Abstract
Background
A large number of studies have investigated whether polymorphisms in the Toll-like receptor (TLR) genes are implicated in susceptibility to tuberculosis (TB) in different populations. However, the results are inconsistent and inconclusive.
Methods
A literature search was conducted using the PubMed, EMBASE, Medline (Ovid), ISI Web of Knowledge and Chinese National Knowledge Infrastructure (CNKI). A meta-analysis on the associations between the TLR1 G1805T, TLR2 T597C, T1350C, G2258A, and TLR6 C745T polymorphisms and TB risk was carried out by comparison using different genetic models.
Results
In total, 16 studies from 14 articles were included in this review. In meta-analysis, significant associations were observed between the TLR2 2258AA (AA vs. AG+AG, OR 5.82, 95% CI 1.30–26.16, P = 0.02) and TLR6 745TT (TT vs. CT+CC, OR 0.61, 95% CI 0.39–0.97, P = 0.04) polymorphisms and TB risk. In the subgroup analysis by ethnicity, Africans and American Hispanic subjects with the TLR1 1805T allele had an increased susceptibility, whereas Asian and European subjects with the TLR2 2258A allele had an increased susceptibility to TB.
Conclusions
The meta-analysis indicated that TLR2 G2258A is associated with increased TB risk, especially in Asians and Europeans. TLR1 G1805T is associated with increased TB in Africans and American Hispanics. TLR6 C745T is associated with decreased TB risk. Our systematic review and meta-analysis reported an interesting preliminary conclusion, but this must be validated by future large-scale and functional studies in different populations.
Introduction
Tuberculosis (TB) is a contagious and potentially fatal disease caused by various strains of mycobacteria, usually Mycobacterium tuberculosis (Mtb) in humans. It can infect almost any part of the body, but manifests mainly as an infection of the lungs and kills more people each year than any other single infectious disease. It is estimated by WHO that about one-third of the current global population is infected asymptomatically with Mtb, of whom 5–10% will develop clinical disease during their lifetime [1]. This fact together with other substantial evidence indicates that variations in host hereditary factors play an important role in susceptibility to TB [2]. The susceptibility to Mycobacterium can result from either inadequate or excessive acute inflammation. Thus, single nucleotide polymorphisms (SNPs) in genes that control the balance of pro- and anti- inflammatory response could confer different TB risk to the host, as exemplified in zebra fish and human studies [3]. Host genotype-specific therapies can optimize the inflammatory response to Mycobacterial infections [4]. This personalized therapy requires the knowledge of the host immune response against Mtb and association of host genotypes with TB risk.
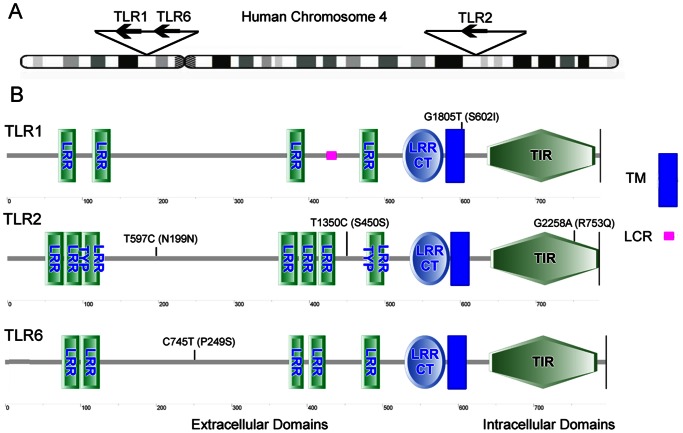
Toll-like receptors (TLRs) can recognize pathogen-associated molecular patterns (PAMPs) of Mtb and initiate signaling pathways that lead to the activation of the innate immune response, cytokines and formation of the adaptive immune response [5]. The major receptors for the Mtb are TLRs-1, -2, -4, -6 and -9 [6]–[10], among them TLR2 and TLR4 play a key role in the reorganization process. TLR2 can form heterodimers with either TLR1 or TLR6 to sense the PAMPs of Mtb and activate macrophages and dendritic cells through adaptor proteins MyD88 and TIRAP [11]. On the other hand, TLR4, together with CD14 and MD2, could recognize the lipopolysaccharide (LPS) and initiate signal transduction in the MyD88-dependent pathway. It can also function via the MyD88-independent pathway involving TRIF-dependent type 1 interferon response [11]. Many studies have reported that polymorphisms in these TLR-2 and -4 pathways can regulate inflammatory response to bacterial components and thus could impact innate immune response and clinical susceptibility to TB [12]–[14]. However, two recent meta-analyses have studied the TB susceptibility and TLR4 Asp299Gly, Thr399Ile, and TIRAP Ser180Leu polymorphisms, which are the most studied SNPs in these two genes [15], [16]. Their results revealed that these polymorphisms were unlikely to substantially contribute to TB susceptibility and raised the question whether the polymorphisms in TLR pathways are associated with the TB risk. Nevertheless, numerous studies have also been performed to investigate other polymorphisms in TLR genes and their association with TB. Among them, TLR1 G1805T, TLR2 T597C, T1350C, G2258A, TLR6 C745T are most studied as potential risk factors for TB. Figure 1 is the schematic representation of these polymorphisms, which showed that TLR2 T597C, T1350C and TLR6 C745T are in the extracellular domains, TLR1 G1850T is in the trans-membrane (TM) domain and TLR2 G2258A is in the Toll-Interleukin-1 receptor (TIR) domain.
Figure 1. Schematic representation of the TLR-1, -2 and -6 genes, proteins and selected sequence variants.
(A) Schematic representation of human chromosome 4 showing location of TLR-1, -2 and -6 genes. (B) Schematic representation of the five selected variants in TLR proteins. TLR1 G1805T is in the TM domain; TLR2 T597, T1350 and TLR6 C745T are in extracellular domains between the LRRs. TLR2 G2258A is in the intracellular TIR domain. LRR: Leucine-rich repeats; LRR TYP: LRR Typical Subfamily; LRR CT: C-terminal LRR Domain; TIR: Toll-Interleukin-1 Receptor Domain; TM: Trans-membrane Region; LCR: Low Complexity Region. Different domains are detected by SMART.
Many studies have reported the association of these polymorphisms with TB; however, the results are inconsistent and inconclusive due to limited sample sizes and different study populations. Therefore, in this article, we chose these polymorphisms as our candidates and performed a systematic review and meta-analysis, based on literature identification until 1st January 2013 to summarize the associations between these polymorphisms and TB susceptibility.
Materials and Methods
Selection of Studies for Analysis
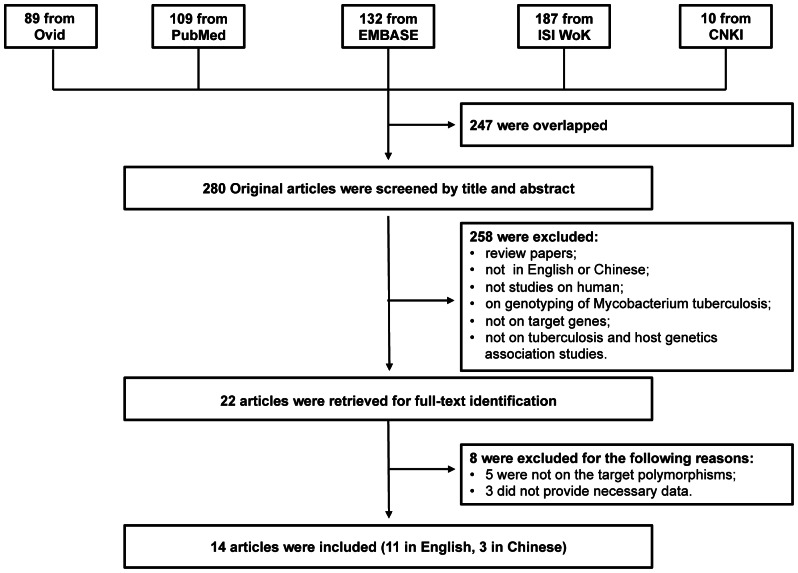
We searched the PubMed, EMBASE, Medline (Ovid), ISI Web of Knowledge and Chinese National Knowledge Infrastructure (CNKI) to identify studies of the association between TB susceptibility and TLRs polymorphisms until 1st January 2013. The key words were: ‘Mycobacterium tuberculosis’ or ‘tuberculosis’ in combination with ‘polymorphism’ or ‘variant’ or ‘genotype’ or ‘allele’ or ‘mutation’, and in combination with ‘toll’ or ‘TLR’ or ‘Toll-like receptor’ or ‘Toll like receptor’. The search results were limited to English and Chinese language articles. All retrieved titles and abstracts were examined for relevant studies on the association between TB and TLR gene polymorphisms. Studies were selected if they met the following criteria: 1) case-control studies of unrelated individuals; 2) evaluation of polymorphisms TLR1 G1805T, TLR2 T597C, T1350C, G2258A, TLR6 C745T and TB susceptibility; 3) genotype distribution in both cases and controls were available. Exclusion criteria were: 1) study design based on family or sibling pairs; 2) genotype frequencies not reported; and 3) data from reviews and abstracts. Additional studies were also identified by hand searching reference lists of original studies and review articles including meta-analysis. The PRISMA Checklist in Table S1 was used as a guide to format this article and the systematic review process was described in the flowchart of Figure 2. The schematic of the SNPs in these genes in Figure 1 are generated based on the protein annotation from SMART database [17].
Figure 2. Flow chart showing the study selection procedure.
Ovid: Ovid MEDLINE; ISI WoK: ISI Web of Knowledge; CNKI: China National Knowledge Infrastructure.
Data Extraction
For all studies, we extracted the following data from original publications: first author and year of publication; distribution of genotypes for each polymorphism among cases and controls; characteristics of the study design and the study population (study base, numbers and mean age of cases and controls, TB diagnosis, HIV status, source of controls, matching criteria and host ethnicity).
Statistical Analyses
Hardy-Weinberg Equilibrium (HWE) was examined in controls by asymptotic Pearson’s Chi-square test for each polymorphism in each study. The association between polymorphism and TB was estimated by means of odds ratios (OR) and corresponding 95% confidence intervals (CI) comparing cases to controls. Between-study heterogeneity was tested by the Q test and I2 test [18], and the heterogeneity was considered significant if P-value was less than 0.05. Fixed-effects models were adopted when P-value is more than 0.05; otherwise random-effects models were used. The funnel plot, Begg’s test and Egger’s test were used to evaluate the publication bias [19], [20]. Statistical analyses were carried out using the Stata/SE 11.0 (College Station, TX, USA) and Review Manager 5.0 software (Oxford, England).
Results
Characteristics of Included Studies
A total of 527 articles were achieved by literature search. As shown in Figure 2, after excluding those overlapped between the databases, 280 abstracts were retrieved for detailed evaluation. Twenty-two studies addressing the association of targeted-gene polymorphisms and TB were identified. Full-text article retrieval excluded 8 of them and the exclusion criteria were shown in Table S2. Finally, 16 studies from 14 articles, 11 in English [14], [21]–[30] and 3 in Chinese [31]–[33] were included in this review and meta-analysis. Among them, there were six studies for TLR1 G1805T, seven studies for TLR2 T597C, six studies for TLR2 T1350C, eleven studies for TLR2 G2258A, and four studies for TLR6 C745T. As shown in Table 1, the study participants were from diverse descents including African, Asian, European and American. The pooled sample size was 7373 (3757 cases and 3616 controls). The genotype and allele distributions of all the polymorphisms are shown in Table 2. In three studies, the genotype distributions in controls were deviated from HWE [24], [25].
Table 1. Characteristics of the 16 studies included in the meta-analysis.
| Author | Year | Country | Ethnicity | Age, years Mean ± SD or Mean (Range) | Samplesn | Genotyping method | ||
| Cases | Controls | Cases | Controls | |||||
| Che, et al. | 2010 | China | Chinese Han | / | / | 115 | 156 | PCR-Sequencing |
| Chen, et al. | 2010 | China | Taiwanese | 56.7±18.7 | 53.9±11.5 | 184 | 184 | PCR-Sequencing |
| Dalgic, et al. | 2011 | Turkey | Turkish | 8.11±4.89 | 8.52±4.55 | 124 | 200 | PCR-RFLP |
| Etokebe, et al. | 2010 | Croatia | Croatian Caucasian | 51.03±18.71 | 41.84±11.90 | 186 | 551 | PCR-Sequencing |
| Jin, et al. | 2007 | China | Chinese Han | 26–63 | 19–55 | 170 | 199 | PCR-SSP |
| Ma, et al. | 2010 | China | Chinese Han | 34.75±16.67 | 38.17±17.39 | 543 | 544 | PCR-RFLP |
| Ma, et al. _a | 2007 | United States | African American | / | / | 339 | 194 | PCR-Sequencing |
| Ma, et al. _b | 2007 | United States | European American | / | / | 180 | 110 | PCR-Sequencing |
| Ma, et al. _c | 2007 | United States | Hispanic American | / | / | 375 | 114 | PCR-Sequencing |
| Ogus, et al. | 2004 | Turkey | Turkish | 35.4±13.5 | 35.9±14.8 | 151 | 116 | PCR-SSP |
| Salvaraj, et al. | 2010 | India | Dravidian | 34.92±11.42 | 32.33±9.75 | 206 | 212 | PCR-RFLP |
| Sanchez, et al. | 2010 | Colombia | Colombian | 39 (26–51) | 42 (25–54) | 499 | 320 | PCR-MS |
| Thuong, et al. | 2007 | Vietnam | Vietnamese | / | / | 358 | 389 | PCR-MS |
| Uciechowski, et al. | 2011 | Germany | German | 58.5±16.8 | 30.5±7.7 | 45 | 49 | PCR-Sequencing |
| Xue, et al. | 2010 | China | Chinese Han | 39.5±17.9 | 26.3±8.5 | 205 | 203 | PCR-Sequencing |
| Yu | 2008 | China | Chinese Han | / | / | 77 | 75 | PCR-RFLP |
PCR = polymerase chain reaction; SSP = sequence-specific primers; RFLP = restriction fragment length polymorphism; MS = mass spectroscopy.
Table 2. Genotype and allele distribution of TLR -1, -2, and -6 polymorphisms in TB and controls.
| SNP | Study | Case | Control | HWE | |||||||||
| GG | GT | TT | G | T | GG | GT | TT | G | T | X2 | P | ||
| TLR1 | Ma, et al. | 510 | 32 | 1 | 1052 | 34 | 509 | 34 | 1 | 1052 | 36 | 0.294 | 0.588 |
| G1805T | Ma, et al. _a | 4 | 63 | 272 | 71 | 607 | 13 | 61 | 120 | 87 | 301 | 1.795 | 0.180 |
| (rs5743618) | Ma, et al. _b | 107 | 61 | 12 | 275 | 85 | 63 | 33 | 14 | 159 | 61 | 6.956 | 0.008 |
| Ma, et al. _c | 20 | 83 | 272 | 123 | 627 | 14 | 39 | 61 | 67 | 161 | 3.518 | 0.061 | |
| Salvaraj et al. | 1 | 9 | 192 | 11 | 393 | 0 | 16 | 189 | 16 | 394 | 0.338 | 0.561 | |
| Uciechowski et al. | 36 | 5 | 4 | 77 | 13 | 24 | 19 | 6 | 67 | 31 | 0.525 | 0.469 | |
| TT | TC | CC | T | C | TT | TC | CC | T | C | X2 | P | ||
| TLR2 | Thuong et al. | 177 | 138 | 35 | 492 | 208 | 205 | 154 | 18 | 564 | 190 | 2.633 | 0.105 |
| T597C | Etokebe et al. | 66 | 84 | 40 | 216 | 164 | 162 | 244 | 83 | 568 | 410 | 0.298 | 0.585 |
| (rs3804099) | Ma, et al. _a | 46 | 165 | 128 | 257 | 421 | 29 | 100 | 65 | 158 | 230 | 0.889 | 0.346 |
| Ma, et al. _b | 55 | 90 | 35 | 200 | 160 | 41 | 47 | 22 | 129 | 91 | 1.562 | 0.211 | |
| Ma, et al. _c | 133 | 191 | 51 | 457 | 293 | 18 | 80 | 16 | 116 | 112 | 18.601 | 0.000 | |
| Sanchez, et al. | 173 | 220 | 72 | 566 | 364 | 95 | 153 | 52 | 343 | 257 | 0.514 | 0.473 | |
| Che, et al. | 52 | 54 | 9 | 158 | 72 | 68 | 68 | 20 | 204 | 108 | 0.214 | 0.644 | |
| TT | TC | CC | T | C | TT | TC | CC | T | C | X2 | P | ||
| TLR2 | Che, et al. | 60 | 48 | 7 | 168 | 62 | 79 | 61 | 16 | 219 | 93 | 0.670 | 0.413 |
| T1350C | Chen, et al. | 131 | 45 | 8 | 307 | 61 | 121 | 55 | 8 | 297 | 71 | 0.297 | 0.586 |
| (rs3804100) | Etokebe, et al. | 159 | 26 | 1 | 344 | 28 | 483 | 67 | 1 | 1033 | 69 | 0.709 | 0.400 |
| Ma, et al. _a | 299 | 38 | 2 | 636 | 42 | 169 | 25 | 0 | 363 | 25 | 0.920 | 0.337 | |
| Ma, et al. _b | 151 | 24 | 5 | 326 | 34 | 101 | 9 | 0 | 211 | 9 | 0.200 | 0.655 | |
| Ma, et al. _c | 312 | 62 | 1 | 686 | 64 | 100 | 14 | 0 | 214 | 14 | 0.488 | 0.485 | |
| GG | GA | AA | G | A | GG | GA | AA | G | A | X2 | P | ||
| TLR2 | Ogus, et al. | 124 | 13 | 14 | 261 | 41 | 107 | 7 | 2 | 221 | 11 | 12.783 | 0.000 |
| G2258A | Ma, et al. _a | 337 | 2 | 0 | 676 | 2 | 194 | 0 | 0 | 388 | 0 | / | / |
| (rs5743708) | Ma, et al. _b | 171 | 9 | 0 | 351 | 9 | 105 | 5 | 0 | 215 | 5 | 0.059 | 0.807 |
| Ma, et al. _c | 374 | 1 | 0 | 749 | 1 | 110 | 4 | 0 | 224 | 4 | 0.036 | 0.849 | |
| Jin, et al. | 99 | 71 | 0 | 269 | 71 | 168 | 31 | 0 | 367 | 31 | 1.420 | 0.233 | |
| Yu | 76 | 1 | 0 | 153 | 1 | 75 | 0 | 0 | 150 | 0 | / | / | |
| Salvaraj, et al | 192 | 1 | 0 | 385 | 1 | 198 | 1 | 0 | 397 | 1 | 0.001 | 0.972 | |
| Sanchez, et al. | 463 | 3 | 0 | 929 | 3 | 296 | 4 | 0 | 596 | 4 | 0.014 | 0.907 | |
| Xue, et al. | 204 | 1 | 0 | 409 | 1 | 202 | 1 | 0 | 405 | 1 | 0.001 | 0.972 | |
| Ma, et al. | 543 | 0 | 0 | 1086 | 0 | 544 | 0 | 0 | 1088 | 0 | / | / | |
| Dalgic, et al. | 93 | 31 | 0 | 217 | 31 | 186 | 14 | 0 | 386 | 14 | 0.263 | 0.608 | |
| CC | TC | TT | C | T | CC | TC | TT | C | T | X2 | P | ||
| TLR6 | Ma, et al. _a | 289 | 47 | 3 | 625 | 53 | 137 | 50 | 7 | 324 | 64 | 0.805 | 0.370 |
| C745T | Ma, et al. _b | 61 | 88 | 31 | 210 | 150 | 38 | 46 | 26 | 122 | 98 | 2.594 | 0.107 |
| (rs5743810) | Ma, et al. _c | 291 | 74 | 10 | 656 | 94 | 78 | 31 | 5 | 187 | 41 | 0.696 | 0.404 |
| Salvaraj, et al. | 0 | 2 | 197 | 2 | 396 | 0 | 3 | 199 | 3 | 401 | 0.011 | 0.915 | |
HWE: Hardy-Weinberg Equilibrium.
Quantitative Data Synthesis
TLR1 G1805T polymorphism
Six case-control studies (1684 cases and 1216 controls) on the relationship between the G1805A polymorphisms and the risk of TB were included in the meta-analysis.
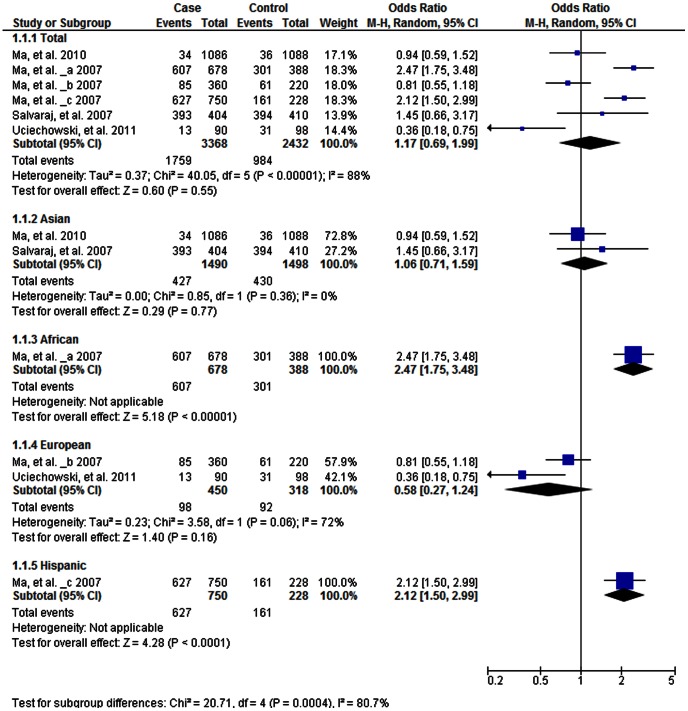
As shown in Table 3, the heterogeneity of T vs. G for all the studies was analyzed. The χ2 value was 40.05, with 5 degrees of freedom (df) and P<0.001 in a random effect model. I 2, another index of the test of heterogeneity, was 88%, suggesting a high heterogeneity. We therefore chose the random-effect model to synthesize the data. The overall OR for the T vs. G alleles was 1.17 (P = 0.55; Figure 3). No association was found in the allelic frequency with TB risk. The publication bias by Begg’s test showed no significant bias (P>0.05; Figure S1). We also performed comparison for the other four genetic models and no associations were found in any of these (Table 3).
Table 3. Summary of different comparative meta-analysis results.
| Polymorphism | Genetic model | Participants | OR [95%CI] | Z | P value | I2 % | P het | Effect model | Begg’s test p>|z| | Egger’s test p>|t| |
| TLR1 G1805T | TT+GT vs. GG | 2900 | 1.13 [0.54, 2.36] | 0.33 | 0.74 | 80 | 0.0001 | R | 0.707 | 0.899 |
| TT vs. GT+GG | 2900 | 1.43 [0.83, 2.48] | 1.29 | 0.20 | 68 | 0.007 | R | 0.260 | 0.147 | |
| TG vs. GG | 1756 | 0.94 [0.51, 1.76] | 0.18 | 0.86 | 68 | 0.008 | R | 1.000 | 0.661 | |
| TT vs. GG | 2445 | 1.31 [0.44, 3.92] | 0.48 | 0.63 | 77 | 0.0006 | R | 1.000 | 0.670 | |
| T vs. G | 2900 | 1.17 [0.69, 1.99] | 0.60 | 0.55 | 88 | <0.001 | R | 0.260 | 0.222 | |
| TLR2 T597C | CC+CT vs. TT | 3754 | 0.90 [0.68, 1.18] | 0.77 | 0.44 | 69 | 0.004 | R | 0.764 | 0.555 |
| CC vs. CT+TT | 3754 | 1.12 [0.93, 1.34] | 1.20 | 0.23 | 42 | 0.11 | F | 0.548 | 0.717 | |
| CT vs. TT | 3108 | 0.87 [0.66, 1.15] | 0.99 | 0.32 | 67 | 0.006 | R | 1.000 | 0.736 | |
| CC vs. TT | 1966 | 1.00 [0.69, 1.44] | 0.01 | 0.99 | 63 | 0.01 | R | 0.764 | 0.646 | |
| C vs. T | 3754 | 0.98 [0.83, 1.15] | 0.24 | 0.81 | 60 | 0.02 | R | 0.764 | 0.689 | |
| TLR2 T1350C | CC+CT vs. TT | 2688 | 1.06 [0.86, 1.31] | 0.55 | 0.58 | 27 | 0.24 | F | 0.133 | 0.024 |
| CC vs. CT+TT | 2688 | 1.01 [0.56, 1.79] | 0.02 | 0.98 | 0 | 0.52 | F | 0.707 | 0.063 | |
| CT vs. TT | 2639 | 1.05 [0.84, 1.30] | 0.41 | 0.68 | 4 | 0.39 | F | 0.133 | 0.071 | |
| CC vs. TT | 2214 | 1.01 [0.56, 1.82] | 0.04 | 0.97 | 0 | 0.52 | F | 0.452 | 0.051 | |
| C vs. T | 2688 | 1.05 [0.87, 1.27] | 0.54 | 0.59 | 43 | 0.12 | F | 0.024 | 0.009 | |
| TLR2 G2258A | AA+AG vs. GG | 5077 | 1.72 [0.89, 3.31] | 1.62 | 0.10 | 61 | 0.006 | R | 0.721 | 0.041 |
| AA vs. AG+GG | 5077 | 5.82 [1.30, 26.16] | 2.30 | 0.02 | / | / | R | / | / | |
| AG vs. GG | 5061 | 1.55 [0.78, 3.09] | 1.25 | 0.21 | 63 | 0.004 | R | 1.000 | 0.041 | |
| AA vs. GG | 4877 | 6.04 [1.34, 27.18] | 2.34 | 0.02 | / | / | R | / | / | |
| A vs. G | 5077 | 1.78 [0.99, 3.20] | 1.92 | 0.05 | 56 | 0.01 | R | 0.592 | 0.052 | |
| TLR6 C745T | TT+CT vs. CC | 1713 | 0.64 [0.38, 1.06] | 1.73 | 0.08 | 72 | 0.03 | R | 0.296 | 0.019 |
| TT vs. CT+CC | 1713 | 0.61 [0.39, 0.97] | 2.10 | 0.04 | 0 | 0.41 | F | 1.000 | 0.890 | |
| CT vs. CC | 1235 | 0.69 [0.40, 1.19] | 1.34 | 0.18 | 74 | 0.02 | R | 0.296 | 0.056 | |
| TT vs. CC | 1372 | 0.57 [0.34, 0.95] | 2.16 | 0.03 | 29 | 0.24 | F | 0.296 | 0.261 | |
| T vs. C | 1713 | 0.66 [0.44, 0.99] | 2.01 | 0.04 | 65 | 0.04 | R | 1.000 | 0.811 |
P het = P value for heterogeneity; OR = odds ratio; CI = confidence interval; F = fixed-effect model; R = random-effect model.
Figure 3. Forrest plot of the association between TLR1 C1805T and TB risk in allele comparison (T vs. C).
Subgroup analysis was performed by ethnicity. OR: odds ratio; CI: confidence interval; df: degrees of freedom.
We also performed a stratified subgroup allelic analysis by ethnicity and different results were found in different ethnic populations. Slight decreased risk among Europeans (OR 0.58 [0.27, 1.24], P = 0.16; Figure 3) and slight increased risk among Asians (OR 1.06 [0.71, 1.59], P = 0.77; Figure 3) were observed. However, one study in Africans (OR 2.47 [1.75, 3.48], P<0.01; Figure 3) and one in Hispanics (OR 2.12 [1.50, 2.99], P<0.01; Figure 3) showed a significant increased risk associated with the 1805T allele.
TLR2 T597C polymorphism
Seven case-control studies (2014 cases and 1740 controls) were included in the meta-analysis on the relationship between the T597C polymorphism and the risk of TB.
As shown in Table 3, the heterogeneity of C vs. T for all the studies was analyzed. The χ2 value was 15.14, with 6 df and P = 0.02 using the random-effect model, while I 2 was 60, suggesting a modest heterogeneity. We therefore chose the random-effect model to synthesize the data. The overall OR for the C vs. T alleles was 0.98 (95% CI 0.83–1.15) and the test for overall effect, Z value, was 0.24 (P = 0.81), indicating no association between this polymorphism and TB. The publication bias by Begg’s test showed no significant publication bias (P>0.05). Comparisons for four other genetic models were also performed, and no associations were found either (Table 3).
TLR2 T1350C polymorphism
Six case-control studies (1379 cases and 1309 controls) were included in the meta-analysis to determine the relationship between the T1350C polymorphism and the risk of TB.
As shown in Table 3, the heterogeneity of C vs. T for all the studies was analyzed. The χ2 value was 8.79, with 5 df and P = 0.12 using the fixed-effect model, while I 2 was 43%, suggesting limited heterogeneity (Table 3). The overall OR for the C vs. T alleles was 1.05 (95% CI 0.87–1.27), and the test for overall effect, Z value, was 0.54 (P = 0.59). Comparisons for four other genetic models were also performed, and no associations were found (Table 3). The publication bias by Begg’s test showed no bias in all the genetic models except the allelic model (P<0.05), indicating that there might be some publication bias in these studies.
TLR2 G2258A polymorphism
Eleven case-control studies (2823 cases and 2254 controls) on the relationship between the TLR2 G2258A were included in the meta-analysis.
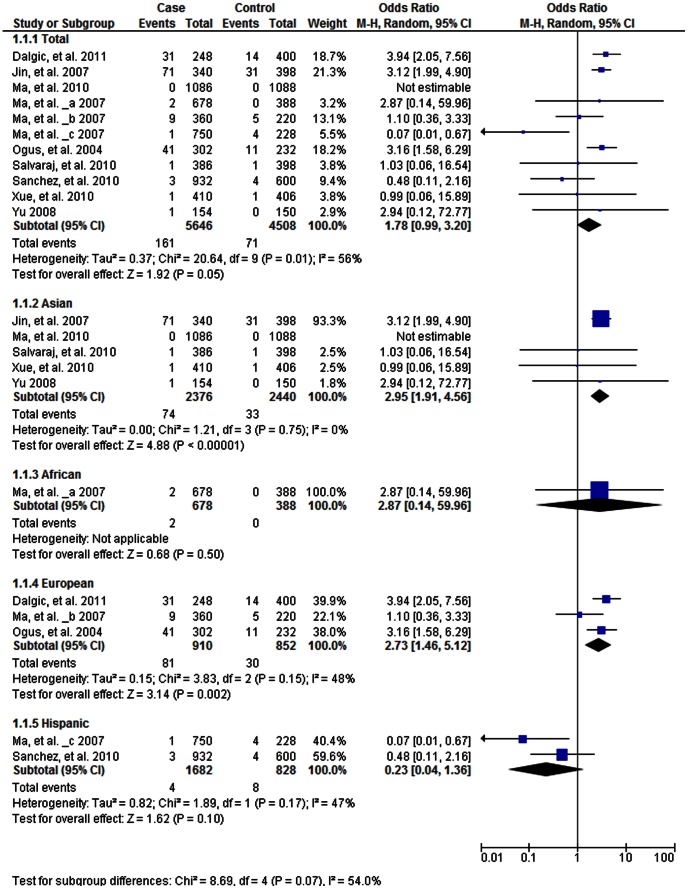
As shown in Table 3, the heterogeneity of A vs. G for all the studies was analyzed. The χ2 value was 20.64, with 9 df and P = 0.01 in a random-effect model and I 2 was 56%, suggesting moderate heterogeneity. The overall OR for the A vs. G alleles was 1.78 (95% CI 0.99–3.20) and the test for overall effect, Z value, was 1.92. (P = 0.05; Figure 4). The results suggest an increased risk in the A alleles. In recessive model analysis, the overall OR for the AA vs. AG+GG was 5.82 (95% CI 1.30–26.16, P = 0.02) using the random effect model, indicating an association of the AA genotype with TB risk. The Begg’s test did not show publication bias in the studies (P>0.05, Figure S2).
Figure 4. Forrest plot of the association between TLR2 G2258A and TB risk in allele comparison (A vs.
G). Subgroup analysis was performed by ethnicity. OR: odds ratio; CI: confidence interval; df: degrees of freedom.
We also performed a stratified analysis by ethnicity for the allelic model (A vs. G). Significantly increased risks were found among Asians (OR 2.95, 95%CI 1.91–4.56, P<0.001; Figure 4) and Europeans (OR 2.73, 95% 1.46–5.12, P = 0.002; Figure 4) and decreased risk was found in Hispanic population (OR 0.23, 95% 0.04–1.36, P = 0.10; Figure 4), while there was a slight but not significant increased risk for A allele in Africans (P = 0.50; Figure 4).
TLR6 C745T polymorphism
Four case-control studies (1093 cases and 620 controls) were included in the meta-analysis on the relationship between the TLR6 C745T polymorphism and the risk of TB.
As shown in Table 3, the heterogeneity of T vs. C for all the studies was analyzed. The χ2 value was 12.64, with 3 df and P = 0.005 using the random-effect model, while I 2 was 76, suggesting a high heterogeneity. The overall OR for the C vs. T alleles was 0.66 (95% CI 0.44–0.99) and the test for overall effect, Z value, was 1.34 (P = 0.04; Figure 5), indicating a protective association between the T allele and TB. In addition, in recessive model analysis, the overall OR for the TT vs. CT+CC was 0.61 (95% CI 0.39–0.97, P = 0.04), also indicating a decreased TB risk in subject with the TT genotype. The publication bias by Begg’s test showed no significant publication bias (P>0.05; Figure S3).
Figure 5. Forrest plot of the association between TLR6 745T and TB risk in allele comparison (T vs. C).
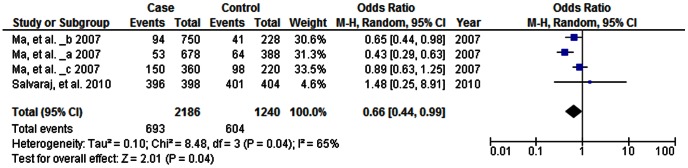
OR: odds ratio; CI: confidence interval; df: degrees of freedom.
Discussion
In this study, we performed a meta-analysis to assess the association between five extensively studied polymorphisms (TLR1 G1805T, TLR2 T597C, T1350C, G2258A and TLR6 C745T) and TB risk reported until 1st January 2013. The meta-analysis revealed an association between the TLR2 2258AA genotype and TLR6 745TT and TB risk. At the same time, different genetic models and ethnicity subgroups comparisons were also performed in TLR1 G1805T, TLR2 G2258A polymorphisms. TLR1 1805T allele was associated with TB in Africans and American Hispanics, and TLR2 2258A allele was associated with TB in Asians and Europeans. However, the TLR2 T597C, TLR2 T1350C polymorphism did not show significant association with TB.
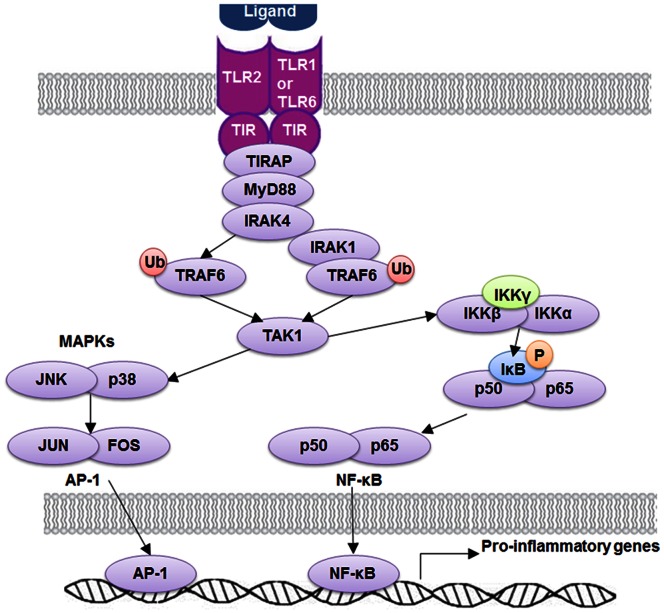
TLR-1, -2, and -6 are critical components in Mtb recognition and signaling [34]–[36]. As shown in Figure 6, ligand recognition leads to TLR2 dimerization with TLR1 or TLR6 that brings together their TIR domains and triggers TLR2 tyrosine phosphorylation. This would serve as docking platforms to enable recruitment of MyD88 [35]. MyD88 interacts with TIRAP via TIR-TIR domain interactions, serving as a scaffold to recruit IL-1 receptor associated kinase (IRAK) 4 and IRAK1. Clustered IRAKs could undergo auto-phosphorylation and kinase activation. Phosphorylated IRAK4 would recruit TNF receptor associated factor 6 (TRAF6), leading to their ubiquitination. Ubiquitinated TRAF6 recruits TGF-β-activated kinase (TAK) 1 by engaging ubiquitin recognition domains within inhibitor of NF-κB (IκB) kinase (IKK)-γ and TAK-interacting proteins [35]. On the other hand, phosphorylated IRAK1 could also undergoes ubiquitination via recruitment of pellinos and TRAF6, resulting in direct recruitment of IKK-γ [35]. Finally, these processes activate TAK1 and the IKK complex and place them into close proximity, promoting TAK-mediating activation of MAPK and IKK-β, resulting in nuclear translocation of transcription factors, such as AP-1 and NF-κB, that induces transcription of inflammatory genes [34]. Because of their essential roles in Mtb recognition process, polymorphisms in TLR-1, -2 and -6 are hypothesized to be associated with susceptibility to TB and other infectious diseases.
Figure 6. Signaling pathway of TLR-1, -2 and -6.
TLR2 ligands induce conformational changes of TLRs that allow interactions of TLR2 and TLR-1, -6. This heterodimer could recruit adaptor proteins MyD88 and TIRAP. MyD88 can activate NF-κB and AP-1 through IRAKs, TRAF6, TAK1, and IKK complex, resulting in induction of pro-inflammatory genes in macrophages and cDCs. TLR: Toll-Like Receptor; TIR: Toll-IL1-Receptor Domain; TIRAP: TIR domain containing adaptor protein; MyD88: Myeloid Differentiation Primary Response Gene 88; IRAK: Interleukin-1 Receptor associated kinase 1; TRAF: TNF Receptor Associated Factor; TAK: TGF-β-Actiated Kinase; IκB: Inhibitor of NF-κB; IKK: IκB kinase.
The G2258A polymorphism in TLR2 was first identified by mutation screen assay in healthy study subjects, which cause the 753 amino acid substitution from Arg to Gln in the conserved TIR domain (Figure 1) [37]. In 293T cells transfected with wild-type or G2258A TLR2 constructs, 2258A allele has a significant decreased NF-κB response against bacterial peptides from B. burgdorferi, T. pallidum and R. akari in comparison to the wild-type, resulting in an attenuated immune response [37], [38]. Recently, another study looked into the mechanisms behind this and demonstrated that G2258A rendered TLR2 signaling-deficient by impairing TLR2 tyrosine phosphorylation, TLR2-TLR6 hetero-dimerization, and recruitment of TIRAP and MyD88 in transfected HEK293 cells, without affecting TLR2 expression [39]. This study also showed that polymorphism in the TIR domain could change the electrostatic potential of the DD loop and α-D region, which interact with the BB loops of TLR1 and TLR6, and thus impair their dimerization. Because of the functional importance of this SNP, numerous studies have investigated the association of this polymorphism with TB, urinary tract infection and other infectious diseases [25], [40], [41]. One of the studies found that atopic dermatitis (AD) patients carrying the 2258A allele showed higher levels of total serum IgE as well as of superantigen-specific IgE and had a higher risk of getting the disease [42]. Another study reported that G2258A TLR2 allele in transfected HEK293 cells had a decreased IL-8 secretion after stimulation by lipoteichoic acid, heat-inactivated Staphylococcus aureus or triacylated lipopeptides, and it was associated with suppressed IL-8 production by monocytes in AD patients as well [43]. Therefore, this SNP might have a significant functional impact on the TLR2 signaling. In our analysis, TLR2 G2258A was linked to TB in the recessive model (AA vs. AG+GG), but other polymorphisms in the TLR2 did not associate with TB. The subgroup analysis by ethnicity showed that G2258A was associated with an increased risk of TB in Asian and European populations. The reasons for this difference of association in different populations were unclear, but the differences in the frequencies of the polymorphisms among different races might contribute to this heterogeneity. In addition, numerous polymorphisms outside the TLR2 gene can contribute to the host susceptibility to TB. These polymorphisms might differ greatly in various ethnic groups with different evolutionary backgrounds. These differences, along with gene-gene interactions and environmental and cultural factors, and even the variations in Mycobacterium strains, made understanding the observed differences between ethnic groups even more complicated.
TLR1 T1805G is a common non-synonymous polymorphisms located in the trans-membrane domain of TLR1 molecule (Figure 1), causing a substitution of isoleucine with serine. Studies showed that this substitution affected the TLR1 surface trafficking and led to an impairment of the inflammation pathway, and it might be the most common SNP affecting TLR function identified in any population [44]. TLR1 T1805G transfected HEK293 cells had substantially greater basal and lipopeptide-induced or Mtb extracts-induced NF-κB signaling [12]. Furthermore, individuals with the 1805TT genotypes produced substantially more IL-6 than those with the 1805GG in a lipopeptide-stimulated whole-blood cytokine assay [12]. In another study, TLR1 1805GG carriers have an impaired cell surface expression of TLR1 and impaired inflammatory response to lipopeptide agonists for TLR1/2 [44]. It was further validated that this effect was caused by T1805G, not a genetically linked allele, by transfection assay [44]. This study also discovered that 1805G was a common SNP and was associated with an increased risk of leprosy [44]. Different groups using different methods have also replicated these findings in various models [45], [46]. In a subgroup analysis of TLR polymorphisms and susceptibility to inflammatory bowel disease, SNP T1805G was associated with Crohn’s disease that was confined to the ileum [47]. These observations together with association studies in other infectious diseases [44], [48] suggested that TLR1 T1805G could potentially impact the innate immune response and clinical susceptibility to TB, which is consistent with our meta-analysis results that 1805G was associated with TB susceptibility in Africans and American Hispanics. The reason why different results were observed in different populations might be that the frequency of this SNP varied widely across different populations [12].
In our analysis, the TLR6 745TT genotype was associated with a reduced TB risk. Given the location of this non-synonymous SNP in the extracellular domain (Figure 1), people have speculated that this SNP might alter ligand recognition. One study showed that this SNP was associated with altered IL-6 secretion from the whole blood samples of the healthy people in response to di-acylated lipopeptide, Mtb lysate and Bacille Calmette-Guerin (BCG) [13]. In addition, this SNP allele was associated with different NF-κB signaling in response to di-acylated lipopeptide, PAM2 or Mtb in an HEK293 cell line reconstitution assay [13]. Another study re-stimulated the whole blood drawn from the newborn treated with BCG and found that individuals with 745T alleles had significantly higher levels of INF-γ, IL-2, and IL-13 [49]. The PBMCs from these individuals also secreted higher amounts of IL-6 and IL-10 after stimulation with lipopeptides, Mtb lysate and BCG [49]. Because inability to produce or respond to IFN-γ in host has been associated with higher susceptibility to disseminated mycobacterial infection [50], the 745C allele that was linked to lower level of IFN-γ response might thus increase TB risk. Therefore, the 745T allele, which is associated with an elevated IFN-γ response against Mtb [49], may confer protection against TB. This is consistent with the results in our meta-analysis.
However, there are still some limitations in our meta-analysis that should be considered when explaining the present results. One of our limitations is the limited number of studies included in this review. Our review reported an interesting preliminary conclusion, but this must be validated by future large-scale and functional studies in different populations. Another limitation is that we mainly focused on the most studied coding variants of candidate genes TLR-1, -2 and -6 and their association with TB risk and excluded the effects of other genetic variants. Other polymorphisms such as TLR2 Arg677Trp polymorphism [51] microsatellite polymorphisms in TLR2 gene might also linked to TB and have effects on our candidate SNPs. [52]. These polymorphisms were not included in this systematic review because of insufficient studies available in the database. In addition, the polymorphisms besides the included ones or even the ones in other genes could also affect our results. Therefore, genome-wide association studies (GWAS) in different ethnic groups are needed to validate our results. To date, four GWAS have been performed to study TB susceptibility [53]–[56]. These studies identified polymorphisms on chromosomes 18q11.2 and 11q13 in African but no polymorphism in Asian to be associated with TB. Meta-analysis of the GWAS study might provide more power to detect SNP linked to complex diseases [57]. Therefore the meta-analysis of these studies and future studies in various populations might provide more information on TB risk loci and polymorphism-polymorphism interactions.
Understanding the relation among the TLR polymorphisms, innate immunity and TB susceptibility is also valuable in translational research and personalized medicine. Although susceptibility to TB is historically ascribed to an inadequate immune response that fails to control infecting mycobacteria, people have find that susceptibility to Mycobacterium can result from either inadequate or excessive acute inflammation [4]. Although lack of TLR signaling might promote the TB development [36], excessive acute inflammation induced by TLR2 activation can lead to severe tissue damages [58]. Recently, Tobin and colleagues have investigated the genetic balance that maintains the host between the two extremes of failed immunity and damaging hyper-immunity [4]. By stratifying tuberculous meningitis patients from a historic randomized trial by ITA4H genotype, they revealed both different outcomes and different responses to dexamethasone, which might promote the personalized application of eicosanoid-targeting drugs [4]. As drugs targeting TLRs are being developed for infectious and inflammatory diseases including TB [59], host genotype-specific therapy based on knowledge of TLRs polymorphisms may optimize the inflammatory response to Mtb infection in the future.
In conclusion, this systematic review summarized the association between TLR-1, -2, and -6 polymorphisms and TB susceptibility. Our results indicate that TLR2 G2258A is associated with increased TB risk, especially in Asians and Europeans. TLR1 G1805T is associated with increased TB in Africans and American Hispanics. TLR6 C745T is associated with decreased TB risk. Whereas, TLR2 T597C and TLR2 T1350C polymorphisms did not show significant association with TB. Our systematic review and meta-analysis reported an interesting preliminary conclusion, but this must be validated by future large-scale studies in different populations. Whether the association between these polymorphisms and the TB risk was due to causal effect need to be further studied by functional studies.
Supporting Information
Funnel plot analysis to detect publication bias for association of TLR1 C1805T and TB risk in allele comparison (T vs. C).
(TIF)
Funnel plot analysis to detect publication bias for association of TLR2 G2258A and TB risk in allele comparison (A vs. G).
(TIF)
Funnel plot analysis to detect publication bias for association of TLR6 C745T and TB risk in allele comparison (T vs. C).
(TIF)
PRISMA 2009 checklist.
(DOC)
Exclusion criteria for excluded studies.
(DOC)
Funding Statement
This work was supported by grants from National Special Sci-Tech Projects (No. 2012ZX10005001-006) and National Natural Science Foundation of China (No. 81072724. No.81273882). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rossman M, Oner-Eyuboglu A (1998) Clinical presentation and treatment of tuberculosis. In: Fishman A, editor. Fishman’s Pulmonary Diseases and Disorders. 3rd ed. New York, NY, USA: McGraww Hill Company. 2483–2501.
- 2. Bellamy R (2003) Susceptibility to mycobacterial infections: the importance of host genetics. Genes Immun 4: 4–11. [DOI] [PubMed] [Google Scholar]
- 3. Tobin DM, Vary JC Jr, Ray JP, Walsh GS, Dunstan SJ, et al. (2010) The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell 140: 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tobin DM, Roca FJ, Oh SF, McFarland R, Vickery TW, et al. (2012) Host genotype-specific therapies can optimize the inflammatory response to mycobacterial infections. Cell 148: 434–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jo EK (2008) Mycobacterial interaction with innate receptors: TLRs, C-type lectins, and NLRs. Curr Opin Infect Dis 21: 279–286. [DOI] [PubMed] [Google Scholar]
- 6. Sanchez D, Rojas M, Hernandez I, Radzioch D, Garcia LF, et al. (2010) Role of TLR2- and TLR4-mediated signaling in Mycobacterium tuberculosis-induced macrophage death. Cell Immunol 260: 128–136. [DOI] [PubMed] [Google Scholar]
- 7. Branger J, Leemans JC, Florquin S, Weijer S, Speelman P, et al. (2004) Toll-like receptor 4 plays a protective role in pulmonary tuberculosis in mice. Int Immunol 16: 509–516. [DOI] [PubMed] [Google Scholar]
- 8. Bafica A, Scanga CA, Feng CG, Leifer C, Cheever A, et al. (2005) TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J Exp Med 202: 1715–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Drage MG, Pecora ND, Hise AG, Febbraio M, Silverstein RL, et al. (2009) TLR2 and its co-receptors determine responses of macrophages and dendritic cells to lipoproteins of Mycobacterium tuberculosis. Cell Immunol 258: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harding CV, Boom WH (2010) Regulation of antigen presentation by Mycobacterium tuberculosis: a role for Toll-like receptors. Nat Rev Microbiol 8: 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O’Neill LA, Bowie AG (2007) The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol 7: 353–364. [DOI] [PubMed] [Google Scholar]
- 12. Hawn TR, Misch EA, Dunstan SJ, Thwaites GE, Lans NTN, et al. (2007) A common human TLR1 polymorphism regulates the innate immune response to lipopeptides. Eur J Immunol 37: 2280–2289. [DOI] [PubMed] [Google Scholar]
- 13. Shey MS, Randhawa AK, Bowmaker M, Smith E, Scriba TJ, et al. (2010) Single nucleotide polymorphisms in toll-like receptor 6 are associated with altered lipopeptide- and mycobacteria-induced interleukin-6 secretion. Genes Immun 11: 561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Etokebe GE, Skjeldal F, Nilsen N, Rodionov D, Knezevic J, et al. (2010) Toll-like receptor 2 (P631H) mutant impairs membrane internalization and is a dominant negative allele. Scand J Immunol 71: 369–381. [DOI] [PubMed] [Google Scholar]
- 15. Miao R, Li J, Sun Z, Xu F, Shen H (2011) Meta-analysis on the association of TIRAP S180L variant and tuberculosis susceptibility. Tuberculosis (Edinb) 91: 268–272. [DOI] [PubMed] [Google Scholar]
- 16. Tian T, Jin S, Dong J, Li G (2013) Lack of association between Toll-like receptor 4 gene Asp299Gly and Thr399Ile polymorphisms and tuberculosis susceptibility: A meta-analysis. Infect Genet Evol 14: 156–160. [DOI] [PubMed] [Google Scholar]
- 17. Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40: D302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Begg CB, Berlin JA (1989) Publication bias and dissemination of clinical research. J Natl Cancer Inst 81: 107–115. [DOI] [PubMed] [Google Scholar]
- 21.Chen YC, Hsiao CC, Chen CJ, Chin CH, Liu SF, et al.. (2010) Toll-like receptor 2 gene polymorphisms, pulmonary tuberculosis, and natural killer cell counts. BMC Med Genet 11. [DOI] [PMC free article] [PubMed]
- 22. Dalgic N, Tekin D, Kayaalti Z, Soylemezoglu T, Cakir E, et al. (2011) Arg753Gln polymorphism of the human Toll-like receptor 2 gene from infection to disease in pediatric tuberculosis. Hum Immunol 72: 440–445. [DOI] [PubMed] [Google Scholar]
- 23. Ma MJ, Xie LP, Wu SC, Tang F, Li H, et al. (2010) Toll-like receptors, tumor necrosis factor-alpha, and interleukin-10 gene polymorphisms in risk of pulmonary tuberculosis and disease severity. Hum Immunol 71: 1005–1010. [DOI] [PubMed] [Google Scholar]
- 24.Ma X, Liu Y, Gowen BB, Graviss EA, Clark AG, et al.. (2007) Full-exon resequencing reveals toll-like receptor variants contribute to human susceptibility to tuberculosis disease. PLoS ONE 2. [DOI] [PMC free article] [PubMed]
- 25. Ogus AC, Yoldas B, Ozdemir T, Uguz A, Olcen S, et al. (2004) The Arg753GIn polymorphism of the human Toll-like receptor 2 gene in tuberculosis disease. European Respiratory Journal 23: 219–223. [DOI] [PubMed] [Google Scholar]
- 26. Sanchez D, Lefebvre C, Rioux J, Garcia LF, Barrera LF (2012) Evaluation of Toll-like receptor and adaptor molecule polymorphisms for susceptibility to tuberculosis in a Colombian population. Int J Immunogenet 39: 216–223. [DOI] [PubMed] [Google Scholar]
- 27. Selvaraj P, Harishankar M, Singh B, Jawahar MS, Banurekha VV (2010) Toll-like receptor and TIRAP gene polymorphisms in pulmonary tuberculosis patients of South India. Tuberculosis 90: 306–310. [DOI] [PubMed] [Google Scholar]
- 28. Thuong NTT, Hawn TR, Thwaites GE, Chau TTH, Lan NTN, et al. (2007) A polymorphism in human TLR2 is associated with increased susceptibility to tuberculous meningitis. Genes Immun 8: 422–428. [DOI] [PubMed] [Google Scholar]
- 29. Uciechowski P, Imhoff H, Lange C, Meyer CG, Browne EN, et al. (2011) Susceptibility to tuberculosis is associated with TLR1 polymorphisms resulting in a lack of TLR1 cell surface expression. J Leukoc Biol 90: 377–388. [DOI] [PubMed] [Google Scholar]
- 30. Xue Y, Zhao ZQ, Wang HJ, Jin L, Liu CP, et al. (2010) Toll-like receptors 2 and 4 gene polymorphisms in a southeastern Chinese population with tuberculosis. Int J Immunogenet 37: 135–138. [DOI] [PubMed] [Google Scholar]
- 31.Yu SL (2008) Expression and Function of Toll-Like Receptor-2 in Mycobacterium Infected Macrophages and Its association with Tuberculosis Susceptibility.
- 32. Jin L, Ding SP, Zhu XF, Zhong JP, Li JC (2007) The Arg753Gln Polymorphism of the Human Toll-like Receptor 2 Gene and Its Association with Tuberculosis Disease in Zhejiang Han Population. Chinese Journal of Cell Biology 29: 229–231. [Google Scholar]
- 33. Che N, Jiang S, Gao T, Li S, Zhang X, et al. (2011) Relationship between toll-like receptor 2 gene polymorphism and pulmonary tuberculosis in Chineses Han population. The Journal of the Chinese Antituberculosis 33: 204–208. [Google Scholar]
- 34. Lee MS, Kim YJ (2007) Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu Rev Biochem 76: 447–480. [DOI] [PubMed] [Google Scholar]
- 35. Kawai T, Akira S (2011) Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34: 637–650. [DOI] [PubMed] [Google Scholar]
- 36. Drennan MB, Nicolle D, Quesniaux VJ, Jacobs M, Allie N, et al. (2004) Toll-like receptor 2-deficient mice succumb to Mycobacterium tuberculosis infection. Am J Pathol 164: 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lorenz E, Mira JP, Cornish KL, Arbour NC, Schwartz DA (2000) A novel polymorphism in the toll-like receptor 2 gene and its potential association with staphylococcal infection. Infect Immun 68: 6398–6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Quevedo-Diaz MA, Song C, Xiong Y, Chen H, Wahl LM, et al. (2010) Involvement of TLR2 and TLR4 in cell responses to Rickettsia akari. J Leukoc Biol 88: 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xiong Y, Song C, Snyder GA, Sundberg EJ, Medvedev AE (2012) R753Q polymorphism inhibits Toll-like receptor (TLR) 2 tyrosine phosphorylation, dimerization with TLR6, and recruitment of myeloid differentiation primary response protein 88. J Biol Chem 287: 38327–38337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Misch EA, Hawn TR (2008) Toll-like receptor polymorphisms and susceptibility to human disease. Clin Sci (Lond) 114: 347–360. [DOI] [PubMed] [Google Scholar]
- 41. Tabel Y, Berdeli A, Mir S (2007) Association of TLR2 gene Arg753Gln polymorphism with urinary tract infection in children. Int J Immunogenet 34: 399–405. [DOI] [PubMed] [Google Scholar]
- 42. Ahmad-Nejad P, Mrabet-Dahbi S, Breuer K, Klotz M, Werfel T, et al. (2004) The toll-like receptor 2 R753Q polymorphism defines a subgroup of patients with atopic dermatitis having severe phenotype. J Allergy Clin Immunol 113: 565–567. [DOI] [PubMed] [Google Scholar]
- 43. Mrabet-Dahbi S, Dalpke AH, Niebuhr M, Frey M, Draing C, et al. (2008) The Toll-like receptor 2 R753Q mutation modifies cytokine production and Toll-like receptor expression in atopic dermatitis. J Allergy Clin Immunol 121: 1013–1019. [DOI] [PubMed] [Google Scholar]
- 44. Johnson CM, Lyle EA, Omueti KO, Stepensky VA, Yegin O, et al. (2007) Cutting edge: A common polymorphism impairs cell surface trafficking and functional responses of TLR1 but protects against leprosy. J Immunol 178: 7520–7524. [DOI] [PubMed] [Google Scholar]
- 45. Barreiro LB, Ben-Ali M, Quach H, Laval G, Patin E, et al. (2009) Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet 5: e1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Uciechowski P, Imhoff H, Lange C, Meyer CG, Browne EN, et al. (2011) Susceptibility to tuberculosis is associated with TLR1 polymorphisms resulting in a lack of TLR1 cell surface expression. Journal of Leukocyte Biology 90: 377–388. [DOI] [PubMed] [Google Scholar]
- 47. Pierik M, Joossens S, Van Steen K, Van Schuerbeek N, Vlietinck R, et al. (2006) Toll-like receptor-1, -2, and -6 polymorphisms influence disease extension in inflammatory bowel diseases. Inflamm Bowel Dis 12: 1–8. [DOI] [PubMed] [Google Scholar]
- 48. Leoratti FM, Farias L, Alves FP, Suarez-Mutis MC, Coura JR, et al. (2008) Variants in the toll-like receptor signaling pathway and clinical outcomes of malaria. J Infect Dis 198: 772–780. [DOI] [PubMed] [Google Scholar]
- 49.Randhawa AK, Shey MS, Keyser A, Peixoto B, Wells RD, et al.. (2011) Association of human TLR1 and TLR6 deficiency with altered immune responses to BCG vaccination in South African infants. PLoS Pathogens 7. [DOI] [PMC free article] [PubMed]
- 50. Casanova JL, Abel L (2002) Genetic dissection of immunity to mycobacteria: the human model. Annu Rev Immunol 20: 581–620. [DOI] [PubMed] [Google Scholar]
- 51. Ben-Ali M, Barbouche MR, Bousnina S, Chabbou A, Dellagi K (2004) Toll-like receptor 2 Arg677Trp polymorphism is associated with susceptibility to tuberculosis in Tunisian patients. Clin Diagn Lab Immunol 11: 625–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xue Y, Jin L, Li AZ, Wang HJ, Li M, et al. (2010) Microsatellite polymorphisms in intron 2 of the toll-like receptor 2 gene and their association with susceptibility to pulmonary tuberculosis in Han Chinese. Clinical Chemistry & Laboratory Medicine 48: 785–789. [DOI] [PubMed] [Google Scholar]
- 53. Thye T, Vannberg FO, Wong SH, Owusu-Dabo E, Osei I, et al. (2010) Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet 42: 739–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thye T, Owusu-Dabo E, Vannberg FO, van Crevel R, Curtis J, et al. (2012) Common variants at 11p13 are associated with susceptibility to tuberculosis. Nat Genet 44: 257–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mahasirimongkol S, Yanai H, Mushiroda T, Promphittayarat W, Wattanapokayakit S, et al. (2012) Genome-wide association studies of tuberculosis in Asians identify distinct at-risk locus for young tuberculosis. J Hum Genet 57: 363–367. [DOI] [PubMed] [Google Scholar]
- 56. Png E, Alisjahbana B, Sahiratmadja E, Marzuki S, Nelwan R, et al. (2012) A genome wide association study of pulmonary tuberculosis susceptibility in Indonesians. BMC Med Genet 13: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Minozzi G, Williams JL, Stella A, Strozzi F, Luini M, et al. (2012) Meta-analysis of two genome-wide association studies of bovine paratuberculosis. PLoS One 7: e32578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Olivera S, Rodriguez-Ithurralde D, Henley JM (2003) Acetylcholinesterase promotes neurite elongation, synapse formation, and surface expression of AMPA receptors in hippocampal neurones. Mol Cell Neurosci 23: 96–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hennessy EJ, Parker AE, O’Neill LA (2010) Targeting Toll-like receptors: emerging therapeutics? Nat Rev Drug Discov 9: 293–307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Funnel plot analysis to detect publication bias for association of TLR1 C1805T and TB risk in allele comparison (T vs. C).
(TIF)
Funnel plot analysis to detect publication bias for association of TLR2 G2258A and TB risk in allele comparison (A vs. G).
(TIF)
Funnel plot analysis to detect publication bias for association of TLR6 C745T and TB risk in allele comparison (T vs. C).
(TIF)
PRISMA 2009 checklist.
(DOC)
Exclusion criteria for excluded studies.
(DOC)