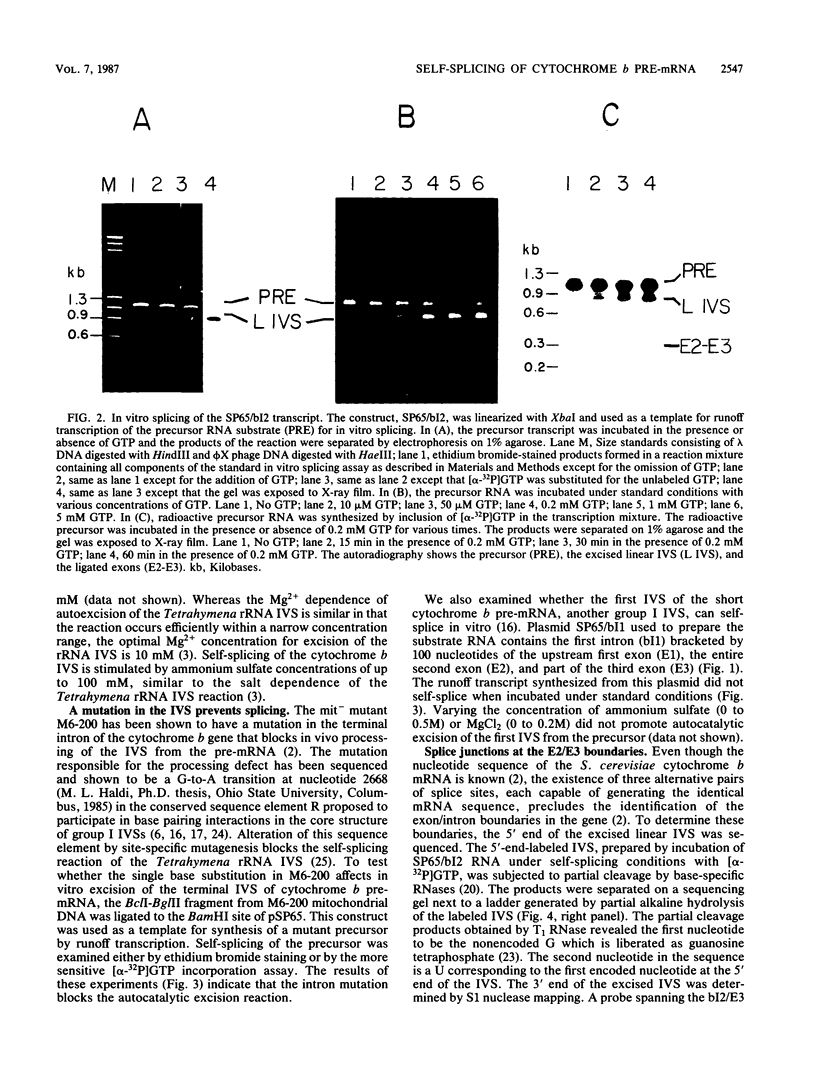
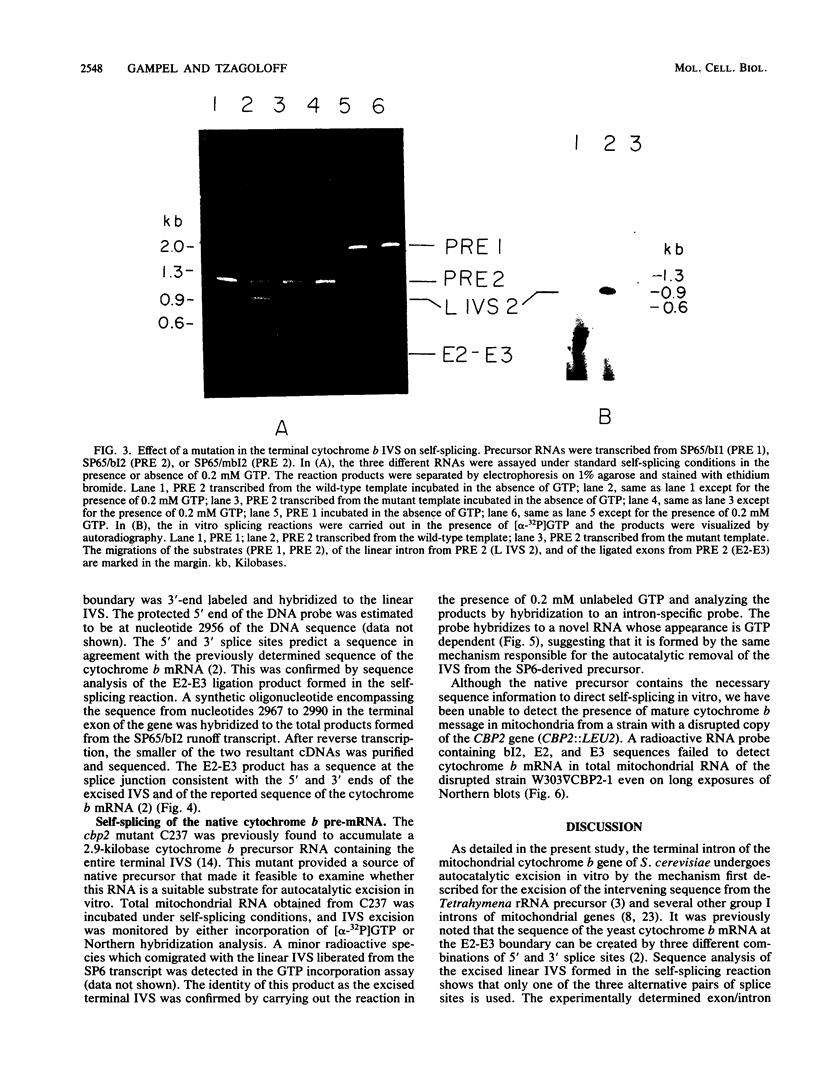
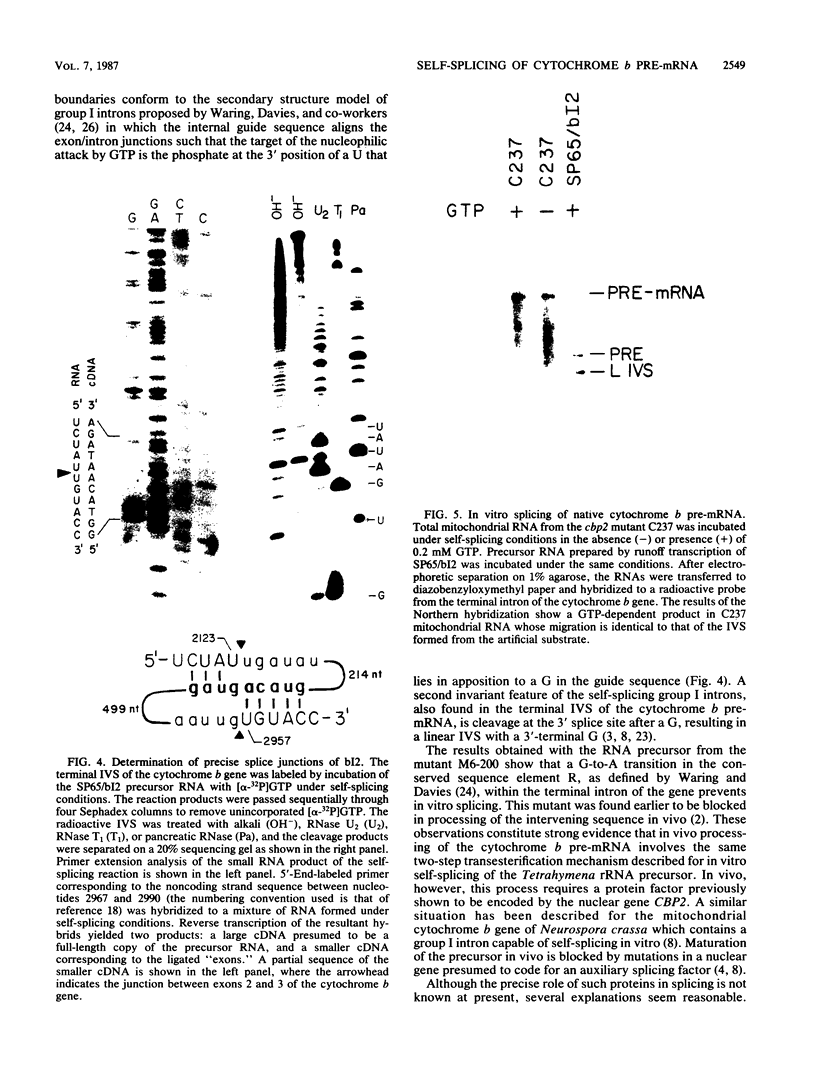
Abstract
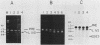
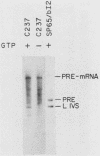
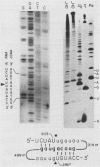
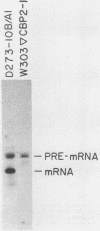
A region of the Saccharomyces cerevisiae mitochondrial cytochrome b gene encompassing the entire terminal intron plus flanking exonic sequences has been cloned in an SP6 vector. A runoff transcript prepared from this construct as well as the native cytochrome b pre-mRNA containing the terminal intervening sequence were found to act as substrates for the autocatalytic excision of the intervening sequence in vitro. This reaction proceeds under conditions previously shown by Cech and co-workers to promote protein-independent excision of the Tetrahymena rRNA intervening sequence. The 5' and 3' termini of the excised intervening sequence, determined by S1 nuclease mapping and sequence analysis, are consistent with the known sequence of the cytochrome b mRNA. The same region of the cytochrome b gene from a yeast mutant, defective in splicing due to a mutation in a critical sequence inside the terminal intron, has also been cloned in an SP6 vector. The mutant transcript fails to self-splice in the in vitro assay. These observations provide strong presumptive evidence that in vivo processing of the terminal intervening sequence of the cytochrome b pre-mRNA occurs by an autocatalytic mechanism analogous to that shown for other group I introns. In vivo processing of the terminal intervening sequence of the cytochrome b pre-mRNA, however, exhibits complete dependence on a protein factor previously shown to be encoded by the nuclear gene CBP2.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonitz S. G., Homison G., Thalenfeld B. E., Tzagoloff A., Nobrega F. G. Assembly of the mitochondrial membrane system. Processing of the apocytochrome b precursor RNAs in Saccharomyces cerevisiae D273-10B. J Biol Chem. 1982 Jun 10;257(11):6268–6274. [PubMed] [Google Scholar]
- Cech T. R., Zaug A. J., Grabowski P. J. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1981 Dec;27(3 Pt 2):487–496. doi: 10.1016/0092-8674(81)90390-1. [DOI] [PubMed] [Google Scholar]
- Collins R. A., Lambowitz A. M. RNA splicing in Neurospora mitochondria. Defective splicing of mitochondrial mRNA precursors in the nuclear mutant cyt18-1. J Mol Biol. 1985 Aug 5;184(3):413–428. doi: 10.1016/0022-2836(85)90291-8. [DOI] [PubMed] [Google Scholar]
- Corkey B. E., Duszynski J., Rich T. L., Matschinsky B., Williamson J. R. Regulation of free and bound magnesium in rat hepatocytes and isolated mitochondria. J Biol Chem. 1986 Feb 25;261(6):2567–2574. [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Ray J. A., Brown T. A., Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982 Dec 23;300(5894):719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- Faye G., Kujawa C., Fukuhara H. Physical and genetic organization of petite and grande yeast mitochondrial DNA. IV. In vivo transcription products of mitochondrial DNA and localization of 23 S ribosomal RNA in petite mutants of saccharomyces cerevisiae. J Mol Biol. 1974 Sep 5;88(1):185–203. doi: 10.1016/0022-2836(74)90304-0. [DOI] [PubMed] [Google Scholar]
- Garriga G., Lambowitz A. M. RNA splicing in neurospora mitochondria: self-splicing of a mitochondrial intron in vitro. Cell. 1984 Dec;39(3 Pt 2):631–641. doi: 10.1016/0092-8674(84)90470-7. [DOI] [PubMed] [Google Scholar]
- HATEFI Y., HAAVIK A. G., GRIFFITHS D. E. Studies on the electron transfer system. XLI. Reduced coenzyme Q (QH2)-cytochrome c reductase. J Biol Chem. 1962 May;237:1681–1685. [PubMed] [Google Scholar]
- Hill J., McGraw P., Tzagoloff A. A mutation in yeast mitochondrial DNA results in a precise excision of the terminal intron of the cytochrome b gene. J Biol Chem. 1985 Mar 25;260(6):3235–3238. [PubMed] [Google Scholar]
- Lazowska J., Jacq C., Slonimski P. P. Sequence of introns and flanking exons in wild-type and box3 mutants of cytochrome b reveals an interlaced splicing protein coded by an intron. Cell. 1980 Nov;22(2 Pt 2):333–348. doi: 10.1016/0092-8674(80)90344-x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw P., Tzagoloff A. Assembly of the mitochondrial membrane system. Characterization of a yeast nuclear gene involved in the processing of the cytochrome b pre-mRNA. J Biol Chem. 1983 Aug 10;258(15):9459–9468. [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Jacquier A., Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982 Oct;64(10):867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- Nobrega F. G., Tzagoloff A. Assembly of the mitochondrial membrane system. DNA sequence and organization of the cytochrome b gene in Saccharomyces cerevisiae D273-10B. J Biol Chem. 1980 Oct 25;255(20):9828–9837. [PubMed] [Google Scholar]
- Sanders J. P., Borst P., Weijers P. J. The organization of genes in yeast mitochondrial DNA II. The physical map of EcoRI and HindII + III fragments. Mol Gen Genet. 1975 Dec 30;143(1):53–64. doi: 10.1007/BF00269420. [DOI] [PubMed] [Google Scholar]
- Simoncsits A., Brownlee G. G., Brown R. S., Rubin J. R., Guilley H. New rapid gel sequencing method for RNA. Nature. 1977 Oct 27;269(5631):833–836. doi: 10.1038/269833a0. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Akai A., Foury F. Assembly of the mitochondrial membrane system XVI. Modified form of the ATPase proteolipid in oligomycin-resistant mutants of Saccharomyces cerevisiae. FEBS Lett. 1976 Jun 15;65(3):391–395. doi: 10.1016/0014-5793(76)80154-8. [DOI] [PubMed] [Google Scholar]
- Waring R. B., Davies R. W. Assessment of a model for intron RNA secondary structure relevant to RNA self-splicing--a review. Gene. 1984 Jun;28(3):277–291. doi: 10.1016/0378-1119(84)90145-8. [DOI] [PubMed] [Google Scholar]
- Waring R. B., Ray J. A., Edwards S. W., Scazzocchio C., Davies R. W. The Tetrahymena rRNA intron self-splices in E. coli: in vivo evidence for the importance of key base-paired regions of RNA for RNA enzyme function. Cell. 1985 Feb;40(2):371–380. doi: 10.1016/0092-8674(85)90151-5. [DOI] [PubMed] [Google Scholar]
- van der Horst G., Tabak H. F. Self-splicing of yeast mitochondrial ribosomal and messenger RNA precursors. Cell. 1985 Apr;40(4):759–766. doi: 10.1016/0092-8674(85)90335-6. [DOI] [PubMed] [Google Scholar]