Abstract
Cryptococcus is a major fungal pathogen that frequently causes systemic infection in patients with compromised immunity. Glucose, an important signal molecule and the preferred carbon source for Cryptococcus, plays a critical role in fungal development and virulence. Cryptococcus contains more than 50 genes sharing high sequence homology with hexose transporters in Saccharomyces cerevisiae. However, there is no report on their function in glucose sensing or transport. In this study, we investigated two hexose transporter-like proteins (Hxs1 and Hxs2) in Cryptococcus that share the highest sequence identity with the glucose sensors Snf3 and Rgt2 in S. cerevisiae. The expression of HXS1 is repressed by high glucose, while the HXS2 expression is not regulated by glucose. Functional studies showed that Hxs1 is required for fungal resistance to oxidative stress and fungal virulence. The hxs1Δ mutant exhibited a significant reduction in glucose uptake activity, indicating that Hxs1 is required for glucose uptake. Heterologous expression of Cryptococcus HXS1 rendered the S. cerevisiae mutant lacking all 20 hexose transporters a high glucose uptake activity, demonstrating that Hxs1 functions as a glucose transporter. Heterologous expression of HXS1 in the snf3Δ rgt2Δ double mutant did not complement its growth in YPD medium containing the respiration inhibitor antimycin A, suggesting that Hxs1 may not function as a glucose sensor. Taken together, our results demonstrate that Hxs1 is a high-affinity glucose transporter and required for fungal virulence.
Introduction
The ability of a pathogen to sense extracellular signals and adapt to the host environment is essential for the establishment of an infection during a host-pathogen interaction. Characterization of extracellular signals and their sensors in a pathogen is central for understanding its pathogenesis. Cryptococcus neoformans is a major human fungal pathogen and the causative agent of the often fatal cryptococcal meningoencephalitis, which is an AIDS-defining illness [1]. Cryptococcus, a haploid yeast pathogen, is an ideal model system to study signal transduction in pathogenic fungi. Several signaling pathways important for Cryptococcus virulence have been identified [2], [3], [4], [5]. However, extracellular signals and their sensors remain largely unknown.
Glucose is the preferred carbon source for yeasts, including C. neoformans. It also functions as a hormone-like signal molecule for the regulation of cellular function and glucose utilization in Saccharomyces cerevisiae [6]. In C. neoformans, glucose sensing and utilization is required for fungal virulence. Host macrophages are the first line of host defense mechanism against cryptococcal infection. The antiphagocytic protein App1 is a fungal virulence factor and inhibits macrophage-mediated phagocytosis via complement receptor 3 (CR3) [7]. Expression of App1 is highly induced during lung infection due to the low glucose concentration (∼ 0.002%) environment in the lung and macrophages [8]. These results demonstrate the importance of glucose as a host signal in regulation of Cryptococcus-macrophage interactions [8]. A fully functional glycolytic pathway for proper glucose utilization is critical for cryptococcal infection and persistence of the fungus in the central nervous system (CNS) [9], [10]. UDP-glucuronic acid as a product of glycolysis is involved in the formation of the extracellular polysaccharide capsule, a major virulence factor [11], [12]. Despite the importance of glucose, it remains unknown how glucose is sensed and how glucose acquisition is regulated in C. neoformans. It has been shown that one G protein-mediated signaling pathway, the Gpa1-cAMP pathway, can be activated by glucose in C. neoformans and plays a central role in fungal virulence [13], [14]. Glucose can no longer activate the cAMP signaling in a gpa1Δ mutant background, indicating the Gpa1 G protein is essential for glucose signaling [15], [16]. However, the cell surface receptor that senses glucose to activate Gpa1 remains to be identified. One possibility is the G protein-coupled receptor (GPCR) family members function as glucose receptors to sense glucose and activate cAMP signaling via Gpa1. We have identified three GPCR proteins (Gpr4, Gpr5, and Ste3a) that are involved in the Gpa1 signaling activation, but none of them is required for glucose sensing [15], [17]. Alternatively, other mechanisms may be involved in the cAMP signaling activation.
Glucose acquisition and utilization systems have been extensively studied in S. cerevisiae, which employs two glucose sensory systems to sense the availability of extracellular glucose and fine-tune the function of a large gene family of hexose transporters as glucose carriers [6], [18], [19], [20]. One GPCR, Gpr1, functions as a glucose sensor to activate a downstream G protein signaling pathway, the Gpa2-cAMP pathway [21], [22], which is parallel to the Gpa1-cAMP pathway in C. neoformans and is required for cell growth and metabolic activity regulation in response to the availability of nutrients. Besides Gpr1, two unusual members of the hexose transporter gene family, Snf3 and Rgt2, can also sense different levels of extracellular glucose. Snf3 and Rgt2 maintain hexose transporter structures and possess unique long C-terminal cytoplasmic tails. However, they cannot transport glucose, and instead function as glucose sensors. Snf3 senses low glucose concentrations and activates high affinity hexose transporters, while Rgt2 senses high levels of glucose to regulate the expression of low affinity transporters [23]. These two sensors can interact with the casein kinase I Yck1/2, which are responsible for phosphorylation of Mth1 and Std1, two transcriptional regulators [24]. Mth1 and Std1 form a complex with another regulator Rgt1 to repress the expression of hexose transporters [19]. Phosphorylated Mth1 and Std1 are subjected for ubiquitination and degradation through an SCF(Grr1) E3 ligase-mediated ubiquitin-proteasome pathway. The degradation of Mth1 and Std1 in the 26S proteasome releases the binding of Rgt1 on promoters of hexose transporters, which in turn activates the expression of a number of hexose transporters for glucose uptake [25].
Besides Rgt2 and Snf3 in S. cerevisiae, glucose sensors have also been identified in the pathogenic yeast Candida albicans (Hgt4) [26] and in the methylotrophic yeast Hansenula polymorpha (Hxs1) [27]. C. neoformans also contains a large group of hexose transporter homologs based on available genome sequences, but how these transporters function in response to glucose availability is unknown. In this study, we identified two hexose transporter candidates (Hxs1 and Hxs2) that share the highest sequence identity with Snf3 and Rgt2 in S. cerevisiae. Hxs1 expression is repressed by glucose, while Hxs2 expression is not regulated by glucose. Functional studies in C. neoformans demonstrate that Hxs1 is required for oxidative stress response and fungal virulence. Heterologous expression of HXS1 in a Saccharomyces strain lacking all hexose transporters showed high glucose uptake activity. Expressing HXS1 in the snf3Δ rgt2Δ double mutant failed to complement the regulatory function of these sensors. Our results indicate Hxs1 is a high-affinity glucose transporter rather than a glucose sensor.
Results
Hxs1 and Hxs2 are Homologs of Snf3 and Rgt2 in S. cerevisiae
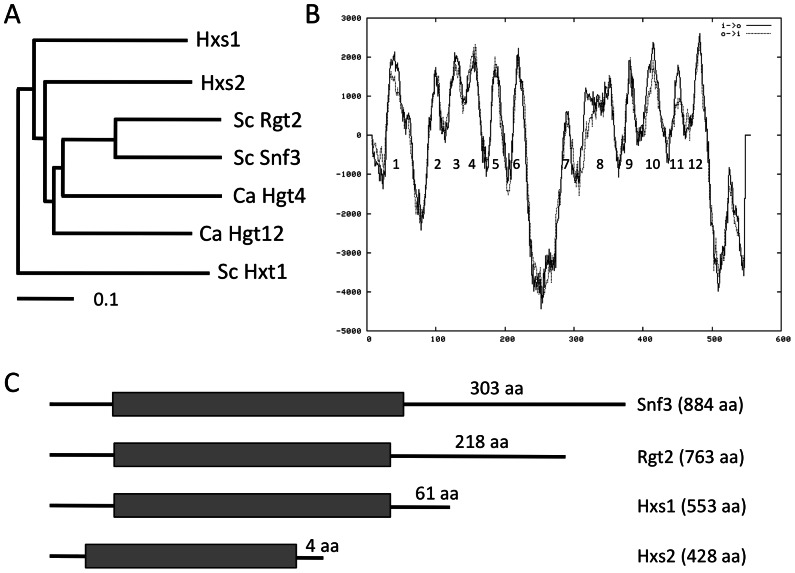
Based on the genome sequence of H99 strain, C. neoformans contains a large gene family of hexose transporter homologs with around 55 members. However, there is no report about their function in glucose sensing or transport. We compared the hexose transporter gene family members among S. cerevisiae, Candida albicans, and C. neoformans, and identified a cluster of proteins showing high sequence similarity with the glucose sensors Snf3 and Rgt2 in S. cerevisiae (Fig. S1 and Fig. 1A). We named two Cryptococcus proteins in this cluster as hexose sensor-like protein 1 and 2 (Hxs1 and Hxs2). Hxs1 contains 12 transmembrane domains, a typical structure of hexose transporters (Fig. 1B). Both Hxs1 (CNAG_03772; 553 amino acids) and Hxs2 (CNAG_04931; 428 amino acids) are much smaller than Snf3 or Rgt2 in S. cerevisiae. Interestingly, neither of them has the long C-terminal tail and the conserved C-terminal domains (R1 and R2) that exist in glucose sensors, including Snf3 and Rgt2 (Fig. 1C) [26], [27].
Figure 1. Identification of Snf3/Rgt2 homologs in Cryptococcus.
A. Phylogenetic tree of proteins from C. albicans (Hgt4 and Hgt12) and C. neoformans (Hxs1 and Hxs2) that share high homology with glucose sensors Snf3 and Rgt2 in S. cerevisiae. The full-length protein sequences were used for alignment using ClustalX. The hexose transporter Hxt1 in S. cerevisiae was used as an out-group. B. Predicted topology of the deduced Hxs1 amino acid sequence based on the TMpred program. Hydropathy values are on the y-axis, and the residue numbers are on the x-axis. The predicted transmembrane domains (TM1 to 12) are numbered. C. Schematic models of the primary protein structures of Snf3, Rgt2, Hxs1 and Hxs2. The black boxes represent transmembrane regions. The total number of amino acids for each protein and its C-terminal tail are indicated.
HXS1 Expression is Repressed by Glucose
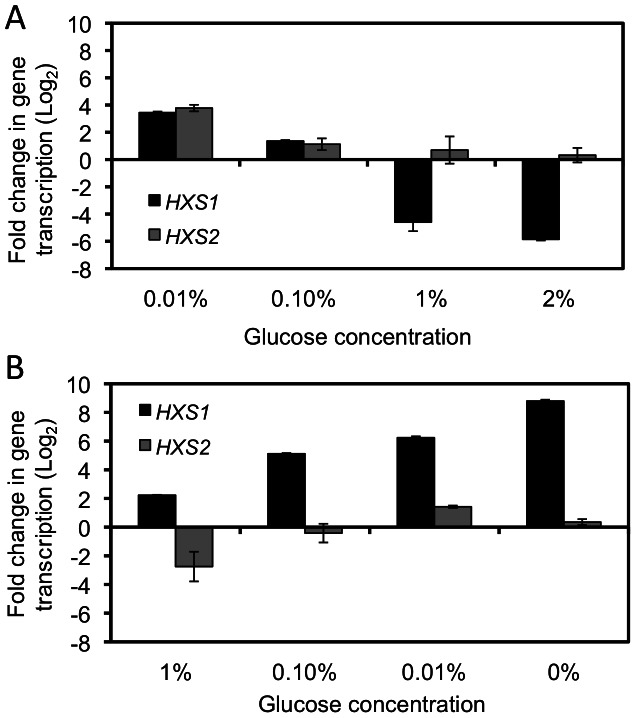
To understand the role of Hxs1 and Hxs2 in glucose utilization, we measured the transcriptional regulation of both Hxs1 and Hxs2 under conditions with different levels of glucose. The culture of the wild type strain H99 growing on medium without glucose but containing 2% galactose (YPG) was switched to medium with 0.01%, 0.1%, 1% or 2% glucose and incubated for 2 hrs, and the expression of HXS1 was measured at transcription level by qRT-PCR (Fig. 2). Our results showed a pattern of decreased HXS1 expression following the increase of glucose concentrations in the medium. When the glucose concentration was 1% or higher, the transcription level of HXS1 decreased significantly (>16 fold) (Fig. 2A). In contrast, when the H99 culture on YPD (2% glucose) was switched to media with lower glucose concentrations (1%, 0.1%, or 0.01%), an significant increase of HXS1 transcription levels was observed (>4 fold) (Fig. 2B). These results showed that the expression of HXS1 is dramatically repressed by the presence of high glucose levels. On the other hand, the expression of HXS2 is very low and is not regulated by glucose concentrations (Fig. 2). We hardly detected the PCR signal of HXS2 even after 35 cycles of amplification (Fig. S2).
Figure 2. The expression of HXS1 is repressed by glucose.
The qRT-PCR method was used to detect the change in transcription levels of HXS1 and HXS2 under growth conditions with different glucose levels. H99 cells were grown on YPG (no glucose) liquid medium, then switched to medium with 0.01%, 0.1%, 1% or 2% glucose (A), or grown on YPD and switched to medium with lower glucose levels (1%, 0.1%, 0.01% or YPG) (B). Cells were collected after 2 hr incubation and RNA prepared for qRT-PCR. Values are expressed as relative expression (log2) of the HXS1 or HXS2 gene, normalized to the GAPDH gene endogenous reference. The changes in gene transcription levels were related to 0-hr time point (H99 overnight liquid cultures on either YPG (A) or YPD (B)). The error bars showed standard deviations of three repeats.
Hxs1 does not Regulate the Expression of HXTs in C. neoformans
We then generated hxs1Δ mutants by homologous recombination to examine the role of Hxs1 in the regulation of glucose utilization in C. neoformans. We failed to generate the gene deletion mutant for the HXS2 gene, which may be due to its telomere location in the genome. The HXS2 gene is the first gene in Chromosome 10 with only 1098 bp away from the beginning of the chromosome, which likely prevented the homologous recombination event from happening to replace the HXS2 gene with a marker. Hence, we focus on the analysis of Hxs1 in this study.
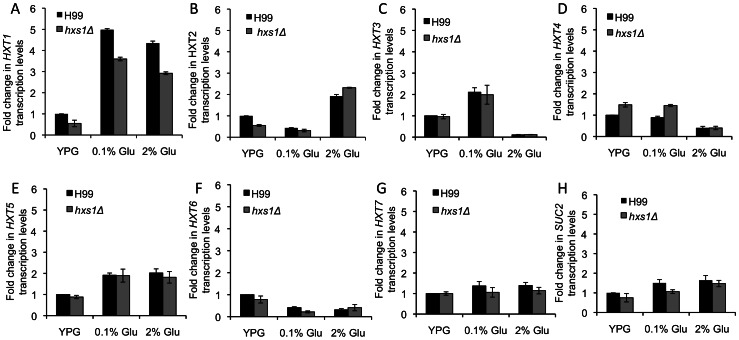
In S. cerevisiae, Hxt1 is a low-affinity glucose transporter, while Hxt2 is a high-affinity glucose transporter. The expression of HXT1 is induced only by high glucose concentration, while HXT2 is induced only by low glucose levels [23]. We compared the expression of Cryptococcus HXT1 and HXT2 homologs in the wild type and the hxs1Δ mutant under different glucose conditions. Because there are multiple protein homologs sharing high sequence identities with Hxt1 and Hxt2 in S. cerevisiae (E score is 0), we selected the first seven hits and investigated their expressions under different glucose conditions. Our qRT-PCR results showed that two gene (CNAG_03438, HXT1; CNAG_04920, HXT5) were induced by both low and high glucose concentrations, with a significantly higher induction in HXT1. Meanwhile, HXT2 (CNAG_06290) was induced only by the high glucose concentration, while HXT3 (CNAG_05387) was induced only by low glucose condition (Fig. 3). The changes of transcription level of other tested HXT homologs, HXT4 (CNAG_06521), HXT6 (CNAG_06963) and HXT7 (CNAG_03432), were not significant (<2 fold). The transcription level of each HXT gene tested was not altered significantly by the lack of HXS1 (<2 fold) when compared to the wild type H99. The changes of HXT1 transcription level between in H99 and the hxs1Δ mutant backgrounds were ∼1.4 folds on both glucose conditions, but was still significantly induced by glucose in both backgrounds. Hence, our results suggest the expression of all seven HXT gene homologs in C. neoformans is independent of Hxs1 (Fig. 3A–G).
Figure 3. Hxs1 is not required for the expression of other hexose transporter homologs.
The qRT-PCR method was used to measure the expression of seven hexose transporter homologs HXT1-7 (A–G) and SUC2 (H) in C. neoformans under YPG, YP with 0.1% glucose, or YPD (2% glucose) growth conditions. Values are expressed as relative expression of these genes, normalized to the GAPDH gene endogenous reference, and relative to HXS1 expression in H99 on YPG medium. Error bar indicates the standard deviation of three repeats.
In S. cerevisiae, the expression of hexose transporters was regulated by the Glucose sensors Rgt2 and Snf3 via the SCF(Grr1) E3 ligase [25]. We have identified an F-box protein, Fbp1, as the Grr1 homolog in C. neoformans [28]. To investigate the potential role of Hxs1 in the regulation of glucose uptake, we examined the expression of FBP1, as well as CCK1, a gene encoding the casein kinase, under low and high glucose conditions in both H99 and hxs1Δ backgrounds. Our qRT-PCR results showed that Hxs1 is not required for the regulation of these two genes in both low and high glucose conditions (data not shown). Because the expression of SUC2, a gene encoding sucrose invertase, is repressed by high glucose in S. cerevisiae and has been used as an indicator for the glucose repression [29], we also measured the expression of SUC2 homolog in C. neoformans using qRT-PCR. Interestingly, the SUC2 homolog in C. neoformans was not repressed by glucose. Hence, our results showed that the expression of this gene was not regulated by Hxs1 either (Fig. 3H). It is possible that the Suc2 protein has either different function or the glucose regulatory mechanism in C. neoformans is different from that of S. cerevisiae.
Hxs1 is Required for Efficient Glucose Uptake and Growth on Low Glucose Conditions
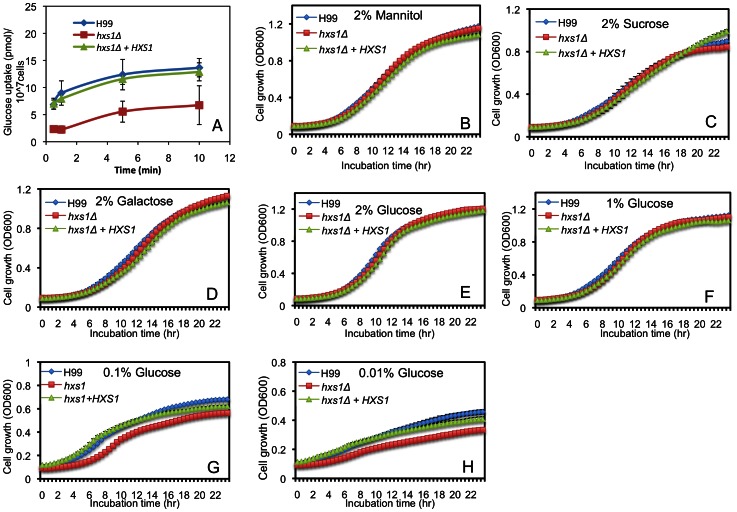
To investigate the potential role of Hxs1 in Cryptococcus glucose uptake, 3H-labeled glucose uptake assays were performed in the wild type, the hxs1Δ mutant and its complemented strain. Our results showed that the hxs1Δ mutant had significantly lower glucose uptake activity than the wild type or the complemented strain, suggesting that Hxs1 plays a major role in glucose uptake (Fig. 4A).
Figure 4. Hxs1 is required for Cryptococcus glucose uptake and cell growth on low glucose media.
A. Glucose uptake assay was performed for Cryptococcus wild type H99, the hxs1Δ mutant or its complemented strain as described in Materials and Methods. Error bar indicates the standard deviation of three repeats. B–H. Cryptococcus cell growth was assayed in 96-well plates. 1×105 cells of each strain were inoculated into the wells containing 100 µl YP supplemented with either 2% mannitol (B), 2% sucrose (C), 2% galactose (D), 2% glucose (E), 1% glucose (F), 0.1% glucose (G), or 0.01% glucose (H). The plates were kept in a PerkinElmer precisely Envision 2014 Multilabel Reader and incubated at 30°C with shaking (350RPM) and OD600 were measured in real time every half hour. Each experiment was performed in triplicates. Error bars indicate standard deviations.
Glucose is the preferred carbon source for Cryptococcus, and also plays an important role in the development of virulence factors. Because Hxs1 is important for glucose uptake, we tested the effect of Hxs1 on the growth of Cryptococcus cells on media with different glucose levels (2%, 1%, 0.1% or 0.001%). We also examined the growth of mutant cells on media with different carbon sources (mannitol, sucrose or galactose). Our results showed that when cells were grown on media with high levels of glucose or other tested carbon source, no growth defect was observed (Fig. 4B–E). However, on media with low glucose levels, the hxs1Δ mutant showed a small but significant growth defect (Fig. 4F–H), indicating that Hxs1 is important for the fungus to survive under conditions with low glucose availability.
Hxs1 is Required for Cell Integrity and Stress Response
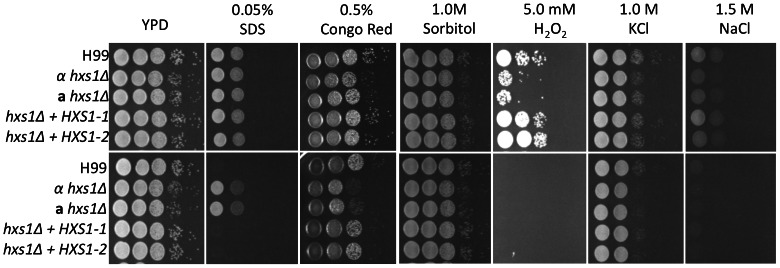
To address the cellular function of Hxs1, we analyzed the potential phenotype of the hxs1Δ mutant under different stress conditions. Phenotypic analyses showed that the hxs1Δ mutant had normal growth on medium with high salt or high osmolarity, indicating that Hxs1 is not required for these stress resistance conditions. However, the hxs1Δ mutant had a growth defect on medium with 5 mM H2O2, indicating Hxs1 is involved in cell resistance to oxidative stress. Interestingly, the hxs1Δ mutant showed a better growth at higher temperature (37°C) on medium with SDS, suggesting it is more resistant to SDS treatment (Fig. 5).
Figure 5. Hxs1 is required for stress response.
Cultures of wild type H99, the hxs1Δ mutant and its complemented strain were inoculated on YPD with 0.05% SDS, 0.5% Congo Red, 1 M Sorbitol, 5 mM H2O2, 1 M KCl, or 1.5 M NaCl, respectively. Plates were incubated at 30°C (upper) or 37°C (lower) for 3 days and photographed.
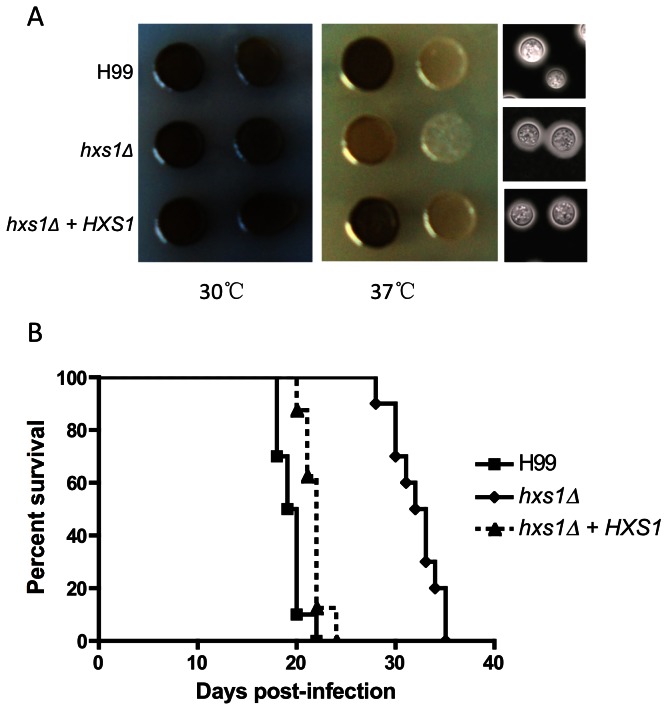
Because glucose sensing regulates capsule and melanin production via the cAMP signaling pathway [2], [15], we also examined the potential involvement of Hxs1 in the development of virulence factors. Our studies showed that the hxs1Δ mutant produced normal melanin on L-DOPA medium at 30°C, but had a modest melanin defect at 37°C. No obvious difference in capsule production was observed between the wild type and the mutant (Fig. 6A).
Figure 6. Hxs1 is required for fungal virulence.
A. Cultures of H99, hxs1Δ and its complement strain were inoculated on DME medium for capsule induction, or on L-DOPA medium for melanin induction. Plates were incubated at 30°C or 37°C for 48 hr and photographed. B. To determine the fungal virulence, female A/Jcr mice were intranasally infected with 105 cells of H99, hxs1Δ and its complement strain. Animals were monitored for clinical signs of cryptococcal infection and sacrificed at predetermined clinical endpoint that predicts imminent mortality.
Hxs1 is Required for Fungal Virulence
Because the importance of glucose for fungal cellular development and the importance of Hxs1 in stress response and melanin production, we examined the potential impact of Hxs1 on fungal virulence using a murine inhalation infection model of cryptococcosis. In accord with previous results [28], all mice infected with 105 yeast cells of wild-type strain H99 had a median survival time of 19.5 days due to lethal infection. In contrast, the hxs1Δ mutant showed significant virulence attenuation (P<0.0001) with a median survival time of 32.5 days (Fig. 6B). This result demonstrates that Hxs1 is required for fungal full virulence.
Hxs1 Showed High-affinity Uptake Activity in a Heterologous Expression System
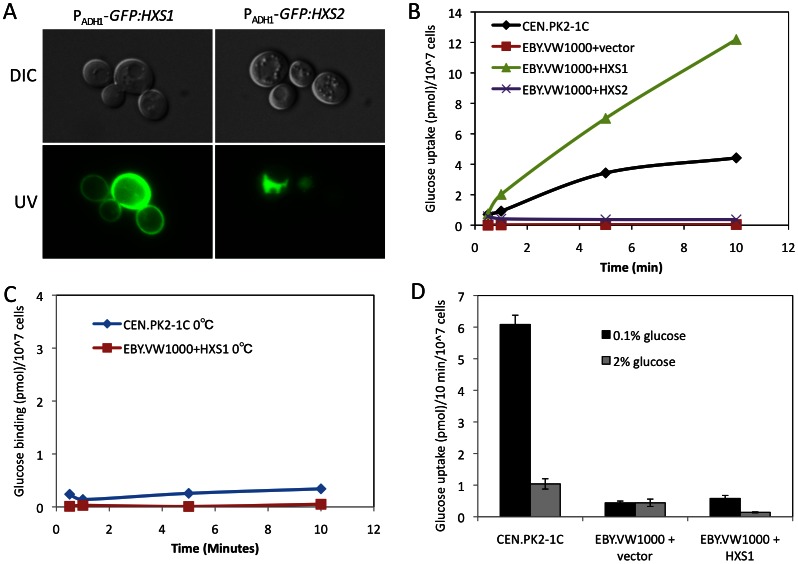
To examine whether Hxs1 and Hxs2 in C. neoformans function as glucose sensors or transporters, GFP:HXS1 and GFP:HXS2 fusion constructs were expressed under the control of the ADH1 promoter in an S. cerevisiae mutant strain EBY.VW1000, in which all 20 hexose transporters are deleted [30]. The EBY.VW1000 strain thus cannot grow on YPD medium. The expression of HXS1 was confirmed by GFP signals and RT-PCR. Our results showed that GFP:Hxs1 was localized on cell plasma membrane as expected (Fig. 7A).
Figure 7. Heterologous expression of HXS1 in a Saccharomyces strain lacking hexose transporters showed high glucose uptake activity.
A. Localization of HXS1, HXS2 in S. cerevisiae was determined by overexpressing a GFP:HXS1 or GFP:HXS2 fusion protein in EBY.VW1000, a mutant lacking all hexose transporters. EBY.VW1000 expressing an empty vector was used as a control. B and C. Glucose uptake activity (B) or glucose binding activity (C) of a Saccharomyces strain expressing HXS1. Cultures of the background strain CEN.PK2.1C and EBY.VW1000 expressing the pTH74 empty vector, the HXS1, or the HXS2 genes were inoculated on SD media lacking Uracil but containing 2% maltose as carbon source. Yeast cells were mixed with 3H-labeled glucose and incubated at 30°C(B) or 0°C (C) for 1, 5, and 10 mins. This assay was repeated twice with similar patterns. D. Glucose uptake assay was performed for S. cerevisiae strains CEN.PK2.1C and EBY.VW1000 expressing the empty vector or HXS1 in the presence of 0.1% or 2% cold glucose. The error bar indicates the standard deviation of three repeats.
Glucose uptake assay was performed to examine the uptake ability of Hxs1 in this heterologous expression strain. We used 1 µCi 3H-glucose for all assays and found that the strain expressing GFP:Hxs1 could transport glucose at a rate much higher than that of the wild type strain, likely due to its overexpression. The wild type strain becomes saturated after 5 mins, while the HXS1-expressing strain remains efficient uptake of 3H-labeled glucose, an indication of impaired regulation on glucose uptake (Fig. 7B). To examine the possibility that the outcome of uptake assays were resulted from glucose binding, instead of transport, we also performed the glucose binding assay at 0°C, and found that each strain only can bind to very limited amount of glucose (Fig. 7C). We also performed glucose uptake assays by addition of cold glucose to compete with the 3H-labeled glucose. In the presence of 0.1% cold glucose, the wild type strain still showed high uptake of labeled glucose, while its uptake signal was significantly reduced when 2% glucose was added in the reaction. In contrast, very low uptake signal was detected in the strain expressing HXS1, by adding either 0.1% or 2% cold glucose (Fig. 7D). Overall, these results demonstrate that Hxs1 is a high-affinity glucose transporter.
We also introduced the GFP:HXS2 overexpression construct into the same strain. To our surprise, even though the construct was prepared exactly same as that of the GFP:HXS1 construct with correct sequence, the GFP signal of the strain expressing GFP:Hxs2 was very weak. For a small population of cells that showed stronger signal, most of the fluorescence signal was localized in the vacuole instead of cell plasma membrane (Fig. 7A). It remains unclear whether the GFP:Hxs2 fusion protein is functional. We also repeated the glucose uptake assay for this strain and found the strain expressing GFP:Hxs2 fusion protein failed to transport glucose, suggesting the Hxs2 protein might not function in glucose uptake (Fig. 7B).
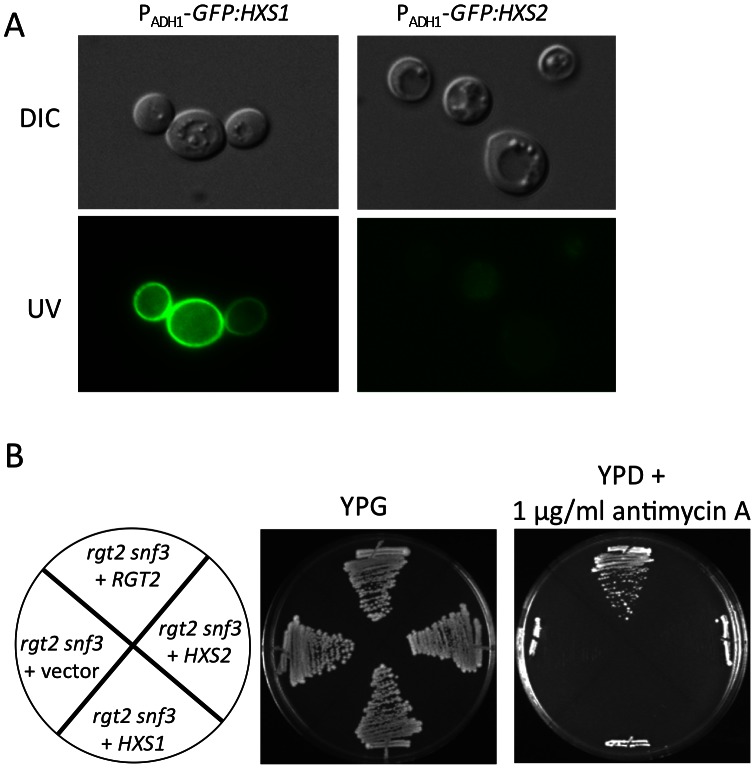
Hxs1 could not Rescue the Growth Defect of the snf3Δ rgt2Δ Double Mutant on YPD with Antimycin A
To further investigate the possibility that Hxs1 and Hxs2 may function as glucose sensors, we generated a S. cerevisiae snf3Δ rgt2Δ double mutant by genetic crossing of a snf3Δ mutant and a rgt2Δ mutant (kindly provided by Dr. Mark Johnston). The pADH1-GFP:HXS1 and pADH1-GFP:HXS2 constructs were introduced in this snf3Δ rgt2Δ double mutant, respectively. Their expressions were confirmed by GFP signals (Fig. 8A). While a strong GFP signal was observed for strains expressing HXS1, only a weak fluorescent signal was observed for strains expressing HXS2, which is consistent with their expression in EBY.VW1000, the strain lacking all hexose transporters (Fig. 7A). Based on previous studies, the double mutant could not grow on YPD medium containing 1 µg/ml antimycin A, which substantially inhibits respiration by blocking electron transfer between cytochromes b and c [26], [31]. We thus tested the growth of these strains expressing HXS1 or HXS2 on YPD medium with 1 µg/ml antimycin A. Consistent with the previous report, the double mutant expressing an empty vector could not grow on this medium, while reintroducing a RGT2 copy rescued the growth defect. However, the double mutant expressing either HXS1 or HXS2 failed to grow on this medium, indicating that neither of them could complement the function of glucose sensors (Fig. 8). Thus, Hxs1 and Hxs2 may not function as glucose sensors. It is likely that Hxs2 is not a functional protein.
Figure 8. Heterologous expression of HXS1 in the snf3Δ rgt2Δ double mutant background failed to complement glucose sensor function.
A. Localization of HXS1, HXS2 in S. cerevisiae was determined by overexpressing a GFP:HXS1, GFP:HXS2, or GFP:RGT2 fusion protein in a Saccharomyces strain lacking both Rgt2 and Snf3 glucose sensors. B. The growth of S. cerevisiae wild type BY4741, the snf3Δ rgt2Δ double mutant expressing empty vector pTH74, HXS1, HXS2, and RGT2 were inoculated on YPG or YPD with 1 µg/ml antimycin A. Plates were incubated at 30°C for 3 days.
Overall, our results showed that Hxs1 is a high-affinity glucose transporter. We have seen no activity of Hxs2 and it is possible that Hxs2 may not function properly in Cryptococcus. Other hexose transporter-like proteins may function as glucose sensors to regulate glucose uptake.
Discussion
Studies in S. cerevisiae revealed a complex network of glucose sensing, regulation of glucose uptakes, and subsequent glucose utilization. There are two major glucose sensory systems in the baker’s yeast, Rgt2/Snf3 and Gpr1, that coordinate the function of the large hexose transporter gene family for optimized glucose utilization [19]. In C. neoformans, how the fungus senses and acquires glucose remains unclear. The genome of Cryptococcus revealed a large gene family with over 50 members that shares high sequence identity with hexose transporters in other yeasts. There is only one previous report that attempted to link one hexose transporter homolog to copper resistance in C. neoformans [32]. How these genes are regulated and whether there is a similar transporter-like glucose sensor remains unknown. The identification of Hxs1 and Hxs2 in this report is the first attempt to study the function of this important gene family in glucose utilization.
Based on the phylogenetic relationship we presented in Figure S1, Hxs1 and Hxs2 have the highest sequence identity with the Rgt2 and Snf3 in S. cerevisiae, also with Hgt4, a glucose sensor in C. albicans [26]. However, neither Hxs1 nor Hxs2 has the long C-terminal tail or the conserved R1 or R2 domains in that region that has been shown important for the sensory function in other glucose sensors. There was no hit when we searched the H99 genome database with either R1 or R2 domains from Snf3 protein sequence, so whether such sequences exist in C. neoformans remain unknown. Hxs1 is repressed by glucose while Hxs2 is very weakly expressed and not regulated by glucose. Our mutagenesis studies showed that Hxs1 plays an important role in fungal virulence. Uptake assays for the hxs1Δ mutant and a S. cerevisiae mutant strain expressing HXS1 demonstrated that Hxs1 is required for glucose transport. Human and animal lungs only contains very low level of glucose [8]. To survive in such an environment, an efficient glucose uptake activity could be important for the pathogen, which may explain why the hxs1Δ mutant showed a significant virulence attenuation. The finding that Hxs1 is a high-affinity glucose transporter further explains its importance during Cryptococcus-host interaction, especially in an environment where glucose is scarce. In addition, we also observed that the hxs1Δ mutant had a defect in melanin production, a major virulence factor, and cell resistance to oxidative stress, which may also contribute to its defect in virulence.
Because we still could not generate a hxs2Δ null mutant even after many repeats, the function of Hxs2 remains to be determined. It is possible that Hxs2 is not functional due to its telomere location. HXS2 is located at the beginning of the chromosome 10, which could be the reason why we could not delete the HXS2 gene. Genes located in the telomere region frequently undergo gene duplication, rearrangement, and transcriptional silencing [33], [34], [35]. Although remaining to be proven, we suspect that HXS2 could be silenced due to the telomere-associated position effect and thus not functional. Although we could amplify the cDNA sequence of HXS2, its amplification was not robust may due to its extreme low expression or silencing effect.
Our heterologous expression assays using the S. cerevisiae strain lacking all hexose transporters showed that Hxs1 functions as a high-affinity transporter. When HXS1 was expressed in an S. cerevisiae mutant lacking both glucose sensors, it did not rescue the growth defect on YPD medium with antimycin A, suggesting that Hxs1 may not have the property of glucose sensing. Thus, whether Cryptococcus has a Rgt2/Snf3-like glucose sensory system remains to be determined. Although our data does not suggest that Hxs1 or Hxs2 plays a role in glucose sensing, we could not completely rule out the possibility that Hxs1 may be a dual function transporter, functioning as both a transporter and a sensor, similar to the ammonium transporter Mep2 [36]. Because there are over 50 hexose transporter-like proteins, although Hxs1 and Hxs2 share the highest sequence similarity with Snf3 and Rgt2, it is possible that additional proteins may function as glucose sensors.
Studies so far suggest there is a difference between Cryptococcus and Saccharomyces in glucose sensing and regulation. In S. cerevisiae, two glucose sensors, Snf3 and Rgt2, regulate other glucose transporter function via a SCF(Grr1) E3 ligase mediated UPS system. Our previous study has identified a Grr1 homolog in C. neoformans, Fbp1, which also functions as a SCF E3 ligase. However, expressing Fbp1 in a grr1Δ mutant failed to rescue the function of Grr1. Our preliminary studies also showed that some Grr1 substrate homologs in C. neoformans did not function as Fbp1 substrates, suggesting they may regulate different biological processes through different substrates (our unpublished data). In S. cerevisiae, Gpr1 functions as a glucose sensor to activate downstream Gpa2 G protein signaling that involves in cAMP levels. In Cryptococcus, although the G protein-cAMP signaling is overall conserved, its glucose receptor that activates this pathway remains unknown. We have identified one GPCR, Gpr4, that shares sequence and structure similarity with Gpr1, but it is not a glucose sensor. Compared with other yeasts, it is likely that Cryptococcus has developed a unique glucose sensing mechanism to accommodate its unique environmental and host conditions, which warrants further investigation.
Materials and Methods
Ethics Statement
The animal studies conducted at University of Medicine and Dentistry of New Jersey (UMDNJ) were in full compliance with all of the guidelines set forth by the Institutional Animal Care and Use Committee (IACUC) and in full compliance with the United States Animal Welfare Act (Public Law 98–198). The UMDNJ IACUCs approved all of the vertebrate studies. The studies were conducted in facilities accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
Strains, Media, and Growth Conditions
C. neoformans and S. cerevisiae strains used in this study are listed in Table 1. Strains were grown at 30°C on yeast extract-peptone-dextrose (YPD) agar medium and synthetic (SD) medium. All other media were prepared as described previously [15].
Table 1. Strains used in this study.
| C. neoformans strains | Genotype | Source/reference |
| H99 | MATα | Perfect et al. (1993) |
| KN99a | MAT a | Nielsen et al. (2003) |
| CUX69 | MAT a hxs1::NEO | This study |
| CUX146 | MATα hxs1::NEO | This study |
| CUX145 | MAT a hxs1::NEO ura5 | This study |
| CUX148 | MAT a hxs1::NEO ura5 HXS1-URA5 | This study |
| S. cerevisiae strains | ||
| YSB4742 | MATα his3Δ1, leu2Δ0, ura3Δ0 | ATCC yeast deletion collection |
| CEN.PK2-1C ( = VW1A) | leu2-3,112 ura3-52 trp1-289 his3Δ1 MAL2-8c SUC2 hxt7Δ | Wieczorke et al. (1999) |
| EBY.VW1000 | CEN.PK2-1C hxt13Δ::loxP hxt15Δ::loxP hxt16Δ::loxP hxt14Δ::loxP hxt12Δ::loxP hxt9Δ::loxP hxt11Δ::loxP hxt10Δ::loxP hxt8Δ::loxP hxt514Δ::loxP hxt2Δ::loxP | Wieczorke et al. (1999) |
| YUX43 | EBY.VW1000 PADH1-GFP | This study |
| YUX44 | EBY.VW1000 PADH1-GFP:HXS1 | This study |
| YUX45 | EBY.VW1000 PADH1-GFP:HXS2 | This study |
| YUX46 | EBY.VW1000 PADH1-GFP:ScHXT1 | This study |
| YM6863 | MATalpha his3Δ1 leu2Δ0 ura3Δ0 MET15 lys2Δ0 rgt2::kanMX::natMX | Mark Johnston |
| FM577 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2 snf3::kanMX | Mark Johnston |
| YUX79 | MATalpha his3Δ1 leu2Δ0 ura3Δ0 MET15 lys2Δ0 rgt2::kanMX::natMX snf3::kanMX | This study |
| YUX82 | MATa his3*1 leu2*0 ura3*0 met15*0 LYS2 rgt2::kanMX::natMX snf3::kanMX PADH1-GFP | This study |
| YUX83 | MATa his3*1 leu2*0 ura3*0 met15*0 LYS2 rgt2::kanMX::natMX snf3::kanMX PADH1-GFP:HXS1 | This study |
| YUX84 | MATa his3*1 leu2*0 ura3*0 met15*0 LYS2 rgt2::kanMX::natMX snf3::kanMX PADH1-GFP:HXS2 | This study |
| YUX85 | MATa his3*1 leu2*0 ura3*0 met15*0 LYS2 rgt2::kanMX::natMX snf3::kanMX PADH1-GFP:RGT2 | This study |
Generation of hxs1Δ Mutants and their Complemented Strains
Mutants for HXS1 and HXS2 were generated in both H99 and KN99a strains background by overlap PCR as previously described. The 5′ and 3′ regions of the HXS1 gene were amplified from H99 genomic DNA with primer pairs JH16929 and JH16930, and 16931 and 16932, respectively (see Table S1 for primer sequences). The 5′ and 3′ regions of the HXS2 gene were amplified from H99 genomic DNA with primer pairs 16922 and 16923, and CX248 and 16925, respectively (see Table S1 for primer sequences). The dominant selectable markers (NEOr) were amplified with the M13 primers (M13F and M13R) from plasmid pJAF1. The dominant selectable markers (NATr) were amplified with the M13 primers (M13F and M13R) from plasmid pPZP-NATcc. Each target gene replacement cassette was generated by overlap PCR with primers 16929 and 16932, 16922 and CX248. Purified overlap PCR products were precipitated onto 10 µl gold microcarrier beads (0.6 µm; Bio-Rad), and strains H99 or KN99a were biolistically transformed as described previously. Stable transformants were selected on YPD medium containing G418 (200 mg/L) or NAT (100 mg/L). To screen for mutants of HXS1 or HXS2 gene, diagnostic PCR was performed by analyzing the 5′ junction of the disrupted mutant alleles with primers 16935 and JH8994, 16928 and JH8994. Positive transformants were identified by PCR screening with primers 16933 and 16934, 16922 and 16927, respectively. While hxs1Δ null mutants were isolated, generation of hxs2Δ deletion mutants was not successful despite extensive effort.
To generate complemented strains of the hxs1Δ mutant, ura5 mutant strains were generated by selecting colonies grown on agar plates containing 0.1% 5-fluoroorotic acid (5-FOA). A genomic DNA fragment that contains a 1.5-kb upstream promoter region, the HXS1 open reading frame (ORF), and its 500-bp downstream region was amplified in a PCR using primers CX213 and CX214. This PCR fragment was digested with XmaI and EcoRI and cloned into the vector pJAF7 containing URA5 selective marker gene. The HXS1-URA5 construct was used to biolistically transform in a hxs1Δ mutant strain. The ectopic expression of the HXS1 gene was confirmed by RT-PCR. Phenotypic assays were performed to identify transformants in which the hxs1Δ phenotype was complemented.
Detection of Gene Expression Using Quantitative Reverse Transcription-PCR (RT-PCR)
The expression of HXT1 and HXT2 genes was measured at mRNA level via quantitative real-time PCR (qRT-PCR) in strains grown with different concentration of glucose. Cultures of C. neoformans var. grubii wild-type strain H99 and hxs1Δ were grown overnight on YPG (2% galactose) liquid medium at 30°C with shaking. Collected cells were washed with distilled H2O (dH2O), resuspended in YP containing 0.1% glucose and YP containing 2% glucose medium and incubated for 2 hrs. Total RNAs were prepared from cells with each treatment and cDNA was synthesized as described below.
We measured HXS1 and HXS2 mRNA levels via quantitative real-time PCR (qRT-PCR) in cells grown with or without glucose. Cultures of C. neoformans var. grubii wild-type strain H99 were grown overnight on YPD (2% glucose) or YPG (2% galactose) liquid medium at 30°C with shaking. Collected cells were washed with distilled H2O (dH2O), resuspended in YPD or YPG medium and incubated for 2 h. Cells were then collected and washed with dH2O. Cells collected from YPD were resuspended in YP containing 0.1% glucose or YPG, while cells collected from YPG were resuspended in YP containing 0.1% glucose or YPD. Both cultures were incubated for 2 hrs. Total RNAs were prepared from cells with each treatment.
Purified RNAs were quantified using a Nanodrop spectrometer (Thermo Scientific) and were used as templates for PCR amplification with primers of glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) to determine potential genomic DNA contamination. First strand cDNAs were synthesized using a Superscript III cDNA synthesis kit (Invitrogen) following the manufacturer’s instructions. Expression levels of HXT1, HXT2, SUC2, and GAPDH were analyzed using SYBR advantage QPCR premix reagents (Clontech) with an Mx4000 QPCR system (Stratagene) as previously described [37], [38]. Gene-expression levels were normalized using the endogenous control gene GAPDH, and the relative levels were determined using the comparative CT method.
Assays for Virulence Factors
Assays for melanin and capsule production were performed as previously described [28]. In brief, melanin production was tested on L-DOPA medium, and incubated at 30°C or 37°C for three days and pigmentation of fungal colonies was assessed and photographed. Capsule production was induced on Dulbecco Modified Eagle’s (DME) agar medium and incubated at 37°C for three days. Capsule size was visualized by India ink staining and observed with an Olympus CX41 microscope.
Assays for Stress Responses and Cell Integrity
Each strain was incubated overnight at 30°C in YPD and sub-cultured in fresh YPD medium to OD600 ∼0.7. The cells were washed, resuspended, and serially diluted (1∶10) in dH2O and spotted (5 µl) on YPD agar plates containing 1.5 M NaCl, or 1.0 M KCl for osmotic shock, or 2.5 mM and 5.0 mM H2O2 for oxidative stress. To test cell integrity, cells were also spotted on YPD agar plates containing 0.05% SDS, 0.5% Congo Red, or 20 µg/ml Calcofluor White (CFW). Plates were incubated at both 30°C and 37°C for two days and photographed.
For growth assay on media with different carbon source, Cultures of wild type H99, the hxs1Δ mutant and its complemented strain were inoculated on YPD for 20 hr. Cells were washed and 1×105 cells of each strain were inoculated into the wells of 96-well plates containing 100 µl YP supplemented with different carbon source. The plates were kept in a PerkinElmer precisely Envision 2014 Multilabel Reader and incubated at 30°C with shaking (350RPM) and OD600 were measured in real time every half hour.
Glucose Uptake Assay
Full-length cDNAs of the HXS1, HXS2 genes were amplified from C. neoformans H99 total cDNA and were cloned into the yeast expression vector pTH74 to generate GFP fusion constructs, under the control of the ADH1 promoter. HXS1 and HXS2 expression plasmids were introduced into an S. cerevisiae strain EBY.VW1000 that lacks all 20 HXT transporters [30]. The expression of Cryptococcus HXS1 or HXS2 in this yeast heterologous system was verified by both GFP localization and RT-PCR using gene-specific primers. Yeast strains were tested for growth on different medium at 30°C.
The S. cerevisiae control strain CEN.PK2.1C, the S. cerevisiae hxtΔ mutant strain EBY.VW1000, and EBY.VW1000 expressing empty vector, GFP-HXS1 or GFP-HXS2 genes from C. neoformans were grown in YP containing 2% maltose liquid cultures overnight at 30°C. Collected cells were washed with dH2O, resuspended in YP containing 2% maltose, and incubated for 2 hrs. Then cells were suspended in PBS at a final concentration of 2×108 cells/ml for uptake assay. Each 100 µl cell suspension was mixed with 100 µl labeled glucose (3H-glucose) solution at room temperature. Samples (100 µl) were removed after 30 s, 1 min, 5 min, and 10 min, and mixed with 1 ml ice-cold water to stop the reactions. Cells were immediately collected on fiber filters, washed three times with 10 ml of ice-cold water, and transferred to scintillation vials for measurement.
Virulence Studies
Survival curves of infected mice in a Cryptococcus murine inhalation model as previously described [38]. Female A/Jcr mice (NCI-Frederick) were inoculated intranasally with the following strains: H99, the hxs1Δ mutant and its complemented strain. Groups of ten mice were infected with 1×105 yeast cells for each strain. Over the course of the experiments, animals that appeared moribund or in pain were sacrificed by CO2 inhalation. Survival data from the murine experiments were statistically analyzed between paired groups using the long-rank test of the PRISM program 4.0 (GraphPad Software, San Diego, CA). P values of <0.001 were considered significant. Infected animals were sacrificed at the endpoint of the experiment according to the UMDNJ IACUC-approved animal protocol.
Supporting Information
Phylogram of hexose transporter homologs in S. cerevisiae, C. albicans , and C. neoformans. The phylogenetic tree was generated using ClustalX 2.0 program and viewed using the TreeView software. A cluster of proteins showed high sequence identity was highlighted.
(TIF)
Expression of the HXS1 and HXS2 under different glucose conditions. C. neoformans wild type H99 was cultured on YPD (2% glucose) or YPG (0% glucose), or cultured on medium containing 2% glucose (YPD) and switched to 0.1% glucose (YP0.1D) or 0% glucose (YPG) and incubated for 2 more hrs. RNAs were extracted and cDNAs synthesized from those cells and were used as templates for qRT-PCR. PCR products amplified for 35 cycles were loaded on 1% agarose gel and photographed. GAPDH gene was used as an internal control.
(TIF)
Primers used in this study.
(DOC)
Acknowledgments
We thank Issar Smith for critical reading of the manuscript and valuable comments for the study. We thank Mark Johnston and Joe Heitman for Saccharomyces mutant strains. We thank David Kaback for assistance on generating the rgt2Δ snf3Δ double mutant and Andy Alspaugh for discussing his unpublished data. We also thank Lydia Chen for her earlier technique assistance on the project.
Funding Statement
This study was supported by American Heart Association grant 12SDG9110034 to C.X.. L. J. was supported by the Key Project of Jiangnan University Independent Scientific Research Plan (JUSRP51313B). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Casadevall A, Perfect JR (1998) Cryptococcus neoformans. Washington, DC: ASM Press.
- 2. Alspaugh JA, Perfect JR, Heitman J (1997) Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev 11: 3206–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang P, Heitman J (1999) Signal transduction cascades regulating mating, filamentation, and virulence in Cryptococcus neoformans . Curr Opin Microbiol 2: 358–362. [DOI] [PubMed] [Google Scholar]
- 4. Idnurm A, Bahn YS, Nielsen K, Lin X, Fraser JA, et al. (2005) Deciphering the model pathogenic fungus Cryptococcus neoformans . Nat Rev Microbiol 3: 753–764. [DOI] [PubMed] [Google Scholar]
- 5. Bahn YS, Xue C, Idnurm A, Rutherford JC, Heitman J, et al. (2007) Sensing the environment: lessons from fungi. Nat Rev Microbiol 5: 57–69. [DOI] [PubMed] [Google Scholar]
- 6. Santangelo GM (2006) Glucose signaling in Saccharomyces cerevisiae . Microbiol Mol Biol Rev 70: 253–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luberto C, Martinez-Marino B, Taraskiewicz D, Bolanos B, Chitano P, et al. (2003) Identification of App1 as a regulator of phagocytosis and virulence of Cryptococcus neoformans . J Clin Invest 112: 1080–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Williams V, Del Poeta M (2011) Role of glucose in the expression of Cryptococcus neoformans antiphagocytic protein 1, App1. Eukaryot Cell 10: 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Price MS, Betancourt-Quiroz M, Price JL, Toffaletti DL, Vora H, et al. (2011) Cryptococcus neoformans requires a functional glycolytic pathway for disease but not persistence in the host. mBio 2: e00103–00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kronstad J, Saikia S, Nielson ED, Kretschmer M, Jung W, et al. (2012) Adaptation of Cryptococcus neoformans to mammalian hosts: integrated regulation of metabolism and virulence. Eukaryot Cell 11: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Griffith CL, Klutts JS, Zhang L, Levery SB, Doering TL (2004) UDP-glucose dehydrogenase plays multiple roles in the biology of the pathogenic fungus Cryptococcus neoformans . J Biol Chem 279: 51669–51676. [DOI] [PubMed] [Google Scholar]
- 12. Doering TL (2009) How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans . Annu Rev Microbiol 63: 223–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. D'Souza CA aHJ (2001) Conserved cAMP signaling cascades regulate fungal developement and virulence. FEMS Microbiology Reviews 25: 349–364. [DOI] [PubMed] [Google Scholar]
- 14. Pukkila-Worley R, Alspaugh JA (2004) Cyclic AMP signaling in Cryptococcus neoformans . FEMS Yeast Res 4: 361–367. [DOI] [PubMed] [Google Scholar]
- 15. Xue C, Bahn YS, Cox GM, Heitman J (2006) G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans . Mol Biol Cell 17: 667–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hicks JK, Bahn YS, Heitman J (2005) Pde1 phosphodiesterase modulates cyclic AMP levels through a protein kinase A-mediated negative feedback loop in Cryptococcus neoformans . Eukaryot Cell 4: 1971–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Okagaki LH, Wang Y, Ballou ER, O'Meara TR, Bahn YS, et al. (2011) Cryptococcal titan cell formation is regulated by G-protein signaling in response to multiple stimuli. Eukaryot Cell 10: 1306–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnston M, Kim JH (2005) Glucose as a hormone: receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae . Biochem Soc Trans 33: 247–252. [DOI] [PubMed] [Google Scholar]
- 19. Kaniak A, Xue Z, Macool D, Kim JH, Johnston M (2004) Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae . Eukaryot Cell 3: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rolland F, Winderickx J, Thevelein JM (2002) Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res 2: 183–201. [DOI] [PubMed] [Google Scholar]
- 21. Xue Y, Batlle M, Hirsch JP (1998) GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. Embo J 17: 1996–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lemaire K, Van de Velde S, Van Dijck P, Thevelein JM (2004) Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae . Mol Cell 16: 293–299. [DOI] [PubMed] [Google Scholar]
- 23. Ozcan S, Johnston M (1999) Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev 63: 554–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moriya H, Johnston M (2004) Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc Natl Acad Sci U S A. 101: 1572–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim JH, Johnston M (2006) Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae . J Biol Chem 281: 26144–26149. [DOI] [PubMed] [Google Scholar]
- 26. Brown V, Sexton JA, Johnston M (2006) A glucose sensor in Candida albicans . Eukaryot Cell 5: 1726–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stasyk OG, Maidan MM, Stasyk OV, Van Dijck P, Thevelein JM, et al. (2008) Identification of hexose transporter-like sensor HXS1 and functional hexose transporter HXT1 in the methylotrophic yeast Hansenula polymorpha . Eukaryot Cell 7: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu TB, Wang Y, Stukes S, Chen Q, Casadevall A, et al. (2011) The F-Box protein Fbp1 regulates sexual reproduction and virulence in Cryptococcus neoformans . Eukaryot Cell 10: 791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ozcan S, Vallier LG, Flick JS, Carlson M, Johnston M (1997) Expression of the SUC2 gene of Saccharomyces cerevisiae is induced by low levels of glucose. Yeast 13: 127–137. [DOI] [PubMed] [Google Scholar]
- 30. Wieczorke R, Krampe S, Weierstall T, Freidel K, Hollenberg CP, et al. (1999) Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae . FEBS Lett 464: 123–128. [DOI] [PubMed] [Google Scholar]
- 31. Ozcan S, Dover J, Johnston M (1998) Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae . EMBO J 17: 2566–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chikamori M, Fukushima K (2005) A new hexose transporter from Cryptococcus neoformans: molecular cloning and structural and functional characterization. Fungal Genet Biol 42: 646–655. [DOI] [PubMed] [Google Scholar]
- 33. Lustig AJ (1998) Mechanisms of silencing in Saccharomyces cerevisiae . Curr Opin Genet Dev 8: 233–239. [DOI] [PubMed] [Google Scholar]
- 34. Tham WH, Zakian VA (2002) Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene 21: 512–521. [DOI] [PubMed] [Google Scholar]
- 35. Yankulov K (2011) Dare to challenge the silence? Telomeric gene silencing revisited. Nucleus 2: 513–516. [DOI] [PubMed] [Google Scholar]
- 36. Forsberg H, Ljungdahl PO (2001) Sensors of extracellular nutrients in Saccharomyces cerevisiae . Curr Genet 40: 91–109. [DOI] [PubMed] [Google Scholar]
- 37. Xue C, Liu T, Chen L, Li W, Liu I, et al. (2010) Role of an expanded inositol transporter repertoire in Cryptococcus neoformans sexual reproduction and virulence. mBio 1: e00084–00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Y, Liu TB, Delmas G, Park S, Perlin D, et al. (2011) Two major inositol transporters and their role in cryptococcal virulence. Eukaryot Cell 10: 618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogram of hexose transporter homologs in S. cerevisiae, C. albicans , and C. neoformans. The phylogenetic tree was generated using ClustalX 2.0 program and viewed using the TreeView software. A cluster of proteins showed high sequence identity was highlighted.
(TIF)
Expression of the HXS1 and HXS2 under different glucose conditions. C. neoformans wild type H99 was cultured on YPD (2% glucose) or YPG (0% glucose), or cultured on medium containing 2% glucose (YPD) and switched to 0.1% glucose (YP0.1D) or 0% glucose (YPG) and incubated for 2 more hrs. RNAs were extracted and cDNAs synthesized from those cells and were used as templates for qRT-PCR. PCR products amplified for 35 cycles were loaded on 1% agarose gel and photographed. GAPDH gene was used as an internal control.
(TIF)
Primers used in this study.
(DOC)