Abstract
Cancer develops through a multistep process in which normal cells progress to malignant tumors via the evolution of their genomes as a result of the acquisition of mutations in cancer driver genes. The number, identity and mode of action of cancer driver genes, and how they contribute to tumor evolution is largely unknown. This study deployed the Mouse Mammary Tumor Virus (MMTV) as an insertional mutagen to find both the driver genes and the networks in which they function. Using deep insertion site sequencing we identified around 31000 retroviral integration sites in 604 MMTV-induced mammary tumors from mice with mammary gland-specific deletion of Trp53, Pten heterozygous knockout mice, or wildtype strains. We identified 18 known common integration sites (CISs) and 12 previously unknown CISs marking new candidate cancer genes. Members of the Wnt, Fgf, Fgfr, Rspo and Pdgfr gene families were commonly mutated in a mutually exclusive fashion. The sequence data we generated yielded also information on the clonality of insertions in individual tumors, allowing us to develop a data-driven model of MMTV-induced tumor development. Insertional mutations near Wnt and Fgf genes mark the earliest “initiating” events in MMTV induced tumorigenesis, whereas Fgfr genes are targeted later during tumor progression. Our data shows that insertional mutagenesis can be used to discover the mutational networks, the timing of mutations, and the genes that initiate and drive tumor evolution.
Introduction
With the advent of next-generation DNA sequencing technologies the mutational landscape of several tumor types has been defined revealing that there are numerous genetic paths to malignancy [1]. Tumor heterogeneity further contributes to this complexity [2], [3], thus complicating our ability to distinguish driver mutations from passengers. Mouse tumor models present a clean, reproducible in vivo system to study the contribution of driver genes to tumorigenesis and to define their underlying biological mechanisms of action [4].
Insertional mutagenesis (IM) employing retroviruses or transposons has been one of the main tools for inducing tumors in mice [5]–[8]. Mouse Mammary Tumor Virus (MMTV) is a slow-transforming retrovirus that has been used to study mammary tumorigenesis in mice. This virus causes mammary tumors by integration of its proviral DNA in or near cancer genes. Repeated cycles of insertional mutation and clonal expansion leads to mammary tumors carrying multiple clonal and sub-clonal MMTV integrations, including mutations linked to both driver genes as well as passenger mutations.
Molecular cloning of the proviral insertions in MMTV-induced mammary tumors led to the discovery of the first MMTV Common Insertion Site (CIS) and the associated gene Wnt1 (originally called Int1) in 1982 [9]. It was found that MMTV insertions near the Wnt1 gene promoted mammary tumor development via activation of Wnt1, the founding member of the Wnt signaling pathway. Soon after the discovery of Wnt1 another oncogene, Fgf3 (originally termed Int2) was identified. Further research showed that this member of the fibroblast growth factor gene family effectively collaborates with Wnt1 in tumor formation [10]. Following the promise of these early studies, the increasing popularity of MMTV as a screening system resulted in the discovery of several additional genes implicated in cancer development [11]–[17].
The heterogeneity and progression of MMTV-induced tumors can be assessed by analyzing the relative abundance (“clonality”) of individual insertions in a given sample. Highly abundant insertions indicate early, initiating insertion events and lowly abundant insertions indicate events that occur later during tumor development, analogous to recent studies of single nucleotide mutations in human tumors [2], [3]. Previously used PCR-based approaches are unable to reliably quantify the clonality of insertion sites due to sequence amplification biases. We therefore developed a method for simultaneous identification and quantitative assessment of clonality of insertional mutations called Shear-Splink [18]. We applied this method to analyze a large set of MMTV-induced tumors from two wild-type mouse strains (BALB/c and FVB/N) and two genetically engineered mouse (GEM) models of breast cancer: the Pten+/− strain [19] and the K14Cre;Trp53 model [20]. We used the resulting dataset to address four key questions: Firstly, can we identify novel MMTV CISs and thereby extend the repertoire of candidate cancer genes associated with these models? Secondly, can we identify genotype specific driver genes in each of the genetic backgrounds? Thirdly, can we identify co-occurring or mutually exclusive relationships between CISs and thus define functional relationships between the associated driver genes? Finally, can we generate a tumor progression model from the clonality information derived from the sequence reads of the individual insertions? Such a model would specify the order of events based on the insertion profile and shed light on functional relationships between genes involved in MMTV-induced mammary tumorigenesis.
Materials and Methods
Mouse models used for MMTV infection
Newborn BALB/c/He/A (denoted BALB/c) mice were infected with MMTV by foster nursing on C3H/A females harboring the milk transmitted MMTV [21]. Infected BALB/c female mice develop mammary tumors with high incidence (>95% before the age of 1 year). The virus-infected animals were denoted BALB/c+ mice. In this study we used two transgenic mouse lines and their wild-type controls: a strain conditionally deficient for Trp53 in mammary epithelial tissue on a BALB/c background (Balb/c K14Cre;Trp53F/F) and a germ-line heterozygous knockout for Pten on an FVB/N background.
The BALB/c K14Cre;Trp53F/F mouse strain was generated by nine consecutive backcrosses of K14Cre;Trp53F/F animals on a mixed 129P2/Ola and FVB/N background [22] to BALB/c mice. The resulting Balb/c K14Cre;Trp53F/F strain showed only a low mammary tumor incidence before the age of 300 days. These conditional Trp53 knockout mice were infected by MMTV through foster nursing by BALB/c+ females.
For comparison of MMTV induced tumorigenesis between wild type and Pten +/− mice, the conditional Pten knockout allele [23] was used to generate Pten +/− FVB/N mice. Pten +/− mice were crossed with FVB/N wild-type mice. The female littermates in the progeny, both wild-type and heterozygous for Pten, were infected with MMTV by foster nursing on BALB/c+ females harboring the milk transmitted MMTV.
All MMTV-infected animals were monitored for the development of mammary tumors which were isolated when approximately 1 cm in diameter. This study was carried out in strict accordance with the Dutch Code of Practice for Research with Laboratory Animal in Cancer Research. The protocol was approved by the local experimental Animal ethics Committee (DEC) (Permit Numbers: 04065, 08061, 03008) and all efforts were made to minimize suffering. All mice were sacrificed when the tumor reached 1 cm3 (end point) by carbon dioxide. Kaplan-Meier plots of mouse life span were plotted and log-rank tests were calculated using the survival package as implemented in the statistical programming language R.
Cre mediated deletion of the floxed Trp53 alleles in tumors from K14Cre;Trp53F/F mice was assessed by Southern blot analysis as described previously 2001 [24]. Briefly, high molecular weight genomic DNA from MMTV induced tumors was digested with BglII. DNA fragments were separated on 0.6% agarose gels and blotted on to nitrocellulose filters. A PCR labeled 700 nt XbaI fragment corresponding to exon 11 of Trp53 was used as probe to detect deleted and non-deleted alleles. Cre mediated deletion of exon 2–10 of the floxed Trp53 allele could be assessed based on a mobility shift of the BglII DNA fragment. Approximately 50% of the tumors showed bi-allelic loss of the floxed exons. Only those tumors were used for IM analysis.
Shear-Splink method for insertion site mapping
To sequence the insertion sites in tumors we used the Shear-Splink protocol as previously described [18]. Briefly, DNA was sheared to 100–1000 bp fragments, which were blunt-ended and ligated to splinkerette adapters. A primary PCR was performed to specifically amplify viral-to-host junction fragments. A second PCR was performed to introduce the barcode sequences and the adapters needed for 454 sequencing. These PCR products were pooled and sequenced by 454 sequencing. The raw sequencing data has been deposited at the Sequence Read Archive under accession number ERP002483.
Data analysis
Data analysis methods are detailed in the Supporting Methods (Text S1).
RNA sequencing
RNA was extracted from 4 tumors carrying Hbegf insertions, and 4 cases carrying a Myb insertion. We determined FPKMs for both Hbegf and Myb using Cufflinks [25] after alignment using BWA [26]
Results
MMTV insertional mutagenesis in wild-type and Trp53 or Pten mutant mice
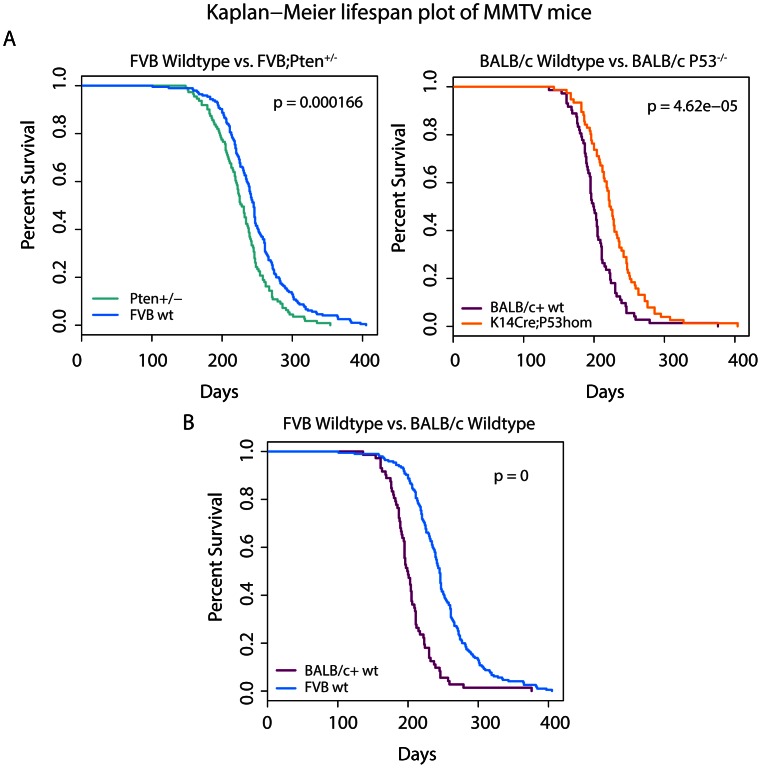
In order to assess tumor heterogeneity and identify cancer gene networks in MMTV-induced mouse mammary tumors, we performed a high-throughput, large-scale Insertional Mutagenesis study in combination with deep sequencing in a large cohort of mice from 4 different genetic backgrounds. A schematic overview of our approach is depicted in Figure S1. We analyzed cohorts of mice of two different genetic backgrounds: FVB/N (hereafter referred to as FVB) and BALB/c. For each genetic background we acquired tumors from both the wild-type strain as well as from specific genetically engineered mouse (GEM) models. Within the FVB background, mammary tumors were harvested from MMTV-infected wild-type mice and Pten+/− heterozygous knockout mice. Within the BALB/c background MMTV-induced tumors were obtained from wild-type animals and K14cre;Trp53F/F mice with epithelium-specific deletion of p53. As can be seen in Figure 1a, there is a lifespan difference between the wild-type controls and the matched GEM models, indicating an interaction between MMTV-induced tumorigenesis and the genetically engineered mutation. Interestingly, the median latency of MMTV-induced tumor development was decreased in the Pten+/− cohort, but increased in the K14cre;Trp53F/F cohort when compared to their wild-type controls. This observation led us to hypothesize that the MMTV insertions might hit genes that collaborate with Pten haploinsufficiency in Pten+/− mice. In contrast, MMTV infection might negatively influence malignant transformation of Trp53−/− mammary epithelial cells or vice versa. There is also a large difference in lifespan between MMTV-infected wild-type FVB/N and BALB/c mice (Figure 1b). This could simply indicate that the virus has slower replication in the FVB strain compared to the BALB/c strain, or it could indicate the presence of BALB/c alleles that promote MMTV-induced tumorigenesis. In support of the latter, BALB/c mice contain a hypomorphic allele of the Cdk2na tumor suppressor gene [27].
Figure 1. Kaplan Meier curves for the four strains of mice used in this study.
Each graph represents a comparison between two cohorts. A pairwise log-rank test was performed for all graphs to determine whether there are significant lifespan differences between the cohorts plotted in each graph. P-values are shown in the upper right corner. A. Within each specific mouse background strain (FVB or BALB/c+) we compared the MMTV-infected wild-type cohort with the infected genetically engineered line (either Pten heterozygous for the FVB cohort or Trp53 deficient for the BALB/c+ cohort). B. The difference in lifespan between the two wild-type strains is shown here.
To map the MMTV insertion sites we used our Shear-Splink protocol [18] to extract and amplify virus-host DNA junction fragments containing the MMTV 5′ LTR as well as the adjacent mouse genomic sequence. We barcoded the fragments allowing us to pool up to 48 individual tumors and sequence them on the 454 Genome Sequencer FLX system. Raw sequences were preprocessed using custom Perl scripts. We identified the genomic sequence from the reads and mapped them to the C57BL/6J (MGSC37) reference genome. For each read we confirmed the presence of the 5′ end of viral sequence as well as the presence of the DNA barcode to de-convolve the pools into individual tumor data. One of the unique aspects of our approach is that we are able to quantify the clonality of insertions by counting the number of unique tumor-host DNA junction fragments marked by unique ligation points (LPs) between the mouse genomic sequence and the splinkerette adapter. Since the number of unique LPs corresponds to the number of cells carrying the corresponding MMTV insertion, the LP count can be used as a measure for the relative clonality of individual MMTV insertions within each tumor [18].
Several additional filtering steps were performed as described in the Methods section and depicted in Figure S1 before the data was entered into the Insertional Mutagenesis Database (iMDB; http://imdb.nki.nl). After filtering we were left with 30942 integrations in 604 tumors. Any insertions that have n unique LPs are guaranteed to have been present in at least n independent cells. To enrich for insertions that had integrated in more than one tumor cell, we disregarded insertions with only one LP. Using this filter we reduced the data to 6605 unique insertions in 600 tumors (4 tumors carried only insertions with 1 LP). This is a reduction of 78% in the total number of insertion sites, but it only constitutes a reduction of 21% in the total number of reads, suggesting that filtering against insertions with single LPs effectively reduces the number of background mutations in our data.
CIS analysis of MMTV integration sites
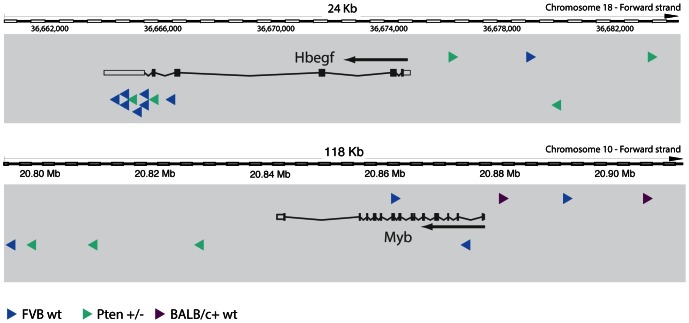
We determined global CISs for all tumors in the dataset. To determine significant CISs we used the Gaussian Kernel Convolution framework [28] implemented in the iMDB. Most of the tumors from wild-type mice contributed at least one insertion to a CIS (Table 1). This percentage is lower for the predisposed backgrounds. This is most probably due to the fact that some of the K14Cre;Trp53F/F and Pten+/− mice develop mammary tumors that are not driven by MMTV insertional mutagenesis. In total, we found 30 significant CISs, of which 18 were already known and 12 were novel (Table 2). All insertions associated with a CIS are accessible via the iMDB. We manually assigned potential target genes to these CISs. As an illustration we show the two most frequent novel CISs and the mapping of the MMTV insertions in these CISs with respect to the target gene in Figure 2. Both Hbegf and Myb are very plausible candidate target genes as the MMTV integrations are very likely to enhance expression (upstream and downstream integrations) or stabilize the mRNA by premature transcription termination and concomitant removal of mRNA destabilizing motifs due to integrations in the 3′ UTR (in the case of Hbegf). Moreover, both Hbegf [29], [30] and Myb [31] have been previously implicated in cancer. Finally, the expression of both Hbegf and Myb is high in samples carrying the integration, showing that these genes are direct targets of the viral integration (Figure S2).
Table 1. Mouse tumors contributing an insertion to a common insertion site.
| Genotype | Strain Background | Number of tumors with insertions in a CIS1 (n (% of total)) |
| Wt | BALB/c | 78 (98%) |
| K14Cre;Trp53f/f | BALB/c | 46 (78%) |
| Wt | FVB | 246 (93%) |
| Pten+/− | FVB | 166 (83%) |
This table gives an overview of the different genotype/strain combinations and the number of tumors that contain at least one.
Common Insertion Site (CIS).
Table 2. Significant known and novel common insertion sites.
| Chromosome | Start (bp) | End (bp) | Curated Target Gene | Tumors with Insertion | CGC | Amplified in Cell Lines (CONAN) | Deleted in Cell Lines | COSMIC (% mutated of samples tested) |
| Known CISs | ||||||||
| chr15 | 98520001 | 98712001 | Wnt1 | 270 | N | 3 | 0 | 0 |
| chr7 | 151923001 | 152121001 | Fgf3 | 195 | N | 28 | 0 | 0 |
| chr19 | 45606001 | 45867001 | Fgf8 | 135 | N | 0 | 0 | 0 |
| chr11 | 59004001 | 59175001 | Wnt3a | 84 | N | 5 | 0 | 1% |
| chr11 | 103557001 | 103779001 | Wnt3 | 59 | N | 1 | 0 | 0 |
| chr7 | 137232001 | 137505001 | Fgfr2 | 51 | Y | 5 | 0 | 3% |
| chr15 | 42954001 | 43134001 | Rspo2 | 48 | N | 6 | 0 | 2% |
| chr18 | 4275001 | 4395001 | Map3k8 | 30 | N | 0 | 0 | 0 |
| chrX | 138084001 | 138234001 | Irs4 | 23 | N | 1 | 1 | 1% |
| chr10 | 29211001 | 29361001 | Rspo3 | 25 | N | 0 | 0 | NA |
| chr7 | 149778001 | 149895001 | Igf2 | 19 | N | 0 | 0 | 0 |
| chr2 | 10242001 | 10353001 | Sfmbt2 | 17 | N | 0 | 0 | 0 |
| chr5 | 75501001 | 75594001 | Pdgfra | 14 | Y | 5 | 0 | 7% |
| chrX | 7464001 | 7554001 | Eras | 13 | N | 0 | 0 | 1% |
| chr18 | 61179001 | 61236001 | Pdgfrb | 7 | Y | 1 | 0 | 1% |
| chr10 | 29490001 | 29529001 | Rspo3 enhancer | 6 | N | 0 | 0 | NA |
| chr6 | 127188001 | 127203001 | Fgf6/Fgf23 | 4 | N | 3 - Fgf6/4 - Fgf23 | 0 | 0.5% - Fgf6/1% Fgf23 |
| chr13 | 119487001 | 119520001 | Fgf10 | 5 | N | 14 | 0 | 1% |
| New CISs | ||||||||
| chr8 | 26397001 | 26667001 | Fgfr1 1 | 30 | Y | 5 | 0 | 1% |
| chr18 | 36621001 | 36714001 | Hbegf | 15 | N | 0 | 0 | 0 |
| chr10 | 20820001 | 20898001 | Myb 2 | 9 | Y | 5 | 0 | 0 |
| chr11 | 121536001 | 121599001 | Metrnl/Ptchd3 | 8 | N | 2 | 0 | 0% - Metrnl/Ptchd3 |
| chr15 | 74901001 | 74943001 | Reg. Feat | 6 | N | NA | NA | NA |
| chr6 | 103575001 | 103623001 | Chl1 | 6 | N | 0 | 0 | 1% |
| chr4 | 124671001 | 124716001 | Rspo1 | 6 | N | 3 | 0 | 0 |
| chr3 | 97782001 | 97821001 | Notch2 | 6 | Y | 2 | 0 | 2% |
| chr5 | 34044001 | 34068001 | Fgfr3 | 5 | Y | 2 | 0 | 26% |
| chr11 | 3051001 | 3072001 | Sfi1 | 5 | N | 0 | 0 | 1% |
| chr14 | 68478001 | 68511001 | Dock5 | 5 | N | 0 | 2 | 4% |
| chr6 | 23181001 | 23217001 | Fezf1 | 5 | N | 10 | 0 | 3% |
This table gives an overview of the significant CISs and their potential target genes.
Although Fgfr1 has not been found as a common insertion site in [11], the authors do mention finding one insertion near the gene.
The Myb CIS is a merge of two overlapping CISs (upstream and downstream of the Myb gene).
Figure 2. MMTV integrations in two novel CISs.
The putative target gene is shown, with the arrow indicating the transcriptional direction. Arrowheads indicate the genomic location of the viral integrations, with the direction of the arrow indicating the viral transcription direction. Colors indicate the cohort from which the integrations were recovered.
Many of the CISs we recovered are canonical CISs for MMTV insertional mutagenesis. In MMTV-induced mammary tumors, proviral insertions are often found near Wnt gene family members (Wnt1, Wnt3 and Wnt3a), growth factor related genes (Fgf and Fgfr genes, Pdgfr genes and Igf2), R-spondin gene family members (Rspo1, Rspo2 and Rspo3) and mitogen signaling pathway genes (Eras and Map3k8) [11]. We identified several novel CISs in these families that were not previously identified: Hbegf, Rspo1 and Fgfr3. These novel targets can only be recovered in a large study since they occur much less frequently compared to the previously identified family members. We also recovered several rare CISs that are likely to be true MMTV hits because the candidate target genes belong to gene families which also include members that are frequent MMTV targets. This finding supports the validity of other rare, but significant novel CISs we identified in our screen, such as Sfi1, Dock5 and Fezf1. Indeed, the human homolog of Fezf1 (known as ZNF312B) has been described as an oncogene in gastric cancer [32]. Furthermore, several known and novel target genes were found to be recurrently amplified in a panel of over 700 human cancer cell lines (http://www.sanger.ac.uk/cgi-bin/genetics/CGP/conan/search.cgi) or listed as mutational targets in the COSMIC database (http://www.sanger.ac.uk/genetics/CGP/cosmic/). This shows that genes associated with CISs in MMTV-induced mammary tumors may also be relevant in human cancer.
Genotype-specific CISs
Besides finding novel MMTV CISs we were interested in finding genotype-specific insertions. MMTV infected Pten +/− mice develop mammary tumors faster than their wild-type controls, suggesting that MMTV insertions might mutate cancer-relevant genes that collaborate with Pten haploinsufficiency in mammary tumorigenesis. If this is the case we would expect to find enrichment for specific CISs in MMTV-induced Pten+/− mammary tumors, compared to control tumors. However, we could not find any significant association with either background in our study (Table 2). This suggests that MMTV integrations occur in or near the same cancer driver genes, regardless of Trp53 or Pten status. There was also a large difference in lifespan between the MMTV-infected FVB and BALB/c wild-type mice (Figure 1b). Also here, we could not find any significant association between CISs and strain background. Although we observed insertions near Hbegf only in the FVB background (Figure 2), this difference was not statistically significant, probably due to the low incidence of insertions near this gene. It does however hint towards the possibility that activation of Hbegf may be oncogenic in FVB mice but not BALB/c mice.
Co-occurrence and mutual exclusivity between CISs
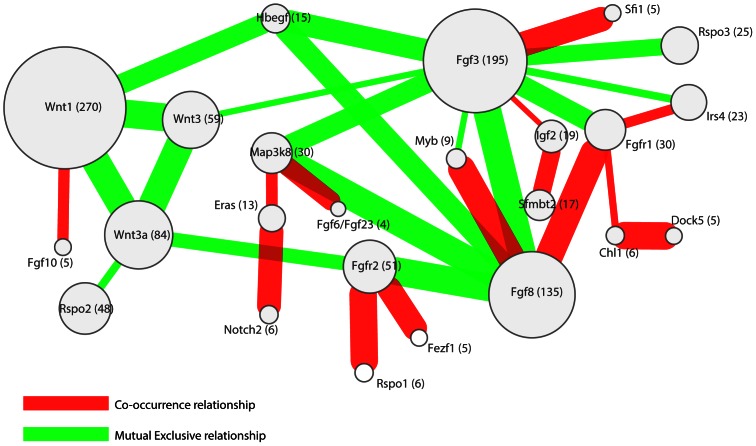
Our analysis of common insertion sites yielded several novel but rarely tagged loci. We analyzed the insertion patterns for all CISs across all tumors to establish relationships between the insertion patterns in these CISs. Insertions can exhibit a mutually exclusive integration pattern, which could signify functional redundancy between the target genes as has been shown for Myc and its paralog N-myc [33]. Conversely, co-occurring insertions may be observed in cases where the oncogenic effect of both insertions is synergistic. To identify such functional relationships, we determined co-occurrence and mutual exclusivity between the statistically significant CISs in our study as described in the Methods section (Figure 3).
Figure 3. A network of significant co-occurring and mutually exclusive common insertion sites (CISs).
CISs are indicated by their manually curated target gene. Red edges indicate a co-occurrence relationship, while green edges indicate a mutually exclusive relationship. The number in parenthesis and the size of the nodes indicate the number of tumors with a viral insertion in the relevant CIS. The thickness of the edges is a measure of the significance of the relationship between the nodes.
Several observations can be made from this analysis. Firstly, significant co-occurrence occurs primarily between infrequent insertions and mutual exclusivity mainly between highly frequent insertions. This bias is most probably due to the way of testing, which is underpowered for low insertion frequencies. Infrequent, mutually exclusive insertions need a larger sample size to become significant, while infrequent, co-occurring insertions can quickly become significant.
Secondly, interesting relationships were found between the members of the Fgf ligand family and their receptors, the Fgfr genes. For example, the FGFR ligands FGF8 and FGF3 appear to have a reciprocal preference for the receptor FGFR1, since Fgf3 insertions are mutually exclusive with Fgfr1 insertions whereas Fgf8 insertions significantly co-occur with Fgfr1 insertions. This might indicate preferential binding partners for the different ligands and a selective advantage for up-regulation of both the ligand and its matched receptor.
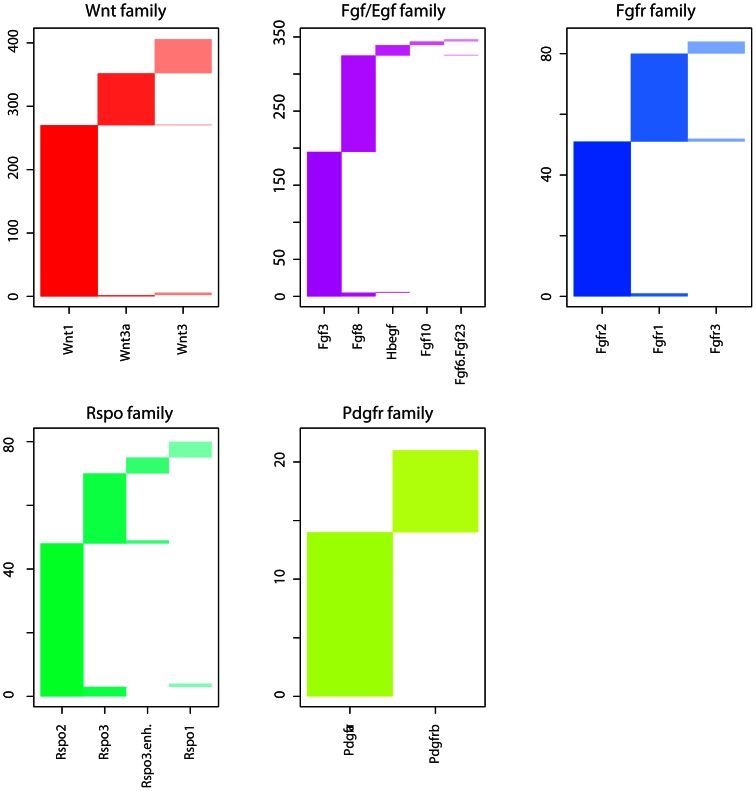
Finally, we observed mutual exclusivity between members of individual gene families (e.g. Wnt genes and Fgf-Hbegf genes). Plotting of cumulative insertion patterns for five distinct families of CIS genes (Wnt, Fgf/Egf, Fgfr, Rspo and Pdgfr) revealed a typical mutually exclusive integration pattern for genes within each family (Figure 4), showing that MMTV infected cells gain little to no selective advantage from MMTV insertions near multiple members of the same gene family. Based on these results we decided to look for relationships between CIS gene families instead of single CIS genes.
Figure 4. Insertion patterns of CIS gene families.
For each of five families the columns indicate which tumors contained an insertion for that specific member of a family. Rows indicate specific tumors.
Model for MMTV Tumor Progression
Our data show that MMTV preferentially targets a rather limited group of genes and gene families. If we restrict ourselves to CIS genes and CIS families that are affected by MMTV insertions in ten or more tumors, we are left with only 11 groups of CIS genes (Table 2). We were interested to see if we could find a difference in clonality between these gene groups. When comparing insertions near two members of these groups within one tumor, the one with more clonal insertions will also have a higher clonality score based on the unique LPs counts. This could indicate an earlier event and/or a more potent hit resulting in stronger positive selection. We tested for all 11 groups of CIS genes all pairs of insertions that occurred in the same tumor in order to test if members of one group have a consistently higher clonality scores than members of another group, when co-mutated in the same tumor (see Methods for details).
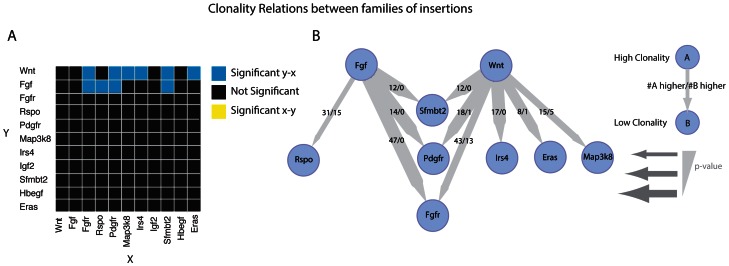
Figure 5a shows a heatmap with the CIS gene/family pairs that had a significant clonality relation according to the binominal test. This analysis reveals significant relationships between the Wnt/Fgf gene families (higher clonality) and Rspo, Sfmbt2, Pdgfr, Fgfr, Irs4, Eras and Map3k8 (lower clonality). In Figure 5b we visualized only these significant relationships together with their directionality. From this data-driven model we can formulate a simple progression model for MMTV-induced mouse mammary tumorigenesis. Tumor-initiating MMTV integrations are most likely to occur near an Fgf or Wnt gene, whereas insertions near other CIS genes are secondary events. For all other relations tested there is either no clear clonality relation or there are no tumors in which they are co-mutated. In all 47 tumors that showed co-mutation of Fgf and its receptor Fgfr, the Fgf insertion was more clonal, showing that the FGF ligand is always activated earlier than the FGF receptor during MMTV tumor progression.
Figure 5. Analysis of clonality between different families of genes.
A. A heatmap of all combinations of gene families and single genes not assigned to a family. Significant difference in clonality for each family are calculated using a binominal test for all samples that are co-inserted in that specific gene (family) pair. Blue squares indicate a significant clonal relation from the group indicated on the Y-axis to the group indicated on the X-axis. Yellow squares indicate a significant clonal relationship from the X-axis to the Y-axis. Black squares indicate no significant relation. B. A network view of the heatmap in A. showing only significant (P<0.05) clonality relationships. An edge points from the more clonal gene(family) to the lesser clonal gene(family). The thickness of an edge is a measure of the significance of the clonality relation. For the fraction displayed on the edges, the numerator represents the number of times the parent node had a higher clonality score while the denominator represents the number of times the child node had a higher clonality score, in a tumor that contained insertions in both nodes.
Discussion
Recent studies have delved into the heterogenic make-up of human tumors [2], [3] showing that a steady accumulation of background mutations covers the fact that only a handful of driving mutations causes oncogenic differentiation. Using the mouse system, we were able to clearly define the driver genes for MMTV-induced mammary tumors and the order in which they get deregulated.
MMTV insertional mutagenesis
In this study we analyzed a large cohort of mice that developed tumors through MMTV insertional mutagenesis. Within a dataset of 6600 non-background MMTV insertions from 600 tumors, we identified 30 common insertion sites encompassing 1271 of the 6600 insertions (19.3%). We used this dataset to address several questions regarding MMTV-induced mouse mammary tumorigenesis. Firstly, we wanted to see if we could identify novel MMTV CISs in this large dataset using our high-throughput Shear-Splink approach. We found that the 30 MMTV CISs target a limited number of well-defined gene families and very few other genes. In total, 53 MMTV CISs have been previously identified (Table S1). The overlap between this list of known MMTV CISs and our list is only 18 CISs. This means that 66% of the previously found CISs could not be confirmed in this study. Although it is possible that CISs with very subclonal insertions are not detected by our Shear-Splink method, we believe that our study is both less biased (through the use of DNA shearing instead of restriction enzyme cleavage) and more robust (since we measure many reads of the same insertion) than older studies based on isolation and Sanger sequencing of individual MMTV insertion sites. Also, the use of the LP score to filter background insertions from the data is a powerful method to limit false positive findings. While it is possible that our method generates false-negatives, it is perhaps more likely that some of the previously identified CISs are passengers or random integration sites. Our data suggest that MMTV-induced mammary tumorigenesis is a very specific disease involving a limited number of cellular target genes that may promote proliferation, survival and/or self-renewal of MMTV-infected mammary epithelial cells. As such, MMTV is not a very flexible insertional mutagenesis system and therefore probably not the best approach to identify novel candidate breast cancer genes in wild-type mice or tumor-predisposed GEM models. This notion is supported by the fact that we cannot find specific MMTV insertions in mammary tumor-prone mice with heterozygous Pten deletion or tissue-specific loss of Trp53, even though MMTV-infected Pten heterozygous mice developed mammary tumors faster than their wild-type counterparts. MMTV-induced tumorigenesis apparently profits from haploinsufficiency for Pten but mutation of specific collaborators is not required for this condition. Our results do not rule out that MMTV might show a different insertion pattern in mice with mammary gland-specific over-expression of a strong oncogene.
Despite the strong bias of MMTV towards activation of Wnt/Fgf family members, we could still identify several novel CISs for MMTV due to the large number of samples we included in our analysis. Some of the new CISs fall within the established target gene families, showing that our method is able to identify true positives even if they only occur in <1% of the samples, as is the case with Fgfr3. It can be expected that additional rare MMTV CISs can be identified by further increasing the sample-size of the study. However, with the diminishing returns and the increasing costs, this strategy is probably not preferable, especially in view of the rapidly decreasing costs of genomic sequencing of spontaneous tumors and the advent of novel transposon systems. Novel CISs that do not map near members of the canonical MMTV gene families, such as Myb and Fezf1, have been previously associated with cancer. Although Myb is a known common target for Mouse Leukemia virus [34] and implicated in human lymphoma [31], it has only recently been associated with breast cancer [31], [35]. Finding Myb as a common target in mouse mammary tumorigenesis adds to the evidence that Myb is a bona fide oncogene in breast cancer.
Our analysis of a large group of MMTV-induced mouse mammary tumors also allowed us to investigate relations of either mutual exclusivity or co-occurrence between the different MMTV CISs. We showed that within the five commonly targeted gene families (Wnt, Fgf, Fgfr, Rspo and Pdgfr) individual members showed a strong mutually exclusive mutation pattern, indicating that an MMTV insertion near one family member abolishes selective pressure for insertions near additional members of the same family. Although overexpressed Fgf and Wnt genes are synergistic in inducing mammary tumors in bitransgenic mice [36], we did not observe a significant co-occurrence of MMTV insertions in Wnt and Fgf family members. While we recovered many tumors with insertions near members of both gene families, we recovered almost equal numbers of tumors with an insertion near members of only one of the two families. These results suggest that combined activation of Fgf and Wnt might not be required for MMTV-induced tumorigenesis or that activation of Fgf and Wnt in MMTV-induced mammary tumors might also occur via mechanisms other than insertional mutations (i.e. point mutations, genomic rearrangements, copy number aberrations and/or epigenetic changes). In support of the latter notion, we could detect increased mRNA expression of Fgf and Wnt family members in several tumors with no detectable MMTV insertions near these genes (data not shown).
The strong mutual exclusivity among members of individual CIS gene families allowed us to group the targeted genes into families and interrogate relationships between insertions in the different CIS groups. We used the LP score that followed from our Shear-Splink analysis to calculate relative clonality in order to determine for all pairs of co-occurring insertions which of the two insertions was present in a higher number of tumor cells. The more clonal insertions have probably occurred earlier during tumor progression. Although the clonality score could be influenced by DNA copy number and the presence of multiple independent tumors in one location, we assume that in most cases the clonality score can effectively distinguish between early and late events. Our pair-wise analysis of the clonality scores showed that insertions near Wnt or Fgf genes are early events compared to insertional mutations near other gene families. Taken together, our observations indicate that MMTV induced mammary tumorigenesis is a very specific disease involving activation of a limited number of cellular target genes.
Conclusions
This study is the largest MMTV insertional mutagenesis screen performed to date. Our high-throughput Shear-Splink method has shown that the pattern of recurrent insertions in MMTV-induced mouse mammary tumors is very specific and dominant over genetic background and tumor-predisposing mutations. Only a handful of gene families are targeted in a specific tumor progression program. What causes the specificity of MMTV towards these genes is unclear but it most likely involves complex interactions between MMTV and the host cell. Target gene specificity seems unlikely to be due to potential integration biases of the MMTV provirus, since the MMTV insertions near the major targets are spread over multiple kilobases and since different family members are targeted in a mutually exclusive fashion. After almost a century of MMTV research [21], MMTV-induced tumorigenesis appears to be a highly defined and potent genetic program.
Supporting Information
Schematic overview of the study. Boxes depict application of protocols and arrows indicate the flow of the resulting products. The gray box indicates that all processes take place inside the insertional Mutagenesis Database.
(TIF)
Expression of Myb and Hbegf in tumors with viral insertions near to these genes. The RPKM gene expression values of the genes Myb and Hbegf have been plotted for 8 mammary tumors. Four of these tumors contained a MMTV insertion near Myb (shown with triangles) and four of the tumors contained a MMTV insertion near Hbegf (shown with circles). The color of the symbols represents the clonality of those insertions.
(TIF)
List of previously identified MMTV Common Insertion Sites. This table lists previously identified MMTV CISs in the literature that were compared to the ones identified in this study.
(XLS)
Supplemental Methods. This document describes the data analysis steps used in this study in more detail.
(DOC)
Acknowledgments
We thank the staff of the animal facility at the Netherlands Cancer Institute for animal husbandry and Wendy Lagcher for her help with this study.
Funding Statement
This work was financially supported by the Dutch Cancer Society (grant 2001-2489 to J.H.), the Association for International Cancer Research (AICR grant 07-585 to J.J.), the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research (NGI/NWO Program grant 050-10-008 to M.v.L.; NGI/NWO Horizon Breakthrough grant 40-41009-98-9109 and NGI/NWO Horizon Zenith grant 40-41009-98-11097 to J.J.), the Netherlands Consortium for Systems Biology (NCSB), the Cancer Genomics Centre (CGC), and the Cancer Systems Biology Center (CSBC). D.J.A. is funded by Cancer Research-UK and the Wellcome Trust (76943). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Stratton MR, Campbell PJ, Futreal PA (2009) The cancer genome. Nature 458: 719–724 doi:10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, et al. (2012) Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481: 506–510 doi:10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, et al. (2012) The Life History of 21 Breast Cancers. Cell 149: 994–1007 doi:10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frese KK, Tuveson DA (2007) Maximizing mouse cancer models. Nat Rev Cancer 7: 654–658. [DOI] [PubMed] [Google Scholar]
- 5. Uren AG, Kool J, Berns A, Van Lohuizen M (2005) Retroviral insertional mutagenesis: past, present and future. Oncogene 24: 7656–7672. [DOI] [PubMed] [Google Scholar]
- 6. Copeland NG, Jenkins NA (2010) Harnessing transposons for cancer gene discovery. Nat Rev Cancer 10: 696–706 doi:10.1038/nrc2916. [DOI] [PubMed] [Google Scholar]
- 7. Kool J, Berns A (2009) High-throughput insertional mutagenesis screens in mice to identify oncogenic networks. Nat Rev Cancer 9: 389–399 doi:10.1038/nrc2647. [DOI] [PubMed] [Google Scholar]
- 8. Hilkens J (2006) Recent translational research: Oncogene discovery by insertional mutagenesis gets a new boost. Breast Cancer Research 8: 102 doi:10.1186/bcr1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nusse R, Varmus HE (1982) Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 31: 99–109. [DOI] [PubMed] [Google Scholar]
- 10. Shackleford GM, MacArthur CA, Kwan HC, Varmus HE (1993) Mouse mammary tumor virus infection accelerates mammary carcinogenesis in Wnt-1 transgenic mice by insertional activation of int-2/Fgf-3 and hst/Fgf-4. Proc Natl Acad Sci USA 90: 740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Theodorou V, Kimm MA, Boer M, Wessels L, Theelen W, et al. (2007) MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat Genet 39: 759–769 doi:10.1038/ng2034. [DOI] [PubMed] [Google Scholar]
- 12. Erny KM, Peli J, Lambert JF, Muller V, Diggelmann H (1996) Involvement of the Tpl-2/cot oncogene in MMTV tumorigenesis. Oncogene 13: 2015–2020. [PubMed] [Google Scholar]
- 13. Lowther W, Wiley K, Smith GH, Callahan R (2005) A new common integration site, Int7, for the mouse mammary tumor virus in mouse mammary tumors identifies a gene whose product has furin-like and thrombospondin-like sequences. J Virol 79: 10093–10096 doi:10.1128/JVI.79.15.10093-10096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chatterjee G, Rosner A, Han Y, Zelazny ET, Li B, et al. (2002) Acceleration of mouse mammary tumor virus-induced murine mammary tumorigenesis by a p53 172H transgene: influence of FVB background on tumor latency and identification of novel sites of proviral insertion. Am J Pathol 161: 2241–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Theodorou V, Boer M, Weigelt B, Jonkers J, Van der Valk M, et al. (2004) Fgf10 is an oncogene activated by MMTV insertional mutagenesis in mouse mammary tumors and overexpressed in a subset of human breast carcinomas. Oncogene 23: 6047–6055 doi:10.1038/sj.onc.1207816. [DOI] [PubMed] [Google Scholar]
- 16. Gallahan D, Callahan R (1997) The mouse mammary tumor associated gene INT3 is a unique member of the NOTCH gene family (NOTCH4). Oncogene 14: 1883–1890 doi:10.1038/sj.onc.1201035. [DOI] [PubMed] [Google Scholar]
- 17. Kapoun AM, Shackleford GM (1997) Preferential activation of Fgf8 by proviral insertion in mammary tumors of Wnt1 transgenic mice. Oncogene 14: 2985–2989 doi:10.1038/sj.onc.1201146. [DOI] [PubMed] [Google Scholar]
- 18. Koudijs MJ, Klijn C, Van Der Weyden L, Kool J, Ten Hoeve J, et al. (2011) High-Throughput Semiquantitative Analysis of Insertional Mutations in Heterogeneous Tumors. Genome Res 21: 2181–2189 doi:10.1101/gr.112763.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stambolic V, Tsao MS, Macpherson D, Suzuki A, Chapman WB, et al. (2000) High incidence of breast and endometrial neoplasia resembling human Cowden syndrome in pten+/− mice. Cancer Res 60: 3605–3611. [PubMed] [Google Scholar]
- 20. Peterse JL, Berns A, Jonkers J, Liu X, Holstege H, et al. (2007) Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proceedings of the National Academy of Sciences 104: 12111–12116 doi:10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bittner JJ (1936) SOME POSSIBLE EFFECTS OF NURSING ON THE MAMMARY GLAND TUMOR INCIDENCE IN MICE. Science 84: 162 doi:10.1126/science.84.2172.162. [DOI] [PubMed] [Google Scholar]
- 22. Hanahan D, Weinberg RA (2011) Hallmarks of Cancer: The Next Generation. Cell 144: 646–674 doi:10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 23. Marino S, Krimpenfort P, Leung C, Van der Korput HAGM, Trapman J, et al. (2002) PTEN is essential for cell migration but not for fate determination and tumourigenesis in the cerebellum. Development 129: 3513–3522. [DOI] [PubMed] [Google Scholar]
- 24. Jonkers J, Meuwissen R, Van der Gulden H, Peterse H, Van der Valk M, et al. (2001) Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet 29: 418–425 doi:10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 25. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology 28: 511–515 doi:10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H, Durbin R (2009) Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 25: 1754–1760 doi:10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang S, Ramsay ES, Mock BA (1998) Cdkn2a, the cyclin-dependent kinase inhibitor encoding p16INK4a and p19ARF, is a candidate for the plasmacytoma susceptibility locus, Pctr1. Proc Natl Acad Sci USA 95: 2429–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. De Ridder J, Uren A, Kool J, Reinders M, Wessels L (2006) Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLoS Comput Biol 2: e166 doi:10.1371/journal.pcbi.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miyamoto S, Yagi H, Yotsumoto F, Kawarabayashi T, Mekada E (2006) Heparin-binding epidermal growth factor-like growth factor as a novel targeting molecule for cancer therapy. Cancer Sci 97: 341–347 doi:10.1111/j.1349-7006.2006.00188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yotsumoto F, Yagi H, Suzuki SO, Oki E, Tsujioka H, et al. (2008) Validation of HB-EGF and amphiregulin as targets for human cancer therapy. Biochem Biophys Res Commun 365: 555–561 doi:10.1016/j.bbrc.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 31. Ramsay RG, Gonda TJ (2008) MYB function in normal and cancer cells. Nat Rev Cancer 8: 523–534 doi:10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 32. Song I-S, Oh NS, Kim H-T, Ha G-H, Jeong S-Y, et al. (2009) Human ZNF312b promotes the progression of gastric cancer by transcriptional activation of the K-ras gene. Cancer Res 69: 3131–3139 doi:10.1158/0008-5472.CAN-08-2240. [DOI] [PubMed] [Google Scholar]
- 33. Cox T, Reinders M, Hubbard TJ, Rogers J, Jonkers J, et al. (2008) Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell 133: 727–741 doi:10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krimpenfort P, Gadiot J, Pritchard C, Lenz J, Lund AH, et al. (2010) Insertional mutagenesis in mice deficient for p15Ink4b, p16Ink4a, p21Cip1, and p27Kip1 reveals cancer gene interactions and correlations with tumor phenotypes. Cancer Res 70: 520–531 doi:10.1158/0008-5472.CAN-09-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stenman G, Andersson MK, Andrén Y (2010) New tricks from an old oncogene: gene fusion and copy number alterations of MYB in human cancer. Cell Cycle 9: 2986–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kwan H, Pecenka V, Tsukamoto A, Parslow TG, Guzman R, et al. (1992) Transgenes expressing the Wnt-1 and int-2 proto-oncogenes cooperate during mammary carcinogenesis in doubly transgenic mice. Mol Cell Biol 12: 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic overview of the study. Boxes depict application of protocols and arrows indicate the flow of the resulting products. The gray box indicates that all processes take place inside the insertional Mutagenesis Database.
(TIF)
Expression of Myb and Hbegf in tumors with viral insertions near to these genes. The RPKM gene expression values of the genes Myb and Hbegf have been plotted for 8 mammary tumors. Four of these tumors contained a MMTV insertion near Myb (shown with triangles) and four of the tumors contained a MMTV insertion near Hbegf (shown with circles). The color of the symbols represents the clonality of those insertions.
(TIF)
List of previously identified MMTV Common Insertion Sites. This table lists previously identified MMTV CISs in the literature that were compared to the ones identified in this study.
(XLS)
Supplemental Methods. This document describes the data analysis steps used in this study in more detail.
(DOC)