 and pVEX2212 that encode Sp and Cam resistance cassettes, respectively. Correct insertion of loxP sites was confirmed by PCR using primers SPI-6_OUT5 and STM0266_VEX_H2_U2 for loxP insertion located in the upstream region of the T6SS cluster, and primers SPI-6_OUT_DOWN and STM0298_VEX_H2_D2 for the downstream loxP insertion. This cluster was excised from the chromosome as a non-replicating circular DNA molecule by specific recombination of loxP sites mediated by the action of Cre recombinase encoded in plasmid pEKA30. This intermediate was captured into the R995-VC6 vector by a homologous recombination event, producing the R995+SPI-6 plasmid. The R995-VC6 plasmid contains a 1,209 bp internal region of homology to the T6SSSPI-6 cluster, cloned by PCR amplification with primers STM_VC_OUT5 and STM_VC_OUT3 ( Table 2 ).
Plasmid R995+SPI-6 was transferred to E. coli strain EC100D pir-116 by conjugation and the presence and structural integrity of the T6SSSPI-6 gene cluster cloned onto R995 was verified by tiling-PCR analysis in order to amplify ten fragments that cover the entire T6SS region (Figure S1). For competitive infections in chickens, the in vivo stability of plasmids R995 and R995+SPI-6 was assessed in each organ at each time point studied. No differences were observed on colony forming units (CFU) indicating that R995 and its derivatives are highly stable in vivo.
Experimental Infections of Chickens
S. Typhimurium strains were grown aerobically at 42°C for 16 hours in LB broth. This temperature of incubation was used because it corresponds to the body temperature of chicks. For single and competitive infections, fifteen 4-day old unsexed White Leghorn chicks were orally inoculated with 109 CFU of a single strain or with an equal mixture of the strains to be tested in a volume of 100 µl of sterile PBS. The inoculum was serially diluted and plated to determine the titer and input ratio. Five birds from the infected group were sacrificed by asphyxiation with CO2 on days 1, 3 and 9 post-infection. Ileum, cecum (including contents), liver and spleen were collected. These organs were homogenized in sterile PBS and serial ten-fold dilutions spread on LB agar plates containing the appropriate antibiotics for determination of CFU. For histopathological analysis, the cecum and liver of experimental animals were fixed in 10% formalin for 24 h followed by incubation in 70% ethanol and then embedded in paraffin. The samples were stained with hematoxylin and eosin and 10 fields per sample were examined and scored by a trained veterinary pathologist to determine histopathological changes.
Statistical Analysis
Data obtained from competitive infection experiments were calculated as a mean ratio of logarithmically converted CFU of mutant to wild type normalized to the input ratio. Error bars indicate standard error. Statistical significance was determined using a two-tailed Students t-test. P values of <0.05 were considered statistically significant (SPSS software, SPSS, Inc., Chicago, IL).
Ethics Statement
All animal experiments in this study were approved by the Texas A&M University Institutional Animal Care and Use Committee (TAMU AUP# 2010-38) and were carried out in accordance with the Guide to the Care and Use of Laboratory Animals, the Public Health Service Policy on the Human Care and Use of Laboratory Animals.
Results
The T6SS Encoded in SPI-6 Contributes to Efficient Colonization of the Avian Host by Salmonella Typhimurium
Single infections and competitive index experiments were performed to determine the contribution of the SPI-6 T6SS to intestinal and systemic colonization of chicks by S. Typhimurium.
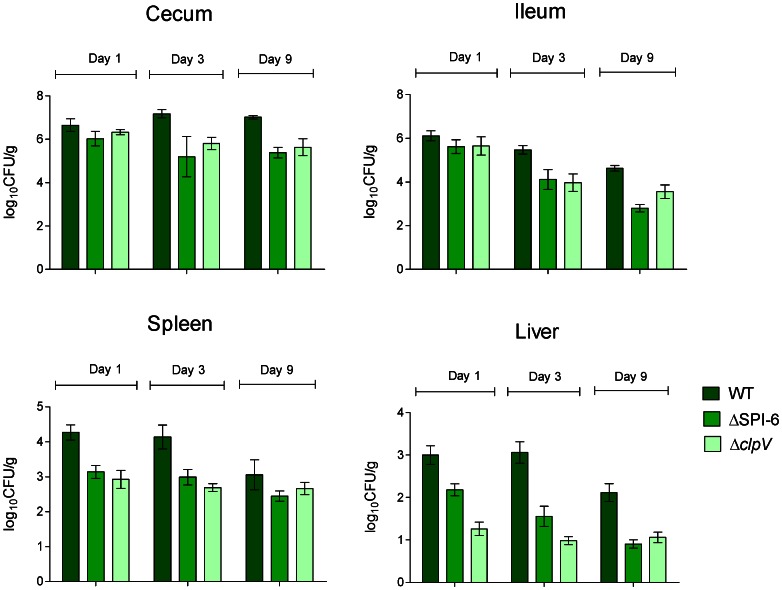
For single infections, White Leghorn chicks were orally-infected with either the wild-type strain, a ΔT6SSSPI-6 mutant (MTM35) or a ΔclpV deletion mutant (MTM2640) and colonization of the cecum, ileum, liver and spleen was evaluated over 9 days of infection. ClpV, a conserved structural component of the T6SS that belongs to Clp/Hsp100 AAA+ of ATPase superfamily, is required for the activity of the secretion system [45], [46]. As shown in Figure 1 , the cecum and ileum of chicks infected with the wild-type strain were heavily colonized at all time points, while the liver and spleen were only lightly colonized, as reported previously [22]. Interestingly, both the ΔT6SSSPI-6 and ΔclpV mutant strains showed an overall lower degree of colonization of the cecum and ileum from day 3 post-infection and of the liver and spleen from day one post-infection, suggesting a role for the SPI-6 T6SS in chick colonization.
Figure 1. Distribution of S. Typhimurium 14028 s and SPI-6 T6SS mutants in the gastrointestinal tract and internal organs of orally infected chickens.
Four-day-old White Leghorn chicks were infected by gavage with 109 CFU of either the wild-type S. Typhimurium 14028s, the ΔT6SSSPI-6 mutant or the ΔclpV mutant strains. After 1, 3 and 9 days post-infection, the chicks were humanely euthanized and the ileum, cecum, liver and spleen were aseptically removed. Tissues were homogenized and viable bacterial counts were determined. Data are mean values of log10 CFU/g of tissue, from five animals at each time point.
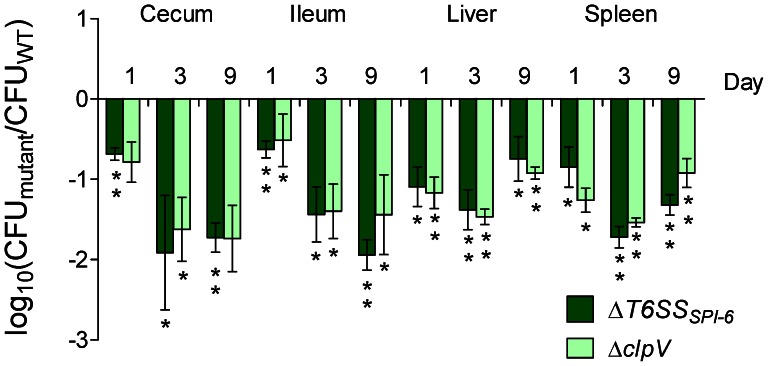
In order to determine the competitive fitness within the host, of each mutant strain, competitive index experiments were performed. White leghorn chicks were orally infected with a mixture of each mutant with the wild-type strain at a 1∶1 ratio and colonization of each organ was evaluated over 9 days of infection. As shown in Figure 2 , a strong colonization defect was observed for both the ΔT6SSSPI-6 and ΔclpV mutants during intestinal and systemic colonization from day 1 post-infection. This markedly attenuated phenotype was more pronounced at the third day post-infection and it was maintained throughout day 9 in each organ analyzed. These results indicate that S. Typhimurium requires a functional T6SS to efficiently colonize the avian host.
Figure 2. In vivo competition between ΔT6SSSPI-6 and ΔclpV deletion mutants and the wild type S. Typhimurium strain 14028 s.
Fifteen four-day-old White Leghorn chicks were infected intragastrically by gavage with 109 CFU of a mixture at a 1∶1 ratio of the respective mutant strain and the wild type S. Typhimurium 14028s. At 1, 3 and 9 days post-infection groups of 5 chicks were sacrificed and organs were excised, homogenized, and serially diluted to determine bacterial loads. Bars represent the geometric mean of the log ratio of the mutant CFU/wild type CFU, normalized to the inoculum ratio. Error bars denote standard error. Statistical significance was determined using a two-tailed Student’s t test, and asterisks indicate that normalized output ratios were significantly statistically different from the equivalent ratio in the inoculum (*P<0.05; **P<0.001).
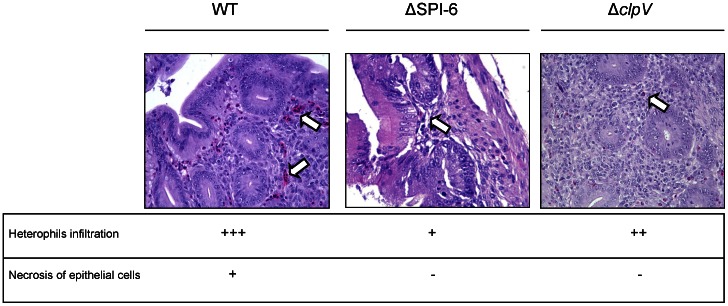
Histopathological analysis of the cecum and liver from infected birds was performed to determine whether or not this attenuated phenotype was accompanied by tissue damage and/or signs of an inflammatory response. Single infections were performed as described above, and 3 days post infection the chicks were sacrificed and each organ tested was excised, fixed, stained with hematoxylin and eosin, and analyzed for histopathological lesions. Significant pathological changes were observed in the cecum of chicks infected with the wild-type strain. Among these changes, focal necrosis of the mucosal epithelial cells and heterophil infiltration were evident, indicating a strong inflammatory response induced by S. Typhimurium 14028s ( Figure 3, left panel). In contrast, chicks infected with either the ΔT6SSSPI-6 or ΔclpV mutant strains showed a considerable lower level of heterophil infiltration in the cecum, with no signs of necrosis of the epithelial cells ( Figure 3 , central and right panels, respectively). No significant histopathological differences were found in livers infected with either the wild-type or the T6SS mutants (data not shown). Absence of lesions in the liver are most probably due to the low levels of bacterial colonization of internal organs by both the wild-type and T6SS mutant strains ( Figure 1 ).
Figure 3. Histopathological changes in the cecum of infected chicks at day 3 post-infection.
Groups of 3 White Leghorn chicks were inoculated intragastrically by gavage with 109 CFU of the wild type S. Typhimurium 14028s strain, the ΔT6SSSPI-6 mutant strain or the ΔclpV mutant strain. At day 3 post-infection the chicks were sacrificed and the ceca were excised, fixed, stained with hematoxylin and eosin, and analyzed for histopathological lesions. Representative images of stained sections (400X) and scores for histopathological lesions in the cecum of infected chicks are shown (-, no changes; +, mild; ++, strong; +++, severe). White arrows indicate heterophil infiltration.
The Colonization Defect of the ΔT6SSSPI-6 Mutant is Complemented by Transfer of the T6SSSPI-6 Gene Cluster
To directly link the absence of the T6SSSPI-6 gene cluster to the phenotype of the ΔT6SSSPI-6 mutant, the complete 35,921 base pair T6SS gene cluster was returned to the mutant on the self-transmissible broad-host range R995 vector. The capture of the entire T6SSSPI-6 gene cluster was performed using the VEX-Capture method [44] and confirmed by tiling PCR analysis (Figure S1).
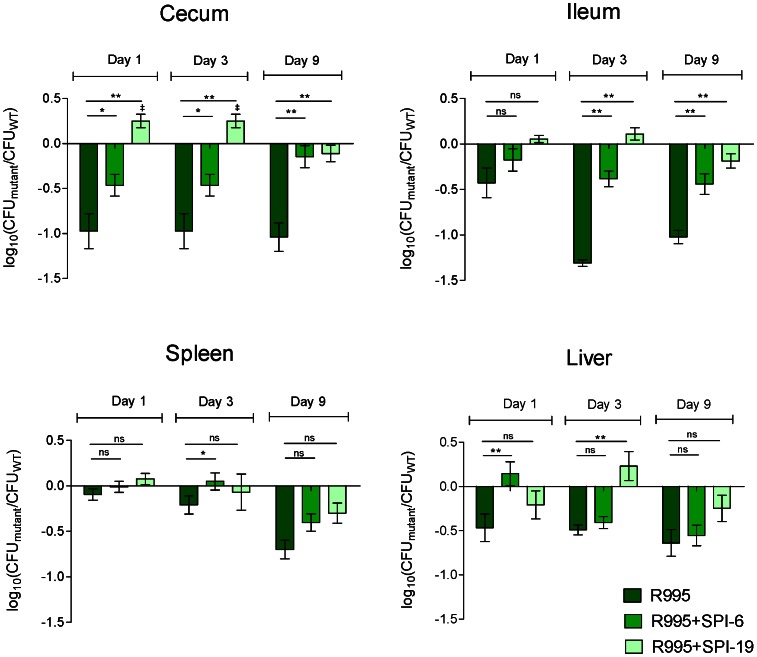
The complemented strain (MTM35R6) was tested in a competition experiment against the ΔT6SSSPI-6 mutant and the wild type, each bearing the empty vector (MTM35/R995 and WT/R995, respectively) and colonization was determined at days 1, 3, and 9 post infection. As shown in Figure 4 , transfer of the T6SSSPI-6 gene cluster to the ΔT6SSSPI-6 mutant restored its ability to colonize the cecum and the ileum at all time points. On the other hand, in the spleen and liver, the results were not conclusive due to a very low and heterogeneous colonization of these deeper tissues by S. Typhimurium harbouring the R995 plasmid (data not shown). Nevertheless, complementation of the defective phenotype of the ΔT6SSSPI-6 mutant in the gastrointestinal tract supports the contribution of T6SSSPI-6 in chicken colonization.
Figure 4. In vivo competition between the ΔT6SSSPI-6 mutants complemented in trans with T6SSSPI-6 or T6SSSPI-19 and the wild type S. Typhimurium 14028 s.
Fifteen four-day-old White Leghorn chicks were orally infected with 109 CFU of a mixture at a 1∶1 ratio of strains WT/R995, ΔT6SSSPI-6/R995+SPI-6 and ΔT6SSSPI-6/R995+SPI-19. At 1, 3 and 9 days post-infection groups of five chicks were sacrificed and the organs were excised, homogenized, and serially diluted for determination of bacterial loads. Bars represent the geometric mean of the log converted ratio of the mutant CFU to the wild type CFU normalized to the equivalent ratio in the inoculum. Error bars denote standard error. Statistical significance was determined using a two-tailed Student’s t test, and asterisks indicate statistically significant differences between normalized output ratios (*P<0.05). ‡Indicate statistically significant differences between normalized output ratios and the equivalent ratio in the inoculum (‡ P<0.05).
The SPI-19 T6SS from S. Gallinarum Restores the Colonization Defect of the SPI-6 T6SS Mutant Strain
In a previous study, we reported that T6SSSPI-19 contributes to efficient colonization of infected chicks by S. Gallinarum 287/91 [42]. T6SSSPI-6 and T6SSSPI-19 have different evolutionary histories, and were probably acquired at different times during Salmonella evolution [35], [36]. Because both T6SS are relevant for Salmonella colonization of infected chicks, we examined the possibility that both T6SS could contribute to chicken colonization in a similar extent. To test whether T6SSSPI-19 can restore the ability of the S. Typhimurium ΔT6SSSPI-6 mutant to efficiently colonize the avian host, the complete T6SSSPI-19 gene cluster captured from S. Gallinarum 287/91 in the R995 plasmid was transferred to S. Typhimurium ΔT6SSSPI-6 by conjugation. The resulting strain (MTM35R19) was tested in a competition experiment with the wild-type S. Typhimurium strain bearing the empty R995 vector (WT/R995). The results showed that introduction of the T6SSSPI-19 complemented the colonization defect of the ΔT6SSSPI-6 mutant in both the cecum and ileum ( Figure 4 ). Interestingly, at days 1 and 3 post-infection, the cross-complemented strain colonized the cecum to higher levels than the wild-type strain. Analysis of the competitive fitness of the complemented strains in the spleen and liver did not show statistically significant differences; this was due to the heterogeneous and low colonization levels of systemic organs reached by Salmonellae in the chicken, as previously reported [22].
Discussion
We previously reported that Salmonella encodes five distinct T6SS differentially distributed among different serotypes [35], [36]. Two of these systems, encoded in the SPI-6 and SPI-19, have been linked to the ability of serotypes Typhimurium and Gallinarum to efficiently infect mice and chickens, respectively [37], [42], [47]. Even though most of our knowledge regarding S. Typhimurium pathogenesis comes from murine models of infection, recent reports have highlighted the limited applicability of this model when it comes to extrapolating conclusions regarding other hosts, including the chicken.
In this work, we evaluated the contribution of T6SSSPI-6 to the ability of S. Typhimurium 14028s to colonize the gastrointestinal tract and internal organs of White Leghorn chicks. Competitive index experiments demonstrated that the T6SSSPI-6 gene cluster was necessary for efficient colonization of the cecum, ileum, spleen and liver from day 1 post-infection. A similar colonization defect was observed for a mutant lacking the T6SS-essential component ClpV. Interestingly, the colonization defects were more pronounced at days 3 and 9 post-infection suggesting that mutants in the ΔT6SSSPI-6 do not persist well.
Histopathological analyses revealed that the attenuated phenotypes of the mutants were accompanied by changes in the inflammatory response in the cecum. Chicks infected with SPI-6 T6SS mutant strains showed considerable less inflammation and necrosis in the cecum in comparison with those infected with the wild-type strain. This could be due to the lower level of colonization of the cecum by the SPI-6 T6SS mutant compared to the wild type, or that this secretion system effectively contributes to the inflammatory response generated by S. Typhimurium infection. Further experiments will be needed to clarify these issues.
To confirm that T6SSSPI-6 was responsible for these phenotypes, the entire gene cluster was cloned and introduced in the ΔT6SSSPI-6 mutant. Although complementation was not observed in the spleen and liver, transfer of the T6SS gene cluster complemented the colonization defect of the mutant in the cecum and ileum throughout infection, suggesting a critical role for T6SSSPI-6 in the gastrointestinal phase of infection. In this context, Sivula et al. have shown that S. Typhimurium preferentially colonize the cecum in order to maintain a long-term persistence in chicks [22]. Therefore, T6SSSPI-6 may be contributing to this critical phase of the infectious process.
A role for T6SS in colonization of the gastrointestinal tract is not unexpected. Several T6SS have been linked to antibacterial killing through delivery of toxins to susceptible Gram-negative bacteria, and several authors have proposed that T6SS could contribute to bacterial adaptation and competition for new niches, including animal hosts [23]–[26], [48]. Therefore, it is possible that the defect observed in colonization of the ileum and cecum of the T6SS mutant is due to an inability of this mutant to compete with normal flora of the chicken gut. Further experiments will be needed to test this hypothesis.
On the other hand, a recent report has pointed out a role for T6SSSPI-6 in the intracellular survival of S. Typhimurium in murine macrophages [37]. Our data indicate that this secretion system is also needed for colonization of the internal organs of the chicken, suggesting a role for T6SSSPI-6 in intracellular survival within avian macrophages. Hence, the T6SSSPI-6 might contribute to both competition with the normal intestinal flora and survival within phagocytic cells.
We have previously reported that a phylogenetically distinct T6SS encoded in SPI-19, is necessary for the efficient colonization of the intestinal tract and systemic organs of chicks, and for survival of serotype Gallinarum in cultured avian macrophages [42], [49]. Because the phenotypes observed for the ΔT6SSSPI-6 mutant were similar to those exhibited by a ΔT6SSSPI-19 mutant of Gallinarum, we hypothesized that both systems could perform similar functions in chicken infection. Transfer of the T6SSSPI-19 gene cluster to the ΔT6SSSPI-6 mutant complemented the colonization defect of this strain in the ileum and cecum. Moreover, it caused an advantage for colonization of cecum at days 1 and 3 post-infection. These results indicate that both T6SS, despite their different evolutionary histories, contribute to a similar extent to chicken colonization by Salmonella. This statement is supported by the fact that both SPI-6 and SPI-19 T6SS have been shown to be required for Salmonella intracellular survival within macrophages [37], [49].
Altogether, we have determined that T6SSSPI-6 contributes to chicken colonization by S. Typhimurium. Also, we show that T6SSSPI-19 from the avian-adapted serotype Gallinarum is able to replace T6SSSPI-6, suggesting a broad role for these secretion systems in Salmonella host colonization. Most interestingly, our results indicate that T6SSSPI-19 confers an advantage to S. Typhimurium to colonize the gastrointestinal tract of the chicks early in infection.
Supporting Information
In vivo cloning of T6SSSPI-6 from S . Typhimurium 14028 s. (A) Scheme of the VEX-Capture procedure: loxP sites were inserted in the chromosome of S. Typhimurium 14028s at each side of the T6SSSPI-6 gene cluster through homologous recombination of PCR products using the Lambda-Red system. In presence of pEKA30, a plasmid that constitutively expresses the Cre recombinase, the T6SS cluster was excised from the chromosome as a non-replicative, circular DNA intermediate that was captured through homologous recombination in R995-VC6, a derivative of R995 plasmid harboring an internal region of homology to T6SSSPI-6. (B) Tiling-PCR analysis of the T6SSSPI-6 gene cluster cloned onto the R995 plasmid. Specific primers were designed to amplify ten fragments that cover the entire T6SSSPI-6 region and whose lengths vary between 3,298 and 4,274 bp.
(TIF)
Acknowledgments
We thank James W. Wilson for generous gift of bacterial strains and plasmids during the implementation of VEX-Capture technique, and Lydia Bogomolnaya, Marissa Talamantes and Claudia Lopez for technical assistance.
Funding Statement
This work was supported by grant 1100092 from Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT), Chile. CJB was supported by Postdoctoral Fellowship 3120175 from FONDECYT. David Pezoa was supported by fellowships from FULBRIGHT, “Beca Doctorado Nacional 2009” CONICYT (N°21090041), “Beca de Apoyo a la Realización de Tesis Doctoral 2012” CONICYT (N° AT-21121297) and from “Beca de Pasantías Doctorales en el Extranjero 2011” CHILE GRANT (N°75110062 BCH-3). CAS was supported by grant 1110172 from FONDECYT. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, et al. (2010) The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50: 882–889. [DOI] [PubMed] [Google Scholar]
- 2.WHO Global Foodborne Infections Network Country Databank www.who.int/salmsurv.
- 3. House D, Bishop A, Parry C, Dougan G, Wain J (2001) Typhoid fever: pathogenesis and disease. Curr Opin Infect Dis 14: 573–578. [DOI] [PubMed] [Google Scholar]
- 4. Jones BD, Falkow S (1996) Salmonellosis: host immune responses and bacterial virulence determinants. Annu Rev Immunol 14: 533–561. [DOI] [PubMed] [Google Scholar]
- 5. Santos RL, Raffatellu M, Bevins CL, Adams LG, Tukel C, et al. (2009) Life in the inflamed intestine, Salmonella style. Trends Microbiol 17: 498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hansen-Wester I, Hensel M (2001) Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect 3: 549–559. [DOI] [PubMed] [Google Scholar]
- 7. Smith RL, Kaczmarek MT, Kucharski LM, Maguire ME (1998) Magnesium transport in Salmonella typhimurium: regulation of mgtA and mgtCB during invasion of epithelial and macrophage cells. Microbiology 144 (Pt 7): 1835–1843. [DOI] [PubMed] [Google Scholar]
- 8. Dorsey CW, Laarakker MC, Humphries AD, Weening EH, Baumler AJ (2005) Salmonella enterica serotype Typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol Microbiol 57: 196–211. [DOI] [PubMed] [Google Scholar]
- 9. Morgan E, Campbell JD, Rowe SC, Bispham J, Stevens MP, et al. (2004) Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol Microbiol 54: 994–1010. [DOI] [PubMed] [Google Scholar]
- 10. Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, et al. (2010) Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc Natl Acad Sci U S A 107: 17733–17738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Galan JE (2001) Salmonella interactions with host cells: type III secretion at work. Annu Rev Cell Dev Biol 17: 53–86. [DOI] [PubMed] [Google Scholar]
- 12. Haraga A, Ohlson MB, Miller SI (2008) Salmonellae interplay with host cells. Nat Rev Microbiol 6: 53–66. [DOI] [PubMed] [Google Scholar]
- 13. Coombes BK, Coburn BA, Potter AA, Gomis S, Mirakhur K, et al. (2005) Analysis of the contribution of Salmonella pathogenicity islands 1 and 2 to enteric disease progression using a novel bovine ileal loop model and a murine model of infectious enterocolitis. Infect Immun 73: 7161–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hapfelmeier S, Ehrbar K, Stecher B, Barthel M, Kremer M, et al. (2004) Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun 72: 795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brawn LC, Hayward RD, Koronakis V (2007) Salmonella SPI1 effector SipA persists after entry and cooperates with a SPI2 effector to regulate phagosome maturation and intracellular replication. Cell Host Microbe 1: 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Steele-Mortimer O, Brumell JH, Knodler LA, Meresse S, Lopez A, et al. (2002) The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell Microbiol 4: 43–54. [DOI] [PubMed] [Google Scholar]
- 17. Kuhle V, Hensel M (2004) Cellular microbiology of intracellular Salmonella enterica: functions of the type III secretion system encoded by Salmonella pathogenicity island 2. Cell Mol Life Sci 61: 2812–2826. [DOI] [PubMed] [Google Scholar]
- 18. Jones MA, Hulme SD, Barrow PA, Wigley P (2007) The Salmonella pathogenicity island 1 and Salmonella pathogenicity island 2 type III secretion systems play a major role in pathogenesis of systemic disease and gastrointestinal tract colonization of Salmonella enterica serovar Typhimurium in the chicken. Avian Pathol 36: 199–203. [DOI] [PubMed] [Google Scholar]
- 19. Porter SB, Curtiss R, 3rd (1997) Effect of inv mutations on Salmonella virulence and colonization in 1-day-old White Leghorn chicks. Avian Dis 41: 45–57. [PubMed] [Google Scholar]
- 20. Dieye Y, Ameiss K, Mellata M, Curtiss R, 3rd (2009) The Salmonella Pathogenicity Island (SPI) 1 contributes more than SPI2 to the colonization of the chicken by Salmonella enterica serovar Typhimurium. BMC Microbiol 9: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Turner AK, Lovell MA, Hulme SD, Zhang-Barber L, Barrow PA (1998) Identification of Salmonella typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect Immun 66: 2099–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sivula CP, Bogomolnaya LM, Andrews-Polymenis HL (2008) A comparison of cecal colonization of Salmonella enterica serotype Typhimurium in white leghorn chicks and Salmonella-resistant mice. BMC Microbiol 8: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, et al. (2011) The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J Bacteriol 193: 6057–6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hood RD, Singh P, Hsu F, Guvener T, Carl MA, et al. (2010) A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7: 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S (2010) The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U S A 107: 19520–19524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, et al. (2010) Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 6(8): e1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schwarz S, Hood RD, Mougous JD (2010) What is type VI secretion doing in all those bugs? Trends Microbiol 18: 531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ma AT, Mekalanos JJ (2010) In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc Natl Acad Sci U S A. 107(9): 4365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang M, Luo Z, Du H, Xu S, Ni B, et al. (2011) Molecular Characterization of a Functional Type VI Secretion System in Salmonella enterica serovar Typhi. Curr Microbiol 63: 22–31. [DOI] [PubMed] [Google Scholar]
- 30. Pukatzki S, McAuley SB, Miyata ST (2009) The type VI secretion system: translocation of effectors and effector-domains. Curr Opin Microbiol 12: 11–17. [DOI] [PubMed] [Google Scholar]
- 31. Burtnick MN, Brett PJ, Harding SV, Ngugi SA, Ribot WJ, et al. (2011) The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei . Infect Immun 79: 1512–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jani AJ, Cotter PA (2010) Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe 8: 2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Pace F, Nakazato G, Pacheco A, de Paiva JB, Sperandio V, et al. (2010) The type VI secretion system plays a role in type 1 fimbria expression and pathogenesis of an avian pathogenic Escherichia coli strain. Infect Immun 78: 4990–4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ (2007) Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A 104: 15508–15513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blondel CJ, Jimenez JC, Contreras I, Santiviago CA (2009) Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics 10: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fookes M, Schroeder GN, Langridge GC, Blondel CJ, Mammina C, et al. (2011) Salmonella bongori Provides Insights into the Evolution of the Salmonellae. PLoS Pathog 7: e1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mulder DT, Cooper CA, Coombes BK (2012) Type VI secretion system-associated gene clusters contribute to pathogenesis of Salmonella enterica serovar Typhimurium. Infect Immun 80: 1996–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chan K, Kim CC, Falkow S (2005) Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice. Infect Immun 73: 5438–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Haneda T, Ishii Y, Danbara H, Okada N (2009) Genome-wide identification of novel genomic islands that contribute to Salmonella virulence in mouse systemic infection. FEMS Microbiol Lett 297: 241–249. [DOI] [PubMed] [Google Scholar]
- 40. Klumpp J, Fuchs TM (2007) Identification of novel genes in genomic islands that contribute to Salmonella typhimurium replication in macrophages. Microbiology 153: 1207–1220. [DOI] [PubMed] [Google Scholar]
- 41. Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, et al. (2006) Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog 2: e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Blondel CJ, Yang HJ, Castro B, Chiang S, Toro CS, et al. (2010) Contribution of the type VI secretion system encoded in SPI-19 to chicken colonization by Salmonella enterica serotypes Gallinarum and Enteritidis. PLoS One 5: e11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wilson JW, Figurski DH, Nickerson CA (2004) VEX-capture: a new technique that allows in vivo excision, cloning, and broad-host-range transfer of large bacterial genomic DNA segments. J Microbiol Methods 57: 297–308. [DOI] [PubMed] [Google Scholar]
- 45. Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ (2012) Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483: 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Silverman JM, Brunet YR, Cascales E, Mougous JD (2012) Structure and Regulation of the Type VI Secretion System. Annu Rev Microbiol 66: 453–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu J, Guo JT, Li YG, Johnston RN, Liu GR, et al. (2012) The type VI secretion system gene cluster of Salmonella typhimurium: required for full virulence in mice. J Basic Microbiol. 52: 1–8. [DOI] [PubMed] [Google Scholar]
- 48. Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, et al. (2012) A Widespread Bacterial Type VI Secretion Effector Superfamily Identified Using a Heuristic Approach. Cell Host Microbe 11: 538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Blondel CJ, Jiménez JC, Leiva LE, Álvarez SA, Pinto BI, et al. (2013) The Type VI Secretion System encoded in SPI-19 is required for Salmonella Gallinarum survival within infected macrophages. Infect Immun. 81: 1207–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Santiviago CA, Reynolds MM, Porwolik S, Choi SH, Long F, et al. (2009) Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice PLoS Pathog. 5(7): e1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vivo cloning of T6SSSPI-6 from S . Typhimurium 14028 s. (A) Scheme of the VEX-Capture procedure: loxP sites were inserted in the chromosome of S. Typhimurium 14028s at each side of the T6SSSPI-6 gene cluster through homologous recombination of PCR products using the Lambda-Red system. In presence of pEKA30, a plasmid that constitutively expresses the Cre recombinase, the T6SS cluster was excised from the chromosome as a non-replicative, circular DNA intermediate that was captured through homologous recombination in R995-VC6, a derivative of R995 plasmid harboring an internal region of homology to T6SSSPI-6. (B) Tiling-PCR analysis of the T6SSSPI-6 gene cluster cloned onto the R995 plasmid. Specific primers were designed to amplify ten fragments that cover the entire T6SSSPI-6 region and whose lengths vary between 3,298 and 4,274 bp.
(TIF)