Summary
Research on ubiquitin (Ub) signaling has focused primarily on homogeneously-linked polyUb. Although polyUb containing different linkages within the same chain exist, their structures and signaling properties are unknown. These mixed-linkage chains could be unbranched (i.e., no more than one lysine- or methionine-linkage per Ub) or branched. Here we examined the structure, dynamics, receptor selectivity, and disasssembly of branched and unbranched tri-Ub containing both K48 and K63-linkages. Each linkage was virtually indistinguishable from its counterpart in homogeneously-linked polyUb. Linkage-selective receptors from hHR23A and Rap80 preferentially bound to the K48- or K63-linkages in the branched trimer. Linkage-selective deubiquitinases specifically cleaved their cognate Ub-Ub linkages in mixed-linkage chains, and the 26S proteasome recognized and processed branched tri-Ub. We conclude that mixed-linkage chains retain the distinctive signaling properties of their K48- and K63-components, and that these multiple signals can be recognized by multiple linkage-specific receptors. Finally, we propose a new, comprehensive notation for Ub and Ub-like polymers.
Introduction
Critical eukaryotic cell functions such as DNA-repair, cell cycle control, protein turnover and receptor-mediated endocytosis depend on the post-translational modification of target proteins by covalent attachment of polyubiquitin (polyUb) (Fushman and Wilkinson; Hershko and Ciechanover, 1998). In ubiquitination, the C-terminus of a “proximal” Ub typically is attached to an εNH2 of a lysine on a substrate protein. Similarly, the C-terminus of another Ub can be ligated to the N-terminus (M1) or, more commonly, to an εNH2 of K6, K11, K27, K29, K33, K48, or K63 of the previously ligated Ub (Xu et al., 2009). This process is tightly regulated in the cell and, once a Ub is linked to a particular lysine (e.g., K48) on another Ub, it is often assumed that subsequent ubiquitination events will propagate this same linkage throughout the polyUb chain (Gregori et al., 1990). However, there is mounting evidence from both in vivo and in vitro studies that more than one linkage type can be present in the same polyUb chain (Ben-Saadon et al., 2006; Crosas et al., 2006; Goto et al., 2010; Kim et al., 2007; Kim et al., 2009; Kirkpatrick et al., 2006; Kravtsova-Ivantsiv and Ciechanover, 2012; Newton et al., 2008). Currently, it is not known if linkage mixing or branching in polyUb confers signaling properties unavailable to homogeneous chains, or if the signaling properties of the individual linkages in mixed-linkage chains are preserved.
Recent mass spectrometry studies have confirmed that Ub–Ub linkages involving all seven lysines as well as M1 exist in vivo (Xu et al., 2009). Additionally, quantitative analyses have determined the relative abundances of each Ub linkage in whole-cell lysates and showed that K48 and K63 linkages occur most frequently by a large margin (Dammer et al., 2011; Ziv et al., 2011). However, these approaches generally fail to differentiate between homogeneous-linkage, mixed-linkage, and branched polyUb chains; therefore, it is unclear if linkages within chains are commonly mixed or branched in the cell. The use of linkage-specific anti-polyUb antibodies has revealed that polyUb is remodeled in several substrates (Dammer et al., 2011; Newton et al., 2008; Seyfried et al., 2010). Precedent for a functional mixed-linkage chain comes from Ring1b, an E3 Ub ligase that requires autoubiquitination by a mixed-linkage chain containing K6, K27, and K48 linkages (Ben-Saadon et al., 2006). In this example, our understanding of the structural and signaling properties is limited because the topology and sequence of Ub–Ub linkages in this novel polyUb chain are unknown. K11 and K63 mixed-linkage polyUb chains have also been suggested to serve a signaling function (Goto et al., 2010). During processing on the 26S proteasome, the substrate's polyUb signal can be remodeled by several deubiquitinases (DUBs) and, in yeast, the E3/E4 Ub ligase Hul5 has been shown to make linkages primarily through K63, but also K11 and K48 (Crosas et al., 2006). Although these and other studies indicate that mixed-linkage forms of polyUb occur in vivo, they do not provide clues about the structures of the chains or whether linkage-mixing promotes specific interactions or processes. Thus, our understanding of the roles and signaling properties of mixed or branched forms of polyUb has been quite limited.
One challenge to studying mixed-linkage polyUb is that, depending on the number of individual Ub units, there could be an enormous number of unique chains (see Table S1). Even discussions of the problem are confounded by the absence of a standardized nomenclature for mixed and branched polyUb. In this study, we have focused on branched and unbranched mixed-linkage chains containing K48 and K63 linkages as a logical starting point. The fact that, when in homogeneous chains, the two linkages are essentially “orthogonal” with respect to their (i) location on the Ub surface, (ii) conformations (i.e., compact versus extended (Varadan et al., 2004; Varadan et al., 2002)), and (iii) signaling properties (i.e., proteolytic versus regulatory (Pickart and Fushman, 2004)) suggests that combinations of K48 and K63 linkages could provide an extreme example of linkage mixing and branching. Additionally, their high relative abundances in the cell suggest that these linkages would predominate in randomly assembled mixed-linkage chains. Although a cellular process that requires both K48 and K63 linkages has yet to be identified, it has been reported that K48 and K63 linkages co-localize in the cell, both linkages have been detected on the same substrate, and at least one DUB, ataxin-3, preferentially cleaves mixed K48 and K63-linkage chains in vitro (Dammer et al., 2011; Newton et al., 2008; Winborn et al., 2008). These studies hint that polyUb chains containing both K48 and K63 linkages form in the cell, but whether they serve a specific function or are simply a mistake that is later “edited” remains to be seen.
For this study, we created the simplest possible model system for mixed-linkage polyUb by limiting chains to just three Ubs with combinations of K48 and K63 linkages. The resulting set of chains (all tri-Ubs) includes a single branched chain, [Ub]2–48,63Ub, and two unbranched mixed-linkage chains, Ub–63Ub–48Ub and Ub–48Ub–63Ub (Fig 1). We used established NMR methods to examine how mixing of the linkages affects the polyUb structurally. Our results show that linkage-specific interdomain contacts observed with conventional, homogeneous chains are preserved in both the unbranched and branched mixed-linkage chains. To address the signaling properties of these chains, we demonstrate their recognition by K48- and K63-linkage-selective receptors, as well as by linkage-specific antibodies. We then show that linkage-selective DUBs can efficiently select and process the respective linkages in these mixed chains. Finally, we monitored the activity of purified yeast proteasomes and observed that [Ub]2–48,63Ub and homogeneous K48 and K63-linked trimers are processed similarly. We conclude that when the K48 and K63 linkages are contained in the same polyUb chain, the properties of each linkage are preserved. Hence, mixed-linkage Ub chains can send mixed messages.
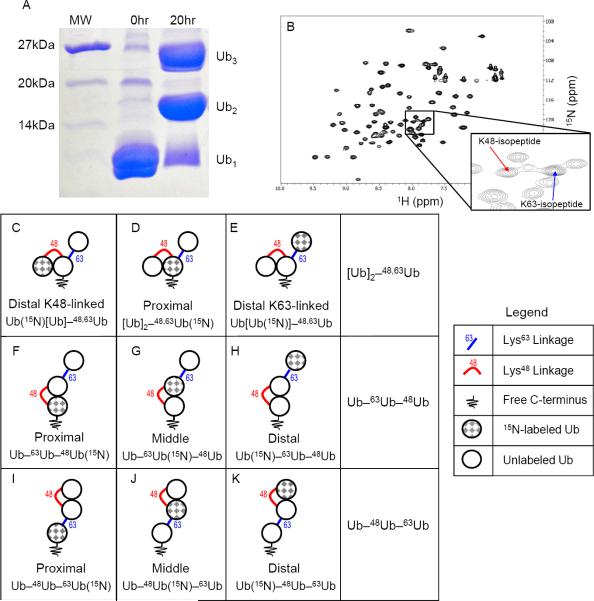
Figure 1.
Assembly and nomenclature of the branched and unbranched mixed-linkage chains used in this study. (A) 15% SDS-PAGE gel showing enzymatic synthesis of [Ub]2–48,63Ub in one step using E2-25K and Ubc13:Mms2 as the E2 ubiquitin-conjugating enzymes. (B) 1H-15N SOFAST-HMQC spectrum of 15N labeled proximal Ub in [Ub]2–48,63Ub(15N). The inset zooms on the region containing characteristic signals from εNH of K48 and K63 as a result of isopeptide bond formation. (C–K) Chain schematics, nomenclature, and unit-specific 15N-enrichment for branched and unbranched mixed-linkage chains studied here. The rows depict isotope labeling schemes for each Ub unit in (C–E) [Ub]2–48,63Ub, (F–H) Ub–63Ub–48Ub, (I–K) Ub–48Ub–63Ub. The name in each box refers to the particular 15N-labeled Ub in the chain (checkered gray); the formal notation indicating the 15N Ub is shown on the bottom of the box. (See also Figs S1– S3)
Results
Notation for polymers of ubiquitin or ubiquitin-like proteins
To facilitate discussion of different forms of polyUb conjugates, we propose a new, systematic notation. This scheme, which can be used to describe unambiguously polymers of Ub assembled from any combination of Ub–Ub linkages and also conjugates that include Ub-like proteins such as SUMO, is described in detail in the Supplemental Materials. Fundamental to our notation is a convention whereby Ub units are connected by an en dash (−), and the polarity of a polyUb chain is indicated by placing the distal-end Ub unit(s) to the left and the proximal Ub (or the target molecule it is conjugated to) to the right. Specific residues (i.e., Ub Met1, or lysines that contribute an ε-amine) in each linkage are indicated as superscripts. Multiple (poly)Ub moieties branching from a single Ub are indicated with brackets that, as needed, may be nested to specify the structure of each branch. Modifications or variants of any Ub within the polymer can be indicated by parentheses that follow the Ub (e.g., Ub with a Lys63 to Arg63 substitution can be written as Ub(K63R), and completely Lys-to-Arg substituted Ub as Ub(K0)). Thus, tri-Ub consisting of a distal-end Ub linked to K48 of a middle Ub that in turn is linked to K63 of a proximal Ub is written as Ub–48Ub–63Ub, whereas Ub[Ub]–48,63Ub or [Ub]2–48,63Ub indicates two distal Ub units linked to K48 and K63 of a proximal Ub; the aforementioned branched tri-Ub would be written as Ub[Ub]–48,63Ub(15N) to indicate isotopic enrichment with 15N specifically in the proximal Ub. If it is necessary to distinguish between the distal moieties in a branched chain, those should be listed in the same order (i.e., left to right) as the superscripts that correspond to the linkage sites. For example, the same branched tri-Ub chain with 15N-enrichment of only the K48-linked or the K63-linked distal Ub would be written as Ub(15N)[Ub]–48,63Ub or Ub[Ub(15N)]–48,63Ub, respectively (see examples in Fig 1). As a final example, tetra-Ub made by conjugation of Ub(K0) to K11, K48, and K63 of a single Ub containing a C-terminal His6-tag would be [Ub(K0)]3–11,48,63Ub(G76-H6). The proposed notation applies as well to homogeneous-linkage chains. While the existing notation for such polymers (e.g., K48-linked Ub2) is straightforward, for consistency we will use the new notation (i.e., Ub–48Ub) throughout this paper.
Assembly of mixed-linkage polyUb chains
Trimeric mixed-linkage unbranched and branched chains, Ub–63Ub–48Ub, Ub–48Ub–63Ub, and [Ub]2–48,63Ub, were produced using enzymatic chain-assembly as detailed in Supplemental Information (Fig S1). Importantly, our method ensured that fully-natural isopeptide linkages were formed with full control of the order of the linkages in each chain and, for NMR studies, which Ub unit is 15N enriched. We found that the K48-linked dimer can accept Ub in a K63-linkage on either the distal or proximal end, and likewise the K63-linked dimer can accept Ub in a K48-linkage at either end. In addition, the K48,K63-branched trimer can be created from appropriate Ub monomers conjugated in one step with both K48- and K63-specific E2s (i.e., E2–25K and Ubc13:Mms2, respectively) in the reaction mixture (Fig 1A).
K48-linked ubiquitins in unbranched or branched mixed-linkage chains form the classical K48 hydrophobic interface
We used NMR chemical shift perturbation (CSP) mapping (e.g., (Varadan et al., 2005a)) to identify interactions between the Ub units in the mixed-linkage chains. In order to unravel monomer-specific contacts, each Ub unit in [Ub]2–48,63Ub, Ub–63Ub–48Ub, and Ub–48Ub–63Ub (see Fig 1 for chain notations) was individually 15N enriched (i.e., one 15N-Ub per chain) resulting in nine distinct 15N-labeled tri-Ub constructs. 1H-15N NMR spectra of each chain (e.g., Fig 1B) were acquired under identical conditions and compared to those of monomeric Ub and of the corresponding Ub unit in Ub–48Ub and Ub–63Ub (see examples in Figs S2 and S3).
Consistent with the absence of detectable non-covalent contacts between Ub units in Ub–63Ub, the only CSPs (versus mono-Ub) observed in that chain (Varadan et al., 2004) were in the C-terminal residues of the distal Ub and those immediately surrounding K63 of the proximal Ub, reflecting the residues involved in the isopeptide linkage between these two units. By contrast, CSP mapping of Ub–48Ub revealed (Varadan et al., 2002) that both the distal and the proximal Ubs exhibited highly specific spectral perturbations in and around the hydrophobic surface patch residues (L8, I44, V70). These CSPs, observed in addition to those in the vicinity of the isopeptide linkage between the C-terminal G76 of the distal Ub and K48 of the proximal Ub, are a clear indicator of the hydrophobic interface between the two Ubs in Ub–48Ub. These distinctive features of the NMR spectra of Ub–63Ub and Ub–48Ub serve as hallmarks of the corresponding linkages and the resulting Ub–Ub contacts.
NMR spectra of [Ub]2–48,63Ub show that the amide resonances in the distal K63-linked Ub are almost identical to those in monomeric Ub with the exception of the C-terminal residues 74–76, where the observed CSPs are caused directly by ligation to K63 of the proximal Ub (Fig 2C). Consistent with this observation, there are virtually no spectral differences between distal K63-linked Ub and the distal Ub in Ub–63Ub, indicating their close structural similarity and the absence of non-covalent contacts with the proximal Ub in the corresponding chains (Fig 2I).
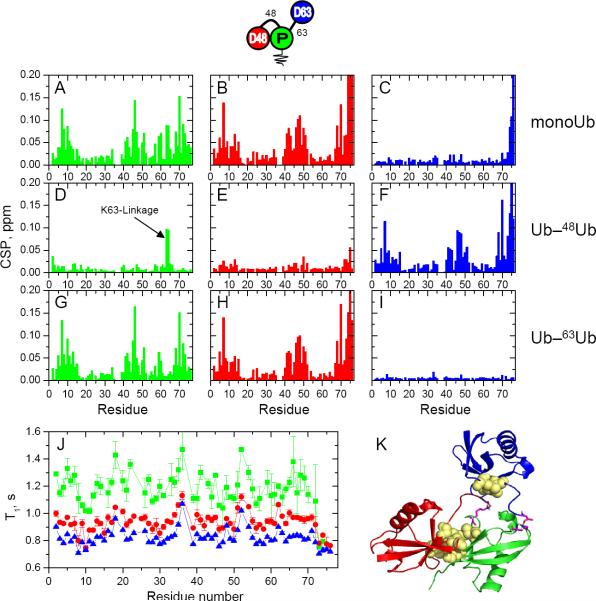
Figure 2.
NMR characterization of [Ub]2–48,63Ub. Data for the proximal Ub are shown in green, for distal K48-linked Ub in red, and for distal K63-linked Ub in blue (see pictograph, top). (A–I) Spectral differences (quantified as CSPs) between each Ub in [Ub]2–48,63Ub and monomeric Ub (top row) or the corresponding Ub units in Ub–48Ub (middle row) or Ub–63Ub (bottom row). Left column: CSPs between the proximal Ub in [Ub]2–48,63Ub and (A) Ub1, (D) proximal Ub in Ub–48Ub, and (G) proximal Ub in Ub–63Ub. Middle column: CSPs between distal K48-linked Ub in [Ub]2–48,63Ub and (B) Ub1, (E) distal Ub in Ub–48Ub, and (H) distal Ub in Ub–63Ub. Right column: CSPs between the distal K63-linked Ub in [Ub]2–48,63Ub and (C) Ub1, (F) distal Ub in Ub–48Ub, and (I) distal Ub in Ub–63Ub. (J) 15N T1 values as a function of the residue number for each Ub in [Ub]2–48,63Ub. (K) A representative structural model of [Ub]2–48,63Ub from the lowest energy cluster. The coloring of the Ub units is the same as above, the isopeptide linkages are shown in magenta, and the hydrophobic-patch residues (L8, I44, V70) are shown as yellow spheres. (See also Fig S4)
A similar comparison of the spectra of the distal K48-linked Ub in [Ub]2–48,63Ub revealed a strikingly different picture. Here we detected large site-specific CSPs between the distal K48-linked Ub and mono-Ub (Fig 2B) and almost negligible spectral differences between the distal K48-linked Ub and the distal Ub in Ub–48Ub (Fig 2E). These results show that the distal K48-linked Ub in [Ub]2–48,63Ub makes essentially the same interdomain contacts (i.e., the hydrophobic interface with the proximal Ub and the isopeptide linkage through G76) as the distal Ub in Ub–48Ub. Predictably, these contacts result in large site-specific spectral differences between the distal K48-linked Ub and the distal Ub of Ub–63Ub (Fig 2H), which are also similar to the CSPs between the distal K48-linked Ub and monomeric Ub (Fig 2E).
Careful analysis of the spectra for the proximal Ub of [Ub]2–48,63Ub revealed two signals originating from the isopeptide εNH groups of K48 and K63 (Fig. 1B and Fig S3). These signals are diagnostic for the isopeptide linkage through the ε-amino group of the corresponding lysine. That they are at the same resonance frequencies as in the respective isolated diubiquitins (K48-or K63-linked) proves the presence of both expected linkages in [Ub]2–48,63Ub and also demonstrates that NMR can be used for linkage-diagnostic purposes. Note in this regard that for both unbranched mixed-linkage tri-Ub chains (see below) we also observed the isopeptide εNH signals that corresponded to each respective linkage.
Because the proximal Ub of [Ub]2–48,63Ub is ubiquitinated at two lysines simultaneously, its NMR spectrum could differ from the proximal-Ub spectrum in the respective isolated di-Ubs. Nevertheless, a clear picture emerged from comparison of the corresponding spectra. For example, a comparison to mono-Ub indicates that the hydrophobic-patch residues of the proximal Ub form an interface with another Ub unit, while its C-terminus is unligated (as in monomeric Ub) unlike those of the distal K48- and K63-linked Ubs (Fig 2A). The spectral differences between the proximal Ubs of [Ub]2–48,63Ub and Ub–48Ub are minimal except for the region around K63 which is linked to another Ub only in the branched trimer (Fig 2D). As with the distal K48-linked Ub (see above), the strong site-specific CSPs with respect to the proximal Ub in Ub–63Ub reflect the hydrophobic-patch contacts between the two K48-linked Ubs (Fig 2G). Based on these and the abovementioned data, we conclude that the two K48-linked Ubs in [Ub]2–48,63Ub form essentially the same hydrophobic-interface contact as in Ub–48Ub whereas only the distal K63-linked Ub behaves as in Ub–63Ub, where no non-covalent contact was detected.
When we rearranged the linkages to form an unbranched mixed-linkage chain, Ub–63Ub–48Ub, the Ub units maintained their expected contacts and spectral properties. The proximal Ub of Ub–63Ub–48Ub has its K48 ligated, and we detected a hydrophobic interface by the large site-specific CSPs relative to mono-Ub and Ub–63Ub (Fig 3A,G). The spectral differences are minimal between the proximal Ub in Ub–48Ub and that in Ub–63Ub–48Ub, which indicates close similarity between the two units ligated via K48 (Fig 3D). The next (i.e., middle) Ub of Ub–63Ub–48Ub has features of both a proximal unit (linked through its K63) and a distal unit linked through its C-terminal G76. The large site-specific CSPs versus mono-Ub (Fig 3B) clearly indicate that the middle Ub participates in a hydrophobic interface. Based on the spectral similarity with the distal Ub of Ub–48Ub (Fig 3E), we conclude that the K48-linked proximal and middle Ubs in Ub–63Ub–48Ub retain the hydrophobic interface characteristic of Ub–48Ub. Note also that the large CSPs observed around K63 in the middle Ub reflect its linkage to the distal Ub.
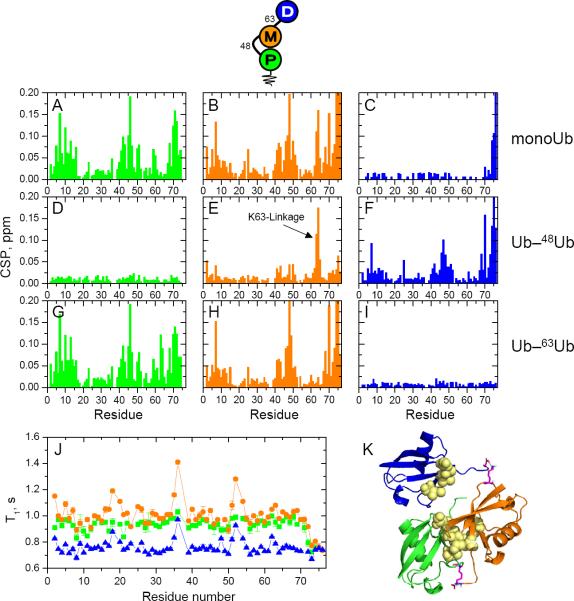
Figure 3.
NMR characterization of the unbranched, mixed-linkage Ub–63Ub–48Ub chain. Data for the proximal Ub are shown in green, for the middle Ub in orange, and for the distal Ub in blue (see pictograph, top). (A–I) Spectral differences (quantified as CSPs) between each Ub in Ub–63Ub–48Ub and monomeric Ub (top row) or the corresponding Ub units in Ub–48Ub (middle row) or Ub–63Ub (bottom row). Left column: CSPs between the proximal Ub in Ub–63Ub–48Ub and (A) Ub1, (D) proximal Ub in Ub–48Ub, and (G) proximal Ub in Ub–63Ub. Middle column: CSPs between the middle Ub in Ub–63Ub–48Ub and (B) Ub1, (E) distal Ub in Ub–48Ub, and (H) distal Ub in Ub–63Ub. Right column: CSPs between the distal Ub in Ub–63Ub–48Ub and (C) Ub1, (F) distal Ub in Ub–48Ub, and (I) distal Ub in Ub–63Ub. (J) 15N T1 values as a function of the residue number for each Ub in Ub–63Ub–48Ub. (K) A representative structural model of Ub–63Ub–48Ub from the lowest-energy cluster. The coloring of the Ub units is the same as above, the isopeptide linkages are shown in magenta, and the hydrophobic-patch residues (L8, I44, V70) are shown as yellow spheres.
The NMR spectra of the distal Ub in Ub–63Ub–48Ub indicate that this unit is excluded from the hydrophobic interface formed between the middle and proximal Ubs. Indeed, the CSPs in distal versus monomeric Ub are localized to the (ligated) C-terminal region (Fig 3C), whereas the spectral differences between the distal Ubs of Ub–63Ub–48Ub and Ub–63Ub are negligible (Fig 3I). This indicates strongly that the distal Ub of Ub–63Ub–48Ub has the same structure and contacts (or absence thereof) as the distal Ub in Ub–63Ub.
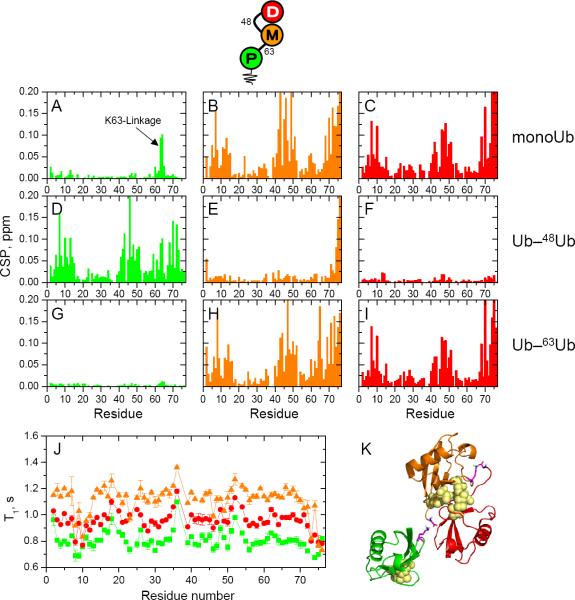
In Ub–48Ub–63Ub, the proximal Ub is expected to be the only unit that does not form a hydrophobic interface. Indeed, this is evident from the strong spectral similarity of that proximal Ub with the proximal Ub of Ub–63Ub (Fig 4G) as well as with mono-Ub (Fig 4A). In contrast to the proximal Ub which exhibits no non-covalent contacts, the middle and distal units of Ub–48Ub–63Ub form a characteristic K48-linked Ub–Ub interface. This is particularly apparent from the minimal CSPs (except for the ligated C-terminus) between the middle Ub of Ub–48Ub–63Ub and the proximal Ub of Ub–48Ub (Fig 4E), and the virtual absence of CSPs between the distal Ubs (Fig 4F). These and the other spectral comparisons in Fig 4 show that the structures and interfaces of the middle and distal units in Ub–48Ub–63Ub are nearly identical to those in Ub–48Ub.
Figure 4.
NMR characterization of the unbranched, mixed-linkage Ub–48Ub–63Ub chain. Proximal Ub is shown in green, middle Ub in orange, and distal Ub in red (see pictograph, top). (A–I) CSPs between each Ub in Ub–48Ub–63Ub versus monomeric Ub (left column), Ub–48Ub (middle column), or Ub–63Ub (right column). (A–I) Spectral differences (quantified as CSPs) between each Ub in Ub–48Ub–63Ub and monomeric Ub (first row) or the corresponding Ub units in Ub–48Ub (middle row) or Ub–63Ub (bottom row). Left column: CSPs between the proximal Ub in Ub–48Ub–63Ub and (A) Ub1, (D) proximal Ub in Ub–48Ub, and (G) proximal Ub in Ub–63Ub. Middle column: CSPs between the middle Ub in Ub–48Ub–63Ub and (B) Ub1, (E) distal Ub in Ub–48Ub, and (H) distal Ub in Ub–63Ub. Right column: CSPs between the distal Ub in Ub–48Ub–63Ub and (C) Ub1, (F) distal Ub in Ub–48Ub, and (I) distal Ub in Ub–63Ub. (J) 15N T1 values as a function of the residue number for each Ub in Ub–48Ub–63Ub. (K) A representative structural model of Ub–48Ub–63Ub from the lowest energy cluster. The coloring of the Ub units is the same as above, the isopeptide linkages are shown in magenta and the hydrophobic-patch residues (L8, I44, V70) are shown as yellow spheres.
Structural models of the mixed-linkage tri-Ub chains
Based on the above NMR data, we generated structural models of the three tri-Ub chains studied here using the biomolecular docking program HADDOCK (de Vries et al., 2010) and site-specific CSPs to guide the docking (see Supplemental Information). Although we observed slightly different overall structures in the top clusters for these chains, all featured a distinct arrangement of the interdomain contacts in which the two K48-linked Ubs formed a well-defined contact mediated by their hydrophobic-patch residues whereas the K63-linked Ub was represented in essentially random positions. For example, the generated structures for [Ub]2–48,63Ub (Fig 2K) clearly show that the proximal and distal K48-linked Ubs form the “canonical” L8, I44, V70 hydrophobic interface, whereas the distal K63-linked Ub samples several different orientations, depending on the HADDOCK cluster (Fig S4). This variation in the K63-linked Ub is not unexpected given that (i) the CSP data show no close non-covalent interactions involving this Ub unit and (ii) the 15N T1 relaxation data (see below) indicate that this unit has greater mobility compared to the other two Ubs. Moreover, the two K48-linked Ubs superimpose quite well (RMSD = 1.72 Å) with the crystal structure (PDB: 1AAR) of Ub–48Ub (Fig S4).
Interdomain mobility supports distinct K48- and K63-linkage behavior
The NMR spectra above clearly indicate that the two K48-linked units in all three tri-Ub chains form a specific interface while the third unit, linked via K63, shows no contacts except for being tethered (via its flexible C-terminus or via K63) and therefore is expected to be more mobile than the other two Ubs. This distinction is further seen in the generated CSP-docked structures of these chains. To test this prediction, we measured and compared 15N longitudinal relaxation times, T1, for each Ub unit in a chain. Generally, T1 senses the overall tumbling of the chain (reflecting its size and shape), the relative intra-chain mobility (on a ns time scale) of the Ub unit under observation (which could depend on its location and intra-chain contacts), and the local polypeptide backbone dynamics within the Ub protomer (which are expected to be similar for all Ubs) (Ryabov and Fushman, 2007). Whereas the overall shape could differ between chains, the T1 values within the same chain are expected to reflect the relative mobility of each Ub unit. For example, if a particular Ub forms a hydrophobic interdomain interface it becomes less mobile and this should result in an increase in T1. Furthermore, a Ub that is linked to two other Ubs should also show an increase in T1 as its reorientation will partially depend on the movement of the other two Ubs. Therefore, one would expect the proximal Ub of [Ub]2–48,63Ub and the middle Ubs of Ub–63Ub–48Ub and Ub–48Ub–63Ub to have the largest T1 values.
Indeed, in [Ub]2–48,63Ub the proximal Ub has the longest T1, whereas of the two distal (singly-linked) Ubs, the distal K63-linked Ub shows the shortest T1 (i.e., the least restricted mobility), and the distal K48-linked Ub has longer T1, consistent with its contact with the proximal Ub (Fig 2J). These results fully agree with the spectroscopic and structural data presented above. The same holds for the two unbranched chains, where the middle Ub shows the longest T1 (most restricted mobility) while the K63-linked Ub is consistently the most mobile (i.e., shortest T1) regardless of whether it is at the distal or the proximal end of the chain (Figs 3J, 4J). K48-linked Ub shows intermediate T1 values, as its tumbling is slowed by the hydrophobic interaction with the middle Ub. Note that in all of these chains the two K48-linked units have different T1 values; thus, despite forming a hydrophobic contact, they are not rigidly locked. This is consistent with our previous findings for Ub–48Ub (Ryabov and Fushman, 2007; Varadan et al., 2002) and indicates dynamic opening and closing of the hydrophobic interface, which is critical for the ability of Ub-receptors to bind to this chain.
pH dependence of the inter-Ub contacts in [Ub]2–48,63Ub supports the classical K48-linked interface
One hallmark of K48-linked di-Ub is that its conformation is pH-dependent: lowering the pH shifts the equilibrium from the predominantly closed conformation at neutral pH to predominantly open, and this manifests itself as nearly complete disappearance of the CSPs at pH 4.5 (Varadan et al., 2002). Indeed, our NMR data (Fig S5) indicate that the pH-dependent behavior of [Ub]2–48,63Ub is nearly identical to that reported for the corresponding Ubs in homogeneous K48- and K63-linked polyUb. This supports the conclusion that interdomain interactions in the branched tri-Ub are governed by the same forces as in the respective homogeneous-linkage chains.
A K48-selective receptor, UBA(2) of hHR23a, selectively binds K48-linked units within the branched [Ub]2–48,63Ub chain
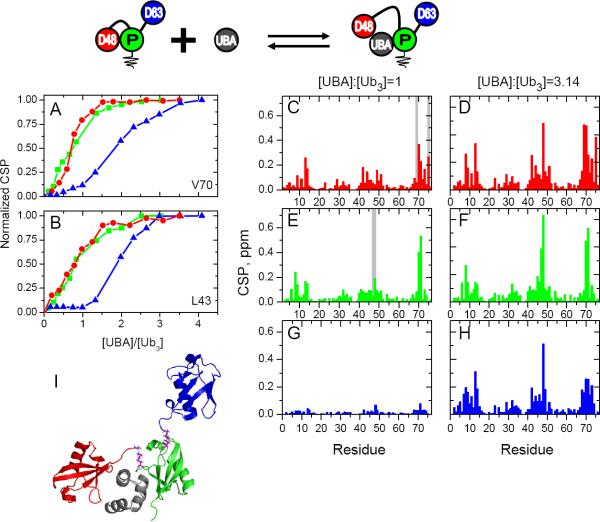
Having established that the branched and unbranched mixed-linkage chains combine the structural properties characteristic of both linkages, we set to examine whether these chains can be recognized by linkage-selective receptors. To determine if the K48-linkage in [Ub]2–48,63Ub retains its receptor selectivity, we titrated [Ub]2–48,63Ub (separately for each Ub unit 15N-enriched; see Fig 1) with the K48-linkage selective receptor UBA(2) from the proteasomal shuttle protein hHR23A. UBA(2) binds Ub–48Ub in a “sandwich” mode and significantly more tightly than to Ub–63Ub or monomeric Ub (Varadan et al., 2005b). We observed strong residue-specific CSPs in both the proximal and the distal K48-linked Ubs (Fig 5); these perturbations center around the hydrophobic-patch residues and the isopeptide linkage, consistent with UBA(2) insertion into the hydrophobic pocket formed by the K48-linked Ubs (Varadan et al., 2005b). We can gauge the extent of binding from titration curves of residue-specific CSPs or by comparing the CSPs for all residues at various points in titration. The CSPs in the proximal and distal K48-linked Ubs of [Ub]2–48,63Ub show little change between [UBA(2)]:[ [Ub]248,63Ub] = 1 and 3 (Fig 5), suggesting that UBA(2) binding to both Ubs reached saturation at approximately 1:1 molar ratio. Additional changes occurring in these Ubs above the 1:1 molar ratio could reflect binding of a second UBA(2) molecule, as observed for Ub–48Ub. Interestingly, the distal K63-linked Ub exhibited an entirely different behavior and showed spectral perturbations only after the distal K48-linked and proximal Ubs were saturated with UBA(2) (Fig 5). Even at [UBA(2)]:[[Ub]2–48,63Ub] = 1, there are virtually no CSPs in the distal K63-linked Ub, and it was not until after this point that we finally observed UBA(2) binding to this Ub (Fig 5G,H). The titration curves for individual residues illustrate this trend from another angle by showing “standard” binding behavior for the distal K48-linked and proximal Ubs, but a “lag-phase” for the distal K63-linked Ub (Fig 5A,B and Fig S6). This strong binding preference for K48-linked Ubs is also supported by our relaxation data that showed a larger increase in 15N T1 of the distal K48-linked Ub compared to distal K63-linked Ub at saturation (Fig S6). These results demonstrate that a K48-linkage receptor can selectively recognize this linkage in a branched polyUb chain using the same binding mechanism as for homogeneous-linkage chains.
Figure 5.
Binding of hHR23A UBA(2) to [Ub]2–48,63Ub. All data are color coded as indicated in the pictograph on the top. Proximal Ub data are show in green, distal K48-linked Ub in red, and distal K63-linked Ub in blue. (A, B) Representative titration curves plotted as normalized CSP vs. [UBA(2)]:[[Ub]2–48,63Ub] for residues V70 and L43 in all three Ub units. (C–H) Residue-specific CSPs in individual Ub units at two points in titration with UBA(2): at the 1:1 molar ratio (left column) and at saturation (right column). Gray bars indicate residues with signals broadened beyond detection at the 1:1 molar ratio. Note that the distal K63-linked Ub exhibits virtually no binding until [UBA(2)]:[[Ub]2–48,63Ub] > 1. (I) A model of the [Ub]2–48,63Ub/UBA(2) complex at the 1:1 molar ratio, based on the observed CSPs and the structure of the Ub–48Ub/UBA(2) complex (PDB: 1ZO6). The coloring of the Ub units is as depicted above; UBA(2) is colored gray. (See also Figs S6, S8)
A K63-selective receptor, Rap80-tUIM, avidly binds across the K63-linkage in the branched [Ub]2–48,63Ub chain
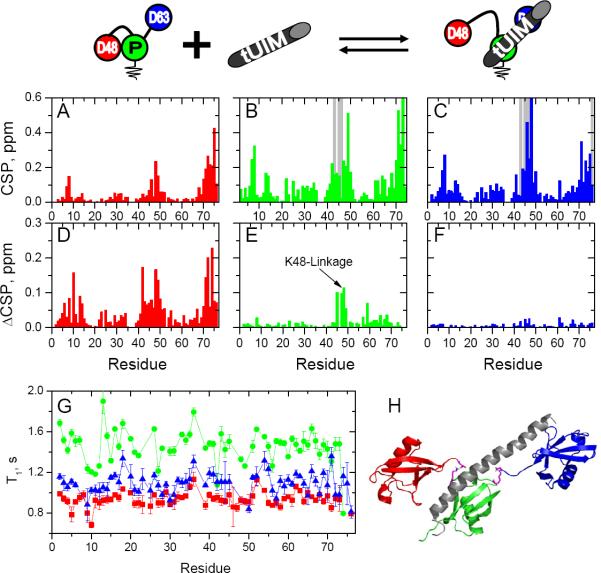
Rap80 contains a tandem Ubiquitin Interacting Motif (tUIM) that shows strong binding preference for Ub–63Ub versus Ub–48Ub (Sims and Cohen, 2009). The tUIM contains a flexible linker region between its two helical UIMs which adopts an alpha-helical conformation upon binding to Ub–63Ub and perfectly aligns the two UIMs' interacting surfaces for binding to adjacent Ubs in a K63 linkage (Sato et al., 2009; Sims and Cohen, 2009). To examine how the tUIM interacts with branched chains, we titrated it into [Ub]2–48,63Ub, as well as Ub–48Ub and Ub–63Ub as controls, and used NMR to monitor the interactions separately for each 15N-enriched Ub unit. As shown in Fig 6, NMR signals of all three Ubs in [Ub]2–48,63Ub were altered by the tUIM. The observed perturbations were site-specific and centered around the hydrophobic-patch residues as well as the Ub C-termini. The distal K48-linked Ub showed noticeably weaker CSPs at saturation (suggesting weaker binding) compared to the other two Ubs. Importantly, during these titrations some residues in the proximal and distal K63-linked Ubs, but not in the distal K48-linked Ub, showed strong attenuations or even disappearance of the NMR signals; this is caused by slow exchange (i.e., slow off-rates) on the NMR chemical shift time scale and is indicative of tighter binding to the K63-linked Ubs. Comparison of the domain-specific CSPs in [Ub]2–48,63Ub with those in the control di-Ub samples (Fig 6D–F), as well as comparison of the signal trajectories upon titration (Fig S7), led us to the following conclusions. First, the distal K63-linked and proximal Ubs of [Ub]2–48,63Ub bind the tUIM in the same mode as their respective units in Ub–63Ub (Fig 6D, E). This is supported by the fact that at saturation with the tUIM virtually all signals in the 1H-15N NMR spectra of the distal K63-linked and proximal Ubs of [Ub]2–48,63Ub overlay perfectly with the corresponding signals of Ub–63Ub. Also, the signal trajectories (in the course of the titration) for the same residue in the K63-linked components of the branched and control chains were nearly superimposable (Fig S7), consistent with identical binding interactions. Second, there is a stark difference between tUIM binding to the distal K48-linked Ub of [Ub]2–48,63Ub and the Ubs of Ub–48Ub. Indeed, our data show spectral differences (ΔCSP) at saturation between the distal K48-linked Ub and either Ub of Ub–48Ub (and even mono-Ub) that are much larger than the differences observed for the K63-linked components (Fig 6).
Figure 6.
Binding of the Rap80 tUIM to [Ub]2–48,63Ub chain. All data are color coded as indicated in the pictograph on the top: proximal Ub data are shown in green, distal K48-linked Ub in red, and distal K63-linked Ub in blue. (A–C) CSP plots for each Ub unit in [Ub]2–48,63Ub at saturation: (A) distal K48-linked Ub at [tUIM]:[[Ub]2–48,63Ub] = 2.43, (B) distal K63-linked Ub at [tUIM]:[[Ub]2–48,63Ub] = 1.52, and (C) proximal Ub at [tUIM]:[[Ub]2–48,63Ub] = 2.00. Residues that showed strong signal attenuation during the titration are represented with gray bars. (D–E) Difference in the CSPs (ΔCSP) for each Ub of [Ub]2–48,63Ub and the corresponding Ub in the respective di-Ub controls at saturation with tUIM: (D) distal K48-linked Ub of [Ub]2–48,63Ub vs. distal Ub of Ub–48Ub, (E) proximal Ub of [Ub]2–48,63Ub vs. proximal Ub of Ub–63Ub, and (F) distal K63-linked of [Ub]2–48,63Ub vs. distal Ub of Ub–63Ub. (G) 15N T1 relaxation time for each amide in the proximal, distal K48-linked, and distal K63-linked Ubs in [Ub]2–48,63Ub at saturation with Rap80 tUIM. (H) A model of the [Ub]2–48,63Ub/tUIM complex at 1:1 molar ratio, based on the observed CSPs and 15N T1 values, and the Ub–63Ub/tUIM complex structure (PDB: 3A1Q). The coloring of the Ub units is the same as above; tUIM is colored gray. (See also Figs S7, S9)
When the tUIM binds to Ub–48Ub it has to disrupt the hydrophobic interface between the two Ubs in order to access the Ub hydrophobic surface. Even though avid tUIM binding to K48-linked di-Ub has not been observed, the reported interaction (Kd = 0.16 mM) was still relatively strong (Sims and Cohen, 2009). In the branched chain, simultaneous tUIM binding to the proximal and distal K63-linked Ubs should displace the distal K48-linked Ub from its hydrophobic contact with the proximal Ub, thus freeing it and making it available for binding to excess tUIM molecules. To verify this model, we measured 15N T1 values for each Ub of [Ub]2–48,63Ub at saturation with the tUIM (Fig 6G) and compared them with those of unbound [Ub]2–48,63Ub (Fig 2J). The tUIM binding significantly increased the T1 values (reflecting slower tumbling) for both the distal K63-linked and the proximal Ub but caused almost no change in the T1 values of the distal K48-linked Ub, rendering it the lowest-T1 unit in the chain (Fig 6G). This confirms that the Rap80 tUIM binds simultaneously to both the proximal and (formerly “free” and the most mobile) distal K63-linked Ubs, thereby restricting their conformational freedom. At the same time, the tUIM binding displaced and freed the distal K48-linked Ub from its contact with the proximal Ub, making it the most mobile Ub in the chain. Thus, we conclude that the tUIM-induced perturbations detected in the distal K48-linked Ub are primarily due to this secondary effect rather than direct interaction with the tUIM. However, we cannot completely rule out that some small fraction of this distal Ub interacts with the excess Rap80 tUIM present at the saturation conditions.
Linkage-specific deubiquitinating enzymes retain linkage selectivity with branched and mixed-linkage polyUb substrates
Given that DUBs are critical to maintain the pool of free Ub and to regulate conjugate levels, we tested if linkage-selective DUBs would still retain their ability to recognize and cleave their cognate Ub–Ub linkage in the branched and unbranched mixed-linkage chains. For this purpose we used OTUB1, which specifically cleaves K48 linkages (Wang et al., 2009), and AMSH, which is specific for K63-linkages (McCullough et al., 2004). As evident in Fig 7, OTUB1 readily reduced [Ub]2–48,63Ub, Ub–63Ub–48Ub, and Ub–48Ub–63Ub to di- and mono-Ub. However, unlike the case with the homogeneously-linked Ub–48Ub–48Ub substrate, OTUB1 alone could not further process the di-Ub species generated from any of the mixed-linkage chains (Fig 7). Only when both OTUB1 and AMSH were present was complete chain disassembly achieved, indicating that the uncleaved di-Ub was linked via K63. An essentially identical behavior was observed for AMSH, which readily cleaved the K63-linkage in all three tri-Ub chains while leaving the K48-linkage intact (Fig 7). Taken together, these results demonstrate that the K48 and K63 linkages in branched and unbranched mixed-linkage polyUb can each individually be processed by linkage-selective DUBs. Recently, it was shown that OTUB1 and AMSH can separately cleave their cognate linkages in a branched tri-Ub construct containing analogs of K48 and K63 linkages formed via a Nο-Gly-L-homothiaLys mimic (which is one C–C bond longer and contains a sulfur atom instead of carbon) of an isopeptide bond (Valkevich et al., 2012). Our study demonstrates this selectivity for fully natural isopeptide bonds.
Figure 7.
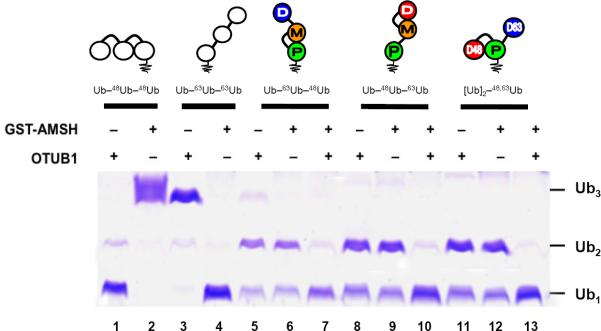
Linkage-selective DUBs cleave their cognate Ub–Ub linkages in homogeneous or mixed-linkage polyUb. The indicated chains were incubated with OTUB1, GST-AMSH, or both and the products evaluated by SDS-PAGE and staining with Coomassie Blue; see Experimental Procedures for details. Lanes 1–4 show reaction products of homogeneous Ub–48Ub–48Ub and Ub–63Ub–63Ub. Lanes 5 and 6, 8 and 9, and 11 and 12 show that alone each DUB can only process one linkage in the unbranched or branched mixed-linkage chains. Only with both DUBs present were the mixed-linkage chains hydrolyzed fully to monomeric Ub (lanes 7, 10 and 13).
The yeast 26S proteasome can process branched chains
One of the most studied outcomes of Ub modification is targeting to the proteasome. In a poorly understood process, proteasome-associated DUBs mainly function to modify or remove the polyUb tag from conjugated substrates (e.g., (Glickman and Adir, 2004; Lee et al., 2011)). The three major DUBs associated with the proteasomes are Rpn11 (Poh1 in humans), Ubp6 (Usp14 in humans), and Uch37 (in humans). Whether and how branched polyUb chains are processed by the proteasome is an open question.
A series of studies has shown that the so-called “forked” polyUb chains that are branched on two closely positioned lysine residues (i.e., K6 & K11, K27 & K29, or K29 & K33) are degraded slowly by purified proteasomes and inhibit proteasomal DUB activity and substrate degradation (Kim and Goldberg, 2012; Kim et al., 2007; Kim et al., 2009). Whether this observation holds for other chains such as those containing the more abundant K48 and K63 linkages was not reported. To evaluate the efficiency of proteasome processing of Ub–48Ub–48Ub, Ub–63Ub–63Ub, or [Ub]2–48,63Ub signals, we incubated these tri-Ubs with purified yeast 26S proteasomes and monitored the cleavage reaction by blotting with anti-Ub and linkage-selective antibodies.
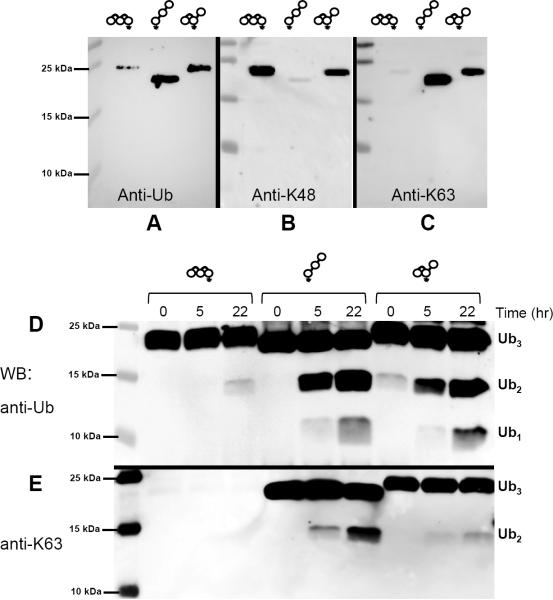
We first established that both the K48 and K63 linkage-selective antibodies recognized their respective Ub–Ub linkages in [Ub]2–48,63Ub (Fig 8B,C). This result in itself clearly demonstrates that chain branching preserves both Ub-specific and linkage-specific epitopes for antibody recognition, and also shows that linkage-specific antibodies can be used to probe if a particular polyUb chain contains multiple linkage types. Next, each chain was incubated with purified 26S proteasome, and reaction products were identified by blotting with anti-Ub antibodies. Processing of the homogenous Ub–63Ub–63Ub chains was faster than that of Ub–48Ub–48Ub (Fig 8D); this result is consistent with previous reports (Cooper et al., 2009; Jacobson et al., 2009). Accordingly, the branched [Ub]2–48,63Ub chain was disassembled faster than Ub–48Ub–48Ub, and generation of di-Ub (by removal of the single K63-linked Ub) was as efficient as from Ub–63Ub–63Ub chains. These observations suggest that (i) the branching did not hinder proteasomal cleavage, and (ii) the presence of a K63 linkage in this chain facilitates its conversion to di-Ub. The latter was verified using a K63 linkage-specific antibody, which revealed that only a minor fraction of the di-Ub products generated from [Ub]2–48,63Ub were K63-linked (Fig 8F). Taken together, these results demonstrate that the 26S proteasome can process branched polyUb chains.
Figure 8.
Disassembly of homogeneous and branched chains by 26S proteasomes and their recognition by linkage-specific antibodies. Upper panel (A–C): 18%-SDS-PAGE blots of Ub–48Ub–48Ub, Ub–63Ub–63Ub, and [Ub]2–48,63Ub with (A) anti-Ub, (B) anti-K48, and (C) anti-K63 antibodies. All three tri-Ub species were detected with anti-Ub. Only the [Ub]2–48,63Ub was detected by both anti-K48 and anti-K63, confirming that both K48 and K63 linkages are present. Lower panel (D–E): Ub–48Ub–48Ub, Ub–63Ub–63Ub, or [Ub]2–48,63Ub were incubated with purified yeast 26S proteasomes and the digests were monitored at 0, 5, and 22 h by SDS-urea PAGE and blotting with (D) anti-Ub and (E) anti-K63 antibodies.
Discussion
Ubiquitination machinery has been shown in some cases to be processive in generating homogeneously-linked chains. We demonstrated here that, when acting together simultaneously or sequentially, two linkage-specific E2 enzymes (K48-specific E2-25K and K63-specific Ubc13:Mms2) can form both branched and unbranched mixed-linkage forms of polyUb. This observation can be generalized further. As shown in Fig S10, adding a K11-specific E2 (Ube2s) to the abovementioned enzymes generates a triply-branched product, [Ub]3–11,48,63Ub, in which three distal Ubs are attached simulateously to K11, K48, and K63 of the same proximal Ub. Altogether, our results indicate that exposure of Ub (or polyUb) to more than one E2 enzyme can result in chains composed of more than one linkage type. A similar outcome could be expected in the case of a single linkage-promiscuous E2 (Kim et al., 2007). Depending on the factors involved (which are currently unclear), the resulting polyUb landscape would include chains of unbranched and branched topologies. This raises the question of why such chains have not been observed more often. Possible reasons are that intracellular DUBs rapidly edit mixed-linkage chains, or that assembly of mixed-linkage chains largely is prevented by regulation (e.g., via compartmentalization) of the various E2 and E3 enzymes in cells. However, because current methods are poorly suited for detection of mixed-linkage chains, such chains may in fact be quite abundant but remain unseen.
This study revealed that K48 and K63 linkages in mixed-linkage polyUb retain structural features of the corresponding homogeneously-linked chains. Our NMR data show unambiguously that the hydrophobic interface characteristic of the K48-linkage is formed across the Ub–48Ub unit in both the branched and unbranched mixed-linkage trimers, and that this interface exhibits the same pH dependence as the “classical” interface in homogeneous K48-linked polyUb. Furthermore, the 15N relaxation data revealed that in the mixed-linkage chains the K63-linked Ub is highly mobile compared to the other two Ubs, which are restricted by their K48-dimer interface contacts.
We found that highly selective receptors for each of the two linkage types bind the branched [Ub]2–48,63Ub chain in a linkage-specific manner; i.e., binding to the cognate Ub–Ub unit was essentially as if the other linkage was not present. It is noteworthy that the proximal Ub in [Ub]2–48,63Ub is adaptable and can bind in both K48 and K63 linkage-selective modes. Furthermore, chain branching also preserves both Ub-specific and linkage-specific epitopes for antibody recognition. Thus, the K48 and K63 linkages within the same polyUb chain can retain their characteristic signaling properties. This conclusion is further strengthened by the fact that linkage-selective DUBs can efficiently process their cognate linkages in both branched and unbranched mixed-linkage chains. Our finding that the 26S proteasome recognizes and cleaves the branched chain suggests that in vivo polyUb containing both K48 and K63 linkages can be disassembled by the proteasome essentially as a homogeneous chain. Taken together, these observations demonstrate that the K48 and K63 signaling properties can be encoded into the same polyUb via linkage mixing or branching, thus allowing the chain to carry two distinct signals. Just as there is little or no steric hindrance in assembling these chains, there appears to be no detectable hindrance in their reading and disassembly.
The reason for targeting conjugates to the proteasome is to degrade a substrate, not to disassemble the polyUb modification, yet inevitably the chain must be disassembled as well. Our observations using yeast 26S proteasomes together with published data for mammalian proteasomes (Cooper et al., 2009; Jacobson et al., 2009) demonstrate that homogeneous K63 linkages are disassembled faster than K48 linkages. This phenomenon holds even for a branched chain. A predicted outcome of the slower cleavage of K48 linkages is a longer residence on the proteasome's 19S regulatory complex, which may be important to allow sufficient time to prepare the substrate for degradation.
In principle, two or more (distal) Ub units attached to the same (proximal) Ub could be positioned to form a recognition surface or Ub–Ub interaction that is only available via branching (see Fig S11). However, in the case of the [Ub]2–48,63Ub system studied here, we detected no interaction between the two distal Ubs. Furthermore, the UBA(2) and Rap80 tUIM receptors selectively engaged Ub–Ub units linked via Ub K48 or K63, respectively, and bound in the same manner as with the di-Ub controls. Nevertheless, the potential exists for receptors, yet to be identified, to bind a branched chain in a completely novel way, e.g., across the juxtaposed distal Ubs (in this case, distal K48- and distal K63-linked Ubs). That this was not observed suggests that, at least for the well-characterized receptors studied here, linkage specificity is maintained in the context of branched polyUb.
In order to explore whether a Ub–Ub interface could be formed between two distal Ubs attached to the same proximal Ub, we generated in silico structural models for all twenty-eight possible branched tri-Ub chains (Table S1). We found that some combinations of linkages (e.g., K11 & K33, or K27 & K29) theoretically could promote close hydrophobic-patch contacts between the distal Ubs while simultaneously freeing the proximal Ub of any non-covalent interactions with the distal Ubs (Fig S11). Although we did not detect such contacts for [Ub]2–48,63Ub, the possibility of the distal units forming a Ub–Ub interface or novel binding interactions with receptors cannot be excluded for other branched chains. It should be emphasized that here we studied only the smallest possible branched chain. It is possible that an extension of a branched tri-Ub from one or both distal ends or from the proximal end could promote additional intra-chain interactions.
Although this study is the first attempt to investigate structurally unbranched and branched polyUb chains of mixed linkages, evidence of physiological relevance of such chains is mounting (Ben-Saadon et al., 2006; Dammer et al., 2011; Goto et al., 2010; Kravtsova-Ivantsiv and Ciechanover, 2012; Newton et al., 2008; Winborn et al., 2008). One cannot discount the possibility that the polyUb signal is constantly subject to editing or remodeling, and some branched or other mixed-linkage chains could simply be accidents that, in analogy to mismatched bases in DNA or misfolded proteins, are eventually corrected by cellular machinery. Nonetheless, as we have demonstrated here, linkage mixing or branching could enhance the signaling capability of polyUb. That mixed-linkage chains can carry multiple recognition signals is an exciting concept opening a new perspective on ubiquitin signaling.
Experimental Procedures
Assembly and purification of [Ub]2–48,63Ub, Ub–63Ub–48Ub, and Ub–48Ub–63Ub
The tri-Ub chains were assembled using enzyme-catalyzed reactions with full control of chain length, linkage composition, and the location of the isotope-enriched Ub unit, as detailed in Supplemental Information.
Preparation of linkage-selective receptors
Rap80-tUIM was prepared as described (Sims and Cohen, 2009). C121 on the C-terminal end of the second UIM was mutated to a tyrosine to prevent disulfide bond formation and to facilitate quantification by absorbance measurement at 280 nm. UBA(2) of hHR23A was expressed as a GST-fusion in E. coli as described (Varadan et al., 2004).
26S proteasome deubiquitination assay
To follow 26S proteasome deubiquitination activity, tri-Ub molecules were incubated with purified Saccharomyces cerevisiae 26S proteasomes in an ATP regenerating system buffer at 30 °C in ~1:10 proteasome/substrate ratio. Samples were taken at indicated time points and denatured in an SDS and 6 M urea loading buffer. Following low voltage (30 V for 2 h) transfer to nitrocellulose membranes, the blots were blocked with 5% low-fat milk powder, and incubated either with anti-Ub (Dako, diluted 1:1000), anti-K63Ub, or anti-K48Ub (Genentech Inc, diluted 1:500), and analyzed by ECL reaction.
Linkage-selective DUB assay
Recombinant human otubain-1 (OTUB1) and GST-AMSH were expressed in E. coli and purified essentially as described (McCullough et al., 2004; Wang et al., 2009). Deubiquitination reactions contained 1 μg/μL of polyUb substrate incubated at 37 °C with OTUB1 (0.04 μg/μL) or GST-AMSH (~ 0.1 μg/μL) in a pH 7.5 buffer (50 mM NaHEPES, 0.05% BSA, and 2.5 mM DTT). After 2.5 h, reactions were stopped with SDS-PAGE sample buffer and analyzed by SDS-PAGE and Coomassie staining.
NMR experiments
All NMR experiments were performed at 23 °C on a Bruker Avance III 600 spectrometer equipped with a cryogenic TCI probe. All experiments at pH 6.8 used protein samples in 20 mM sodium phosphate buffer, pH 6.8, containing 0.02% (w/v) NaN3 and 5% D2O. For experiments at pH 4.5, proteins were exchanged into buffer containing 20 mM sodium acetate, 0.02%(w/v) NaN3, and 5% D2O, adjusted to pH 4.5 with acetic acid. See Supplemental Information for details on the NMR experiments.
Chemical shift perturbations
Differences between 1H-15N NMR spectra of two species (A and B) were quantified as chemical shift perturbations, defined as follows: CSP = [(δHA − δHB)2 + ((δNA − δNB)/52]1/2, where δH and δN are chemical shifts of 1H and 15, respectively, for a given backbone N-H group. The same equation was used to quantify spectral perturbations upon titration; in this case, A refers to the unbound species, and B corresponds to various steps in the titration.
Supplementary Material
Highlights
K48 & K63-linkages in branched and unbranched tri-Ub retain their structural features
Linkage-selective receptors recognize K48- and K63-linkages in the branched tri-Ub
Linkage-specific DUBs cleave their cognate Ub-Ub linkages in the mixed-linkage chains
The 26S proteasome recognizes and processes branched K48-and-K63-linked tri-Ub
Acknowledgements
This work was supported by NIH grants GM065334 to D.F. and GM097452 to R.E.C. and in part by a joint grant from the United States - Israel Binational Science Foundation (BSF) to D.F. and M.H.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A. The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol Cell. 2006;24:701–711. doi: 10.1016/j.molcel.2006.10.022. [DOI] [PubMed] [Google Scholar]
- Cooper EM, Cutcliffe C, Kristiansen TZ, Pandey A, Pickart CM, Cohen RE. K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J. 2009;28:621–631. doi: 10.1038/emboj.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D, Gearing M, Rees H, Lah JJ, Levey AI, et al. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J Biol Chem. 2011;286:10457–10465. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- Fushman D, Wilkinson KD. Structure and recognition of polyubiquitin chains of different lengths and linkage. F1000 Biol Rep. 3:26. doi: 10.3410/B3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Adir N. The proteasome and the delicate balance between destruction and rescue. PLoS Biol. 2004;2:E13. doi: 10.1371/journal.pbio.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto E, Yamanaka Y, Ishikawa A, Aoki-Kawasumi M, Mito-Yoshida M, Ohmura-Hoshino M, Matsuki Y, Kajikawa M, Hirano H, Ishido S. Contribution of lysine 11-linked ubiquitination to MIR2-mediated major histocompatibility complex class I internalization. J Biol Chem. 2010;285:35311–35319. doi: 10.1074/jbc.M110.112763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregori L, Poosch MS, Cousins G, Chau V. A uniform isopeptide-linked multiubiquitin chain is sufficient to target substrate for degradation in ubiquitin-mediated proteolysis. J Biol Chem. 1990;265:8354–8357. [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Jacobson AD, Zhang NY, Xu P, Han KJ, Noone S, Peng J, Liu CW. The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26 s proteasome. J Biol Chem. 2009;284:35485–35494. doi: 10.1074/jbc.M109.052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Goldberg AL. Formation of nondegradable forked ubiquitin conjugates by ring-finger ligases and its prevention by S5a. Methods Mol Biol. 2012;832:639–652. doi: 10.1007/978-1-61779-474-2_45. [DOI] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Uchiki T, Gygi SP, Goldberg AL. S5a promotes protein degradation by blocking synthesis of nondegradable forked ubiquitin chains. EMBO J. 2009;28:1867–1877. doi: 10.1038/emboj.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Kravtsova-Ivantsiv Y, Ciechanover A. Non-canonical ubiquitin-based signals for proteasomal degradation. J Cell Sci. 2012;125:539–548. doi: 10.1242/jcs.093567. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol Cell Proteomics. 2011;10:R110, 003871. doi: 10.1074/mcp.R110.003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Ryabov YE, Fushman D. A model of interdomain mobility in a multidomain protein. J Am Chem Soc. 2007;129:3315–3327. doi: 10.1021/ja067667r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 2009;28:2461–2468. doi: 10.1038/emboj.2009.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfried NT, Gozal YM, Dammer EB, Xia Q, Duong DM, Cheng D, Lah JJ, Levey AI, Peng J. Multiplex SILAC analysis of a cellular TDP-43 proteinopathy model reveals protein inclusions associated with SUMOylation and diverse polyubiquitin chains. Mol Cell Proteomics. 2010;9:705–718. doi: 10.1074/mcp.M800390-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell. 2009;33:775–783. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkevich EM, Guenette RG, Sanchez NA, Chen YC, Ge Y, Strieter ER. Forging isopeptide bonds using thiol-ene chemistry: site-specific coupling of ubiquitin molecules for studying the activity of isopeptidases. J Am Chem Soc. 2012;134:6916–6919. doi: 10.1021/ja300500a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Fushman D. Using NMR spectroscopy to monitor ubiquitin chain conformation and interactions with ubiquitin-binding domains. In Methods Enzymol. 2005a:177–192. doi: 10.1016/S0076-6879(05)99012-5. [DOI] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell. 2005b;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J Mol Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- Wang T, Yin L, Cooper EM, Lai MY, Dickey S, Pickart CM, Fushman D, Wilkinson KD, Cohen RE, Wolberger C. Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of otubain 1. J Mol Biol. 2009;386:1011–1023. doi: 10.1016/j.jmb.2008.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winborn BJ, Travis SM, Todi SV, Scaglione KM, Xu P, Williams AJ, Cohen RE, Peng J, Paulson HL. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J Biol Chem. 2008;283:26436–26443. doi: 10.1074/jbc.M803692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv I, Matiuhin Y, Kirkpatrick DS, Erpapazoglou Z, Leon S, Pantazopoulou M, Kim W, Gygi SP, Haguenauer-Tsapis R, Reis N, et al. A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol Cell Proteomics. 2011;10:M111.009753. doi: 10.1074/mcp.M111.009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.