Summary
Enzymatic oxidation of cholesterol generates numerous distinct bile acids that function both as detergents that facilitate digestion and absorption of dietary lipids, and as hormones that activate four distinct receptors. Activation of these receptors alters gene expression in multiple tissues leading to changes not only in bile acid metabolism, but also in glucose homeostasis, lipid and lipoprotein metabolism, energy expenditure, intestinal motility and bacterial growth, inflammation, liver regeneration and hepato-carcinogenesis. This review covers the roles of specific bile acids, synthetic agonists and their cognate receptors in controlling these diverse functions, as well as their current use in treating human diseases.
An Overview and Brief History of Bile Acids
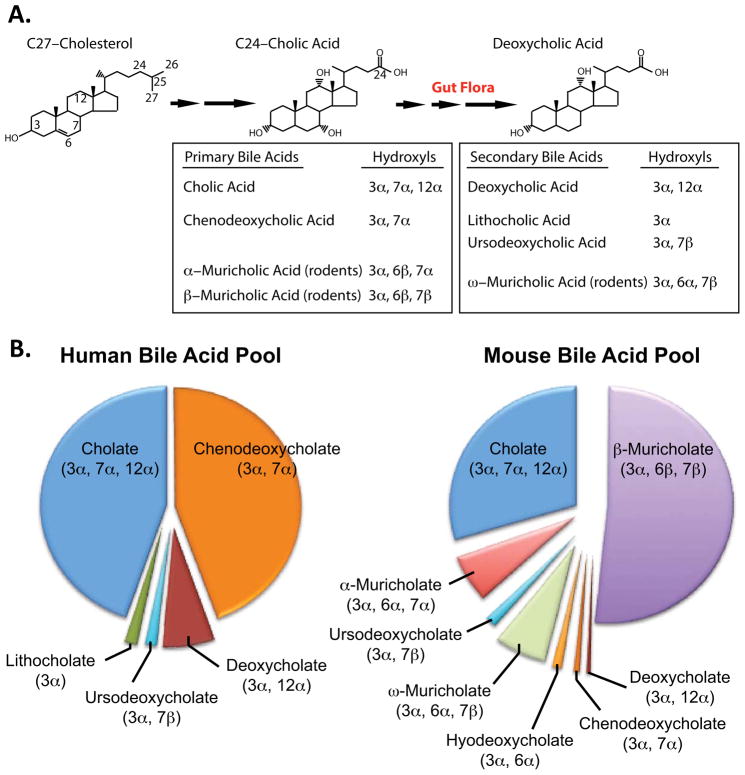
The first description of a bile acid was made in 1848 when cholic acid (CA) was discovered in ox-gall. Subsequent studies in the early 1900s identified additional bile acids that included lithocholic acid (LCA), chenodeoxycholic acid (CDCA), ursodeoxycholic acid (UDCA) and muricholic acid (MCA, #113) from ox, goose, bear and rodents respectively, as described by Wieland in his 1928 Nobel Lecture (Wieland, 1966). More sophisticated methodologies subsequently led to the identification of multiple additional species of bile acids, including deoxycholic acid (DCA), that contribute to the “bile acid pool” (2–4 g in humans) (Figure 1A, B). The relative concentrations of individual bile acids within the bile acid pool of different mammals can vary significantly (Figure 1B) and may affect bile acid-dependent signaling.
Figure 1. Synthesis of Bile Acids and the Bile Acid Pool.
(A) The conversion of the 27 carbon cholesterol to primary and secondary 24 carbon bile acids by liver enzymes and the microbiome/gut flora is shown. Representative primary and secondary bile acids together with the orientation of the hydroxyls are indicated.
(B) Relative bile acid composition in the human (modified from Hofmann, 2010) and mouse bile acid pool (modified from Kerr et al., 2002).
Bile acids are known to play a number of roles in lipid metabolism. First, bile acids are essential for the formation of mixed micelles in the small intestine that facilitate solubilization, digestion, and absorption of dietary lipids and fat-soluble vitamins. Second, the micelles present in the gall bladder serve to solubilize cholesterol in bile, thus impairing cholesterol crystallization and gallstone formation. Third, bile salts induce bile flow from hepatocytes into the bile canaliculi and then gall bladder. Fourth, the hepatic conversion of cholesterol to bile acids and the subsequent excretion of bile acids in the feces represent the major route for cholesterol excretion that is important in whole body sterol homeostasis. Bile is also thought to have a bacteriostatic function that maintains sterility in the biliary tree. Consistent with these roles, disruption of normal bile acid synthesis and metabolism is associated with cholestasis, gallstones, inflammation, malabsorption of lipids and fat-soluble vitamins, bacterial overgrowth in the small intestine, atherosclerosis, neurological diseases and various inborn errors such as Progressive Familial Intrahepatic Cholestasis types I–III (PFIC I–III).
The discovery that specific bile acids differentially activate three nuclear receptors, namely farnesoid X receptor (FXR), pregnane X receptor (PXR) and vitamin D receptor (VDR) and one G-protein-coupled receptor (TGR5), identified bile acids as hormones that alter multiple metabolic pathways in many tissues. The synthesis and use of specific agonists for FXR or TGR5 in rodents, together with preliminary clinical findings with FXR agonists, suggest that such agonists may prove useful in the treatment of a number of diseases.
In this review, we emphasize the mechanisms that maintain bile acid homeostasis through the regulation of bile acid synthesis and transport, and the diverse roles that these bile acids have on the four bile-acid responsive receptors. We also briefly discuss the current clinical uses of bile acids and FXR agonists to treat human diseases. We cite only a small fraction of the appropriate references as the literature on these topics is extraordinarily extensive. We refer those interested to many excellent related reviews on bile acids, FXR, or TGR5 (Angelin et al., 2012; Chiang, 2009; Keitel and Haussinger, 2012; Lefebvre et al., 2009; Matsubara et al., 2012; Pols et al., 2011b; Porez et al., 2012a; Russell, 2003, 2009; Thomas et al., 2008).
An Overview of Bile Acid Structure and Function
Bile acids are soluble products derived from the catabolism of highly insoluble cholesterol (Figures 1A, 2A). In general, bile acids are composed of four steroid rings forming a hydrocarbon lattice with a convex hydrophobic face and a concave hydrophilic face containing hydroxyl groups, and a short 5 carbon acidic side chain that is subsequently amidated with taurine or glycine (Figure 1A). This amphipathic structure gives bile acids the detergent properties that allow for micelle formation and facilitates the digestion and absorption of dietary lipids and fat-soluble vitamins A, D, E and K from the small intestine. The presence or absence of hydroxyl groups in the α or β orientation at position 3, 6, 7 and 12 on the steroid backbone (Figure 1A) affects both their solubility and hydrophobicity (rank order from hydrophobic to hydrophilic: LCA>DCA>CDCA>CA>UDCA>MCA). These small structural differences also have significant effects in the specificity of activation of the four bile acid-responsive receptors (Table 1). Surprisingly, none of the bile acid-responsive receptors have been shown to be activated by muricholic acid (MCA, #113), a major bile acid in rodents (Figure 1).
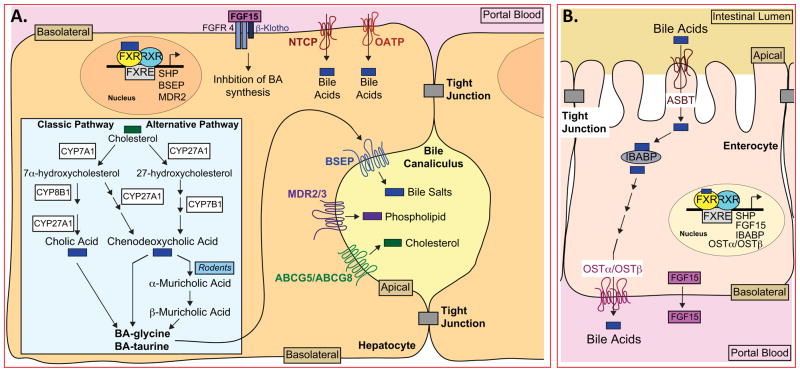
Figure 2. Bile Acid Metabolism in Liver and Intestine.
(A) A cartoon of hepatocytes showing the classic (neutral) and alternative (acidic) bile acid biosynthetic pathways that generate glycine- or taurine- conjugated bile acids (BA), including cholic, chenodeoxycholic and muricholic acids (inset). The location of the lipid transporters BSEP, MDR2/3 and ABCG5/ABCG8 on the apical membrane, and the location of NTCP and OATP transporters that facilitate recovery of bile acids from the blood are also shown. FGF15, secreted into the portal blood from the intestine, is shown bound to FGFR4/β-Klotho, leading to suppression of bile acid (BA) synthesis. Three representative FXR target genes (SHP, BSEP and MDR2) are shown in the nucleus.
(B) A cartoon of enterocytes in the distal ileum showing bile acid absorption from the lumen occurring via ASBT and bile acid efflux out of the cell via OSTα/OSTβ. Representative FXR target genes (SHP, FGF15, IBABP, OSTα and OSTβ) are shown as well as the secretion of FGF15 into the portal blood.
Table 1. Functions and Sites of Action of Bile Acids.
Summary of known functions and sites of action of bile acids present in the rodent and human bile acid pool.
| Function | Site of Action |
|---|---|
| Micelles prevent cholesterol crystallization | Gall bladder and bile duct |
| Micelles facilitate lipid digestion by pancreatic enzymes | Intestinal lumen |
| Micelles facilitate vitamin A, D, E and K absorption | Intestinal lumen |
| Impair bacterial over-growth | Intestinal lumen |
| Maintain intestinal barrier to infection | Intestine |
| FXR activation: CDCA>DCA>LCA≫CA | Liver, Ileum, kidney, adrenal gland |
| TGR5 activation: LCA>DCA>CDCA>CA (Taurine conjugation reduces EC50) | Gall bladder, intestinal neuroendocrine cells, enteric neurons, brown adipose tissue, macrophages, brain |
| PXR activation: LCA~3-keto-LCA | Intestine, liver |
| VDR activation: LCA~3-keto-LCA | Ileal enterocytes |
Bile Acid Synthesis and the Enterohepatic Circulation
At least 17 enzymes are involved in the modification of the cholesterol steroid ring, cleavage of the side chain and subsequent conjugation with glycine or taurine to form primary bile salts (Figure 2A) (Russell, 2003, 2009). The finding that patients with inborn errors of nine of these genes exhibit various diseases, including neonatal cholestasis, neurological defects and malabsorption of fat and fat-soluble vitamins, emphasizes the critical requirement for cholesterol catabolism to bile acids (Heubi et al., 2007).
The classic or neutral bile acid pathway is regulated by cholesterol 7α-hydroxylase (CYP7A1), a cytochrome P450 enzyme that converts cholesterol to 7α-hydroxycholesterol (Figure 2A, inset). The latter intermediate is either converted to CDCA, or is converted to CA by a pathway that depends on 12-hydroxylation of the steroid ring by CYP8B1 (Figure 2A). Deletion of Cyp7a1 in mice suggested that this pathway contributes approximately 75% of total bile acid synthesis (Schwarz et al., 1996). The alternative, or acidic, pathway generates CDCA and accounts for approximately 9% and 25% of the total bile acid synthesis in humans and mice, respectively (Duane and Javitt, 1999; Schwarz et al., 2001) (Figure 2A, inset). Although initially identified in humans (Anderson et al., 1972), Russell and colleagues characterized the alternative pathway when they discovered that Cyp7a1−/− mice had high post-natal lethality, but the few surviving mice actually synthesized bile acids (Ishibashi et al., 1996; Russell, 2009). Subsequent studies demonstrated that this latter pathway was dependent upon oxidation of the cholesterol side chain at C27 by CYP27A1, prior to hydroxylation of the steroid ring by oxysterol 7α-hydroxylase (CYP7B1) (Figure 2A inset). C25- and C24-(S)-hydroxycholesterols can also enter the alternative pathway to generate bile acids (Schwarz et al., 1996). Most of the oxysterols that feed into the alternative bile acid synthetic pathway are derived from the liver or macrophages (Russell, 2003). However, the brain and lung also contribute oxysterols to the alternative pathway as they synthesize 24-, 25- or 27-hydroxycholesterols that are subsequently transported to the liver for metabolism by the alternative bile acid pathway (Bjorkhem et al., 2010).
Prior to secretion, bile acids are conjugated with taurine or glycine (Figure 2A), a process that lowers their pKa and increases their solubility, thereby facilitating micelle formation in the acidic environment of the duodenum. However, unlike non-conjugated bile acids that can diffuse across membranes, bile salts require a transmembrane transporter to move them across membranes. The secretion of bile salts from hepatocytes into the canaliculi requires the bile salt export protein (BSEP; ABCB11), while transport of phospholipids requires ABCB4 (also known as MDR3 in humans or Mdr2 in mice) (Figure 2A). Although cholesterol efflux into bile requires ABCG5/ABCG8, there is an additional requirement for ABCB4, consistent with phospholipids in the bile canaliculi functioning as a sink to accept cholesterol that has been flipped across the apical membrane by ABCG5/ABCG8 (Figure 2A) (Graf et al., 2003; Nicolaou et al., 2012; Wang et al., 2009a). The importance of these transporters can be seen in patients or mice that have inactivating mutations or gene deletions leading to loss of function. For example, loss of BSEP results in progressive familial intrahepatic cholestasis, a disease wherein bile salts accumulate to toxic levels (Nicolaou et al., 2012).
The bile salts together with phospholipids and cholesterol are passed into the gall bladder where they are concentrated to form bile which is composed of 85% water. The remaining solute is a complex mixture of bile salts (67%), phospholipids (22%) and cholesterol (4%), together with electrolytes, minerals, minor levels of proteins, plus bilirubin and biliverdin pigments, which gives it a yellow-green or even orange hue (Dawson, 2010). Small amounts of mucus and secretory IgA (sIgA) may contribute to the bacteriostatic functions of bile (Sung et al., 1992). Bile salts and phospholipid micelles play a key role in solubilizing cholesterol in bile, thus preventing cholesterol crystallization and the formation of cholesterol-gallstones (Wang et al., 2009a).
The presence of dietary fat in the duodenum causes the secretion of cholecystokinin (CCK) from the intestinal mucosa into the circulation which in turn promotes contraction of smooth muscle cells of the gall bladder and relaxation of the sphincter of Oddi, thus allowing bile to enter the duodenum (Chandra and Liddle, 2007). In the lumen the bile salt-containing mixed micelles facilitate absorption of the fat-soluble vitamins A, D, E and the metabolism of dietary lipids by pancreatic enzymes, prior to their absorption. The gall bladder itself is not essential, since rats, which lack a gall bladder, and patients who have undergone cholecystectomy (removal of the gall bladder), are still able to absorb lipids from the diet as a result of direct secretion of bile into the duodenum.
Following secretion of bile salts into the duodenum, most (~95%) are reabsorbed in the distal ileum via the apical sodium-dependent transporter (ASBT) present on the enterocyte brush border (Figure 2B). Intestinal bile acid-binding protein (IBABP/fatty acid-binding protein subclass 6/FABP6) may facilitate transport of bile salts across the enterocyte to the basolateral membrane where they are effluxed into the blood by the heterodimeric transporter OSTα/OSTβ (Figure 2B) (Chiang, 2009; Dawson et al., 2010). However, a small percentage of the bile salts escape resorption, and are deconjugated by bacterial flora before either being absorbed or converted into secondary bile acids (Figure 1A). Secondary bile acids may be absorbed by passive processes, or excreted in the feces. The absorbed primary and secondary bile acids and salts are transported back to the liver where most, but not all, are actively transported into hepatocytes by sodium (Na+)-taurocholate cotransporting polypeptide (NTCP/SLC10A1) and organic anion-transporters (OATPs; e.g. Oat1b2) that mediate the uptake of bile salts and bile acids respectively (Figure 2A) (Chiang, 2009; Thomas et al., 2008). In the liver, bile acids are re-conjugated and then re-secreted together with newly synthesized bile salts to complete one cycle of the enterohepatic circulation. One exception is the secondary bile acid LCA that is present at low levels and is hepatotoxic at elevated concentrations. The small amount of LCA that is returned to the liver is sulfated prior to secretion into bile and excreted in the feces (Wang et al., 2009a).
In humans, the bile acid pool contains approximately 2–4 g of bile acids. Recycling of bile acids/salts between the liver and intestine occurs 6–10 times each day and transports 20–40 g bile acids. However, approximately 0.2–0.6 g of bile acids are excreted in the feces each day, an amount that must be replenished by de novo synthesis from cholesterol (Dawson, 2010). It is notable that the hepatic recovery of bile acids from the portal vein (bile acid concentration 10–80 μM) is incomplete (Angelin et al., 1982), thus accounting for the presence of low levels of bile acids (~2–10 μM) in the peripheral circulation of humans and mice (Angelin et al., 1982; Zhang et al., 2011b). As might be expected, the concentrations of individual bile acids in the portal vein and systemic blood vary with food consumption as resorption of bile acids is greatest in the post-prandial period (Angelin et al., 1982). The relatively high concentrations of bile acids in the tissues involved in the enterohepatic circulation (liver, bile ducts, gall bladder and intestine) is sufficient to activate receptors present in these tissues (Table 1; see below). However, since the total concentration of bile acids in the peripheral circulation is low under normal physiological conditions, additional studies are necessary to determine the importance of specific bile acids in activating TGR5 and FXR in peripheral tissues.
Bile Acids Affect the Microbiome and Vice Versa
Intestinal microbial organisms play an important role in bile acid metabolism as they readily deconjugate and 7α-dehydroxylate primary bile salts that escape reabsorption in the distal ileum and convert them to secondary bile acids (LCA, DCA, UDCA and, in rodents, ω–MCA) (Figure 1A) (Ridlon et al., 2006). However, these secondary bile acids synthesized by the microbiome comprise only a small percent of the normal bile acid pool (Figure 1B).
Recent studies have shown that the microbiome affects not only the composition of the bile acid pool but also the expression of genes controlled by the bile acid activated receptor FXR (Swann et al., 2011). As compared to rats with normal flora, germ-free rats were shown to have reduced levels of bile acids in various tissues, an almost total loss of unconjugated and glycine- conjugated bile acids, a significant increase in taurine-conjugated bile salts, and changes in the hepatic expression of genes regulated by the bile acid-responsive nuclear receptor FXR (Swann et al., 2011). More modest changes were observed following treatment of rats with antibiotics that partially reduce the microbiome (Swann et al., 2011). In contrast, bile acid levels were significantly increased in the livers of germ-free C57BL/6 mice (Selwyn and Klaassen, 2012). Although the reasons for these differences between germ-free rats and mice remain to be elucidated, both studies suggest that the microbiome is an important modulator of bile acid metabolism and function.
Dietary components can also affect the microbiome, the bile acid pool and intestinal inflammation. For example, administration of saturated milk-derived fatty acids to mice specifically increased taurocholate levels that provided a selective advantage in growth of Bilophilia wadsworthia, a sulphite-reducing pathobiont (Devkota et al., 2012). Consistent with this proposal, oral administration of taurocholate but not glycocholate increased the growth of B wadsworthia and increased inflammation and colitis in genetically susceptible mice (Devkota et al., 2012). Additional studies will be required to determine whether these latter findings in rodents are applicable to humans.
Farnesoid X Receptor, the First Bile Acid-responsive Receptor
The identification of the nuclear receptor Farnesoid X Receptor (FXRα, NR1H4, hereafter called FXR) (Forman et al., 1995; Seol et al., 1995), and the subsequent demonstration that bile acids were the endogenous ligands for FXR (Makishima et al., 1999; Parks et al., 1999; Wang et al., 1999) were critical to our understanding of the molecular mechanisms that control bile acid metabolism. The rank order of potency of bile acids as FXR agonists is shown in Table 1. However, the synthesis of numerous FXR specific agonists, including GW4064, 6α-ethyl-chenodeoxycholic acid (6-ECDCA; INT-747;obeticholic acid), fexaramine and GSK2324 (Porez et al., 2012b) has proven particularly important in overcoming the broader effects of bile acids on metabolism.
FXR is one of 48 members of the human nuclear receptor superfamily of transcription factors. Rodents express one additional member, FXRβ (NR1H5), which is activated by lanosterol but insensitive to bile acids (Sonoda et al., 2008). In contrast, human FXRβ is a pseudogene. FXR, like many of the nuclear receptors, contains a DNA binding domain (DBD), a ligand binding domain (LBD) as well as additional activation domains (Calkin and Tontonoz, 2012; Sonoda et al., 2008). There are four human and mouse FXR isoforms that result from alternative promoter usage and alternative RNA splicing (Zhang and Edwards, 2008). The relative expression of these isoforms differs in FXR-rich tissues/cells that are limited to hepatocytes, enterocytes in the ileum and renal tubules, all of which are known to be involved in bile acid metabolism (Forman et al., 1995). In contrast, although FXR is highly expressed in the adrenal gland, this organ has no known role in bile acid metabolism. It also remains to be established whether the very low levels of FXR reported to be present in peripheral tissues such as adipose tissue, heart, pancreatic β-cells and the artery wall are sufficient to directly affect gene expression and metabolism in vivo. Generation of mice with specific deletion of FXR in these latter tissues might help determine the importance of this nuclear receptor in these tissues.
FXR binds to specific DNA response elements (FXRE) as an obligate heterodimer with RXR (Kalaany and Mangelsdorf, 2006). Genome-wide analysis of FXR binding sites revealed that only 10% of FXREs are present in proximal promoters of target genes while most FXRE (90%) were in introns or intergenic regions (Chong et al., 2010; Thomas et al., 2010). Further analysis indicates that FXR regulates genes involved in many metabolic pathways, in addition to the well-established role of FXR in regulating bile acid homeostasis (Figure 2).
The Role of FXR in Regulating the Enterohepatic Circulation
The first description of the bile acid-dependent negative feedback loop was provided in the 1960s, when oral administration of bile acids to rodents was shown to repress bile acid synthesis by inhibiting Cyp7a1 (Reviewed in Russell, 2003). With a better understanding of the many genes involved in the enterohepatic circulation we now have a detailed, but incomplete understanding of the molecular mechanisms involved in this intricate and elegant negative feedback loop.
Repression of Cyp7a1 mRNA levels by bile acids was largely attenuated in Fxr−/− mice, demonstrating a critical role of FXR in bile acid synthesis (Sinal et al., 2000). This feedback mechanism leading to repression of both Cyp7a1 and Cyp8b1 involves at least two distinct mechanisms. The first involves bile acid-activation of hepatic FXR, leading to increased expression of small heterodimer partner (SHP, NR0B2), an unusual member of the nuclear hormone receptor family that lacks a DNA binding domain and functions as a transcriptional repressor (Zhang et al., 2011a). In the liver, SHP antagonizes the effects of liver receptor homologue 1 (LRH-1) and hepatocyte nuclear factor 4 alpha (HNF4α) that positively regulate the expression of Cyp7a1 and Cyp8b1, the rate-limiting enzymes in the synthesis of CDCA and CA (Figure 2A) (Chiang, 2009). Unexpectedly, treatment of Shp−/− mice with bile acids resulted in partial repression of Cyp7a1 and Cyp8b1 mRNA levels, suggesting the presence of additional, SHP-independent mechanism(s) of repression (Kerr et al., 2002; Wang et al., 2002).
A second mechanism for the feedback inhibition of bile acid synthesis involves Fibroblast Growth Factor 15 (FGF15 in mouse) or the human homologue FGF19 (Inagaki et al., 2005). FGF15 and 19 are unusual members of the FGF family since they are secreted into the blood and require a membrane-bound coreceptor protein (β-Klotho) in order to activate the FGF receptor FGFR4 (Figure 2A and B) (Angelin et al., 2012; Jones, 2012; Potthoff et al., 2012).
Initial studies revealed that treatment of human hepatocytes with an FXR agonist led to a robust induction and secretion of FGF19 (Holt et al., 2003; Jones, 2012). However, subsequent studies indicated that levels of FGF19 mRNA in normal human or mouse livers were low/undetectable (Schaap et al., 2009). Nonetheless, the finding that FGF19 mRNA levels were detected in the livers of patients with extrahepatic cholestasis caused by a pancreatic tumor, suggests a possible autocrine role for the hepatic FGF19, at least under these conditions (Schaap et al., 2009).
In contrast, the expression of FGF15/19 is high in enterocytes lining the distal ileum (Figure 2B) (Inagaki et al., 2005; Jones, 2012). Numerous experimental approaches indicate that activation of intestinal FXR by reabsorbed bile salts induces the expression and subsequent secretion of FGF15/19 into the circulation. FGF15/19 then binds to the FGFR4/β–Klotho complex on the hepatocyte plasma membrane to initiate an intracellular signaling cascade involving JNK that represses Cyp7a1 mRNA and bile acid synthesis (Figure 2A and B) (Inagaki et al., 2005; Jones, 2012; Kong et al., 2012). Support for this model comes from genetic studies showing that Fgfr4−/− or β–Klotho−/− mice have elevated Cyp7a1 and Cyp8b1 mRNA levels and increased, unregulated bile acid synthesis (Ito et al., 2005; Yu et al., 2000), and from the observation that treatment of intestinal Fxr−/− mice with specific FXR agonists fails to fully repress Cyp7a1 and Cyp8b1 expression (Kong et al., 2012). Further support for the model comes from studies showing that treatment of Cynomolgus monkeys with an FGF19 antibody leads to increased bile acid synthesis and excretion (Pai et al., 2012). Furthermore, in humans, plasma FGF19 levels exhibit a circadian rhythm that modulates hepatic bile acid synthesis (Galman et al., 2011; Lundasen et al., 2006). Thus, extensive data support the hypothesis that intestinally derived FGF15/19 controls hepatic bile acid synthesis in rodents and primates. Nonetheless, quantification of plasma levels of FGF15 protein in mice under various conditions would provide a missing piece of the puzzle supporting the role of FGF15 in bile acid metabolism, gall bladder filling and glucose metabolism (Angelin et al., 2012). Although both SHP and FGF15/19 clearly play important roles in the repression of bile acid synthesis, the relative physiological importance of these two pathways in humans remains to be determined.
An additional role for FGF15, and by extension, bile acids, was identified when it was shown that gall bladders of Fgf15−/−, Fgfr4−/− and β-Klotho−/− mice are smaller than those of wild type mice (Choi et al., 2006; Kong et al., 2012; Potthoff et al., 2012). These observations suggest that FGF15 protein, secreted from intestinal enterocytes following activation of FXR, binds to FGFR4/β-Klotho on the smooth muscle cells of the gall bladder to promote increased intracellular signaling and relaxation, thereby allowing the refilling of the gall bladder in a timely manner.
Bile acid-dependent activation of FXR also regulates other aspects of the enterohepatic circulation. In the liver, activated FXR induces the expression of the transporters BSEP/ABCB11, MDR3/Mdr2/ABCB4, and ABCG5/ABCG8 that function to efflux bile salts, phospholipids and cholesterol, respectively, into the biliary canaliculi (Figure 2A). At the same time, the expression of the bile acid importer NTCP is also repressed, thus limiting the transport of bile salts from the blood into the hepatocytes (Figure 2A). The mechanism for NTCP repression appears to depend on expression of the transcriptional repressor SHP (Denson et al., 2001). Thus, activation of hepatic FXR promotes the increased efflux of bile salts, coupled with decreased bile acid synthesis and decreased re-uptake of bile salts from the blood. Together, these changes maintain bile acid levels in the biliary tree and intestine appropriate for lipid absorption, while at the same time limiting the hepatic accumulation of bile acids to toxic levels.
In the postprandial state, the bile salts in the lumen of the small intestine are resorped via the transporter ASBT on the surface of enterocytes in the distal ileum (Figure 2B). Within the nucleus of the enterocytes, the bile salts activate FXR leading to induction of intestinal FXR target genes that include Ibabp, Ostα, Ostβ, Shp and Fgf15/19 (Figure 2B) (Matsubara et al., 2012). Repression of ASBT, likely via increased levels of SHP (Dawson et al., 2003), limits excessive uptake of potentially toxic bile acids. Studies with gene-targeted mice, or humans with inactivating mutations in ASBT, indicate that both ASBT and the OSTα/OSTβ heterodimer are essential for transporting bile salts from the intestinal lumen into the portal blood (Figure 2B) (Dawson et al., 2003; Oelkers et al., 1997; Rao et al., 2008). In conclusion, FXR present in hepatocytes and enterocytes functions to monitor cellular bile acid/salt levels and to alter gene expression in order to maintain the enterohepatic circulation of bile salts/acids while at the same time preventing cellular accumulation to toxic levels.
Renal FXR and Bile Acid Conservation
Although not part of the enterohepatic circulation, the kidney also plays a role in conserving bile acids (Dawson et al., 2010; Wang et al., 2009b). Indeed, FXR is highly expressed in the proximal and distal collecting tubules where it is thought to regulate genes involved in bile acid resorption, lipid metabolism and inflammation (Wang et al., 2009b). Cells of the proximal collecting tubules also express ASBT on their apical surface that likely functions to resorb and thus conserve bile acids under normal conditions (Zollner and Trauner, 2006). Presumably, the transmembrane heterodimer OSTα/OSTβ that is induced following activation of renal FXR (Higashiyama et al., 2008; Lee et al., 2006; Zollner and Trauner, 2006), transports these bile acids back into the blood (Boyer et al., 2004; Dawson et al., 2010). Under certain conditions, the capacity for renal reabsorption of bile acids may be limiting, as patients with cholestasis have elevated levels of bile acids both in the urine and peripheral blood (Trottier et al., 2011). It is not known whether bile acids play a more specific role in the kidney.
FXR Activation Modulates Several Distinct Metabolic Pathways
Studies in which humans were administered bile acids and/or wild type or Fxr−/− mice were treated with FXR-specific agonists, indicate that FXR regulates the metabolism of plasma lipoproteins, glucose, steatosis, inflammation, bacterial growth and liver regeneration, in addition to the regulation of bile acid homeostasis. Evidence from limited clinical trials suggests that FXR agonists may have beneficial effects in humans.
i) Lipoprotein Metabolism
The original characterization of Fxr−/− mice suggested that FXR also controls lipid metabolism as these mice had elevated hepatic lipids, as well as increased plasma triglycerides, cholesterol, free fatty acids and lipoproteins (VLDL and LDL) (Sinal et al., 2000). Conversely, treatment of diabetic (db/db), obese (ob/ob) or wild-type mice, but not Fxr−/− mice, with bile acids or specific FXR agonists lowered plasma triglyceride, fatty acids and cholesterol, and decreased hepatic lipid levels/steatosis (Watanabe et al., 2004; Zhang et al., 2006). The decrease in plasma lipoprotein levels appears to be due in part to FXR-dependent induction of hepatic genes involved in lipoprotein clearance from the plasma. These include SR-B1 (Scarb1; the HDL receptor), the VLDL receptor, syndecan-1 and apoC-II, a cofactor for lipoprotein lipase that hydrolyses plasma triglycerides (Calkin and Tontonoz, 2012; Zhang and Edwards, 2008). Activated FXR also leads to repression of hepatic SREBP-1c, a critical transcription factor necessary for the synthesis of fatty acids and triglycerides, thus decreasing synthesis of these lipids (Watanabe et al., 2004; Zhang and Edwards, 2008). Interestingly, decades before the identification of the FXR pathway, patients who were given oral doses of CDCA over many months to gradually dissolve cholesterol gallstones exhibited reduced levels of plasma triglyceride (Iser and Sali, 1981). In retrospect, it seems likely that the reduction in plasma triglycerides were a result of activation of hepatic FXR by the CDCA.
ii) Glucose Metabolism
Bile acid-activation of FXR regulates glucose metabolism by improving insulin sensitivity and repressing hepatic gluconeogenesis. The earlier finding that Fxr−/− mice are insulin resistant suggested a role for FXR in glucose metabolism (Cariou et al., 2006; Ma et al., 2006; Zhang et al., 2006). Consistent with this observation, treatment of wild type or diabetic db/db or insulin resistant ob/ob mice, but not Fxr−/− mice, with CA or FXR-specific agonists improved insulin sensitivity and reduced plasma glucose levels (Cariou et al., 2006; Ma et al., 2006; Zhang et al., 2006). These changes appear to result partly from the FXR-dependent repression of hepatic Pepck and G6Pase expression and gluconeogenesis, as well as an FXR-dependent increase in glycogen synthesis (Reviewed in Calkin and Tontonoz, 2012). More recent studies indicate that FGF15, secreted from the ileum following activation of FXR, has insulin-like actions including inhibition of hepatic gluconeogenesis (Kir et al., 2011; Potthoff et al., 2011; Potthoff et al., 2012; Schaap, 2012). As discussed below, bile acids also reduce plasma glucose levels and increase insulin sensitivity following activation of the G-protein coupled receptor TGR5 (Choi et al., 2006; Potthoff et al., 2012).
iii) Cholestasis, Inflammation and Hepatoprotection
Cholestasis results in the intrahepatic accumulation of cytotoxic bile acids leading to increased inflammatory cytokines and liver injury (Zhang et al., 2012). The elevated levels of intracellular bile acids are expected to activate both FXR and PXR (see below). Activation of FXR was shown to provide hepatoprotection from bile duct-ligation or alpha-naphthylisothiocyanate, both of which have been used as models of cholestasis (Liu et al., 2003), or from a challenge with the high levels of acetaminophen (Lee et al., 2010). Such protection likely results from FXR-dependent induction of several phase I-III genes involved in xenobiotic metabolism (Lee et al., 2010; Matsubara et al., 2012). Very recently, Bennett and colleagues showed that bile acids induce the expression of Flavin-monooxygenase 3 (FMO3) in a mechanism that requires FXR. FMO3 is an enzyme previously thought to be part of the detoxification pathway, but was shown to be required for the synthesis of trimethylamine-N-oxide (TMAO), a metabolite that is strongly associated with atherosclerosis in mice and humans (Bennett et al., 2013).
Bile Acids Activate the Pregnane X Receptor and the Vitamin D Receptor
In addition to FXR, two other nuclear receptors, the pregnane X receptor (PXR; NR1I2) and the vitamin D receptor (VDR; NR1I1) are activated by specific bile acids.
i) PXR
PXR is highly expressed in the liver and intestine with lower levels in many other tissues. The functional PXR/RXR heterodimer is promiscuous as it is activated by numerous xenobiotics, pharmaceutical drugs, and specific bile acids. In contrast to FXR which is activated by a number of bile acids (CDCA>DCA>LCA≫CA; unresponsive to LCA) (Makishima et al., 1999; Parks et al., 1999), PXR is activated by the hepatotoxic bile acid LCA and 3-keto-LCA (EC50 10 μM) (Table 1) and is unresponsive to CDCA, DCA or CA (Staudinger et al., 2001). Activation of PXR induces phase I-III genes involved in metabolism, transport and excretion of many compounds including xenobiotics and toxic bile acids such as LCA. However, the relative importance of hepatic and intestinal PXR activation in response to either LCA or 3-keto-LCA in vivo remains to be determined. A recent report demonstrated that intestinal PXR provided protection against inflammatory bowel disease as a result of attenuated expression of NFκB and reduced expression of inflammatory cytokines (Cheng et al., 2012). Whether, bile acids provide a similar PXR-dependent protective effect against inflammatory bowel disease will require additional investigations.
ii) Vitamin D Receptor (VDR)
VDR is expressed in multiple tissues including the kidney, intestine and macrophages where it controls numerous physiological and pharmacological processes, which include not only bone and calcium metabolism, but also immunity, cellular growth and differentiation. The most potent endogenous agonist of VDR is 1,25-dihydroxyvitamin D3 (1,25diOHD3). Unexpectedly, the toxic bile acid lithocholic acid (LCA) as well as 3-keto LCA were shown to activate VDR target genes, including CYP3A family members, which are involved in the metabolism of LCA and other toxins to less toxic and biologically inactive products (Makishima et al., 2002) (Table 1).
Vitamins A and D are both lipid-soluble vitamins that depend on bile salts for normal absorption. Unexpectedly, these two vitamins were shown to be part of different feedback mechanisms that repress bile acid synthesis. Schmidt et al. reported that activation of VDR in enterocytes resulted in increased secretion of FGF15 and subsequent repression of hepatic Cyp7a1 by a process that required an intact FXR pathway (Schmidt et al., 2010). Consistent with these results, these authors reported that Vdr−/− mice have increased levels of hepatic Cyp7a1 and Cyp8b1, an increased bile acid pool size and decreased Fgf15 expression in enterocytes. At the present time it is not known whether the decrease in Fgf15 noted in Vdr−/− mice is sufficient to impair the FGF15-dependent filling of the gallbladder. However, these results indicate that VDR in the intestine can modulate hepatic bile acid synthesis.
Bile Acid-responsive G-protein Coupled Receptor (TGR5)
Interest in bile acids as hormones was further stimulated a decade ago when two groups independently identified a plasma membrane bound G protein coupled receptor that was activated by low levels (1–10 μM) of bile acids (Kawamata et al., 2003; Maruyama et al., 2002). This bile acid-responsive receptor has been referred to as membrane-type bile acid receptor (M-BAR), G protein-coupled bile acid receptor 1 (GPBAR1), GPR131, as well as the most commonly used name TGR5. In contrast to FXR, hydrophobic bile acids are the most potent activators of TGR5, with taurine conjugation further increasing their potency (EC50 of 0.29–10 μM) (Table 1) (Keitel and Haussinger, 2012; Maruyama et al., 2002; Pols et al., 2011b; Sato et al., 2008). Identification and characterization of the specific roles of TGR5 in vivo have been greatly aided by the synthesis of specific TGR5 agonists, such as INT-777 (6-methyl-23-methyl-CDCA ; EC50=0.58 μM) (Sato et al., 2008), the discovery that oleanolic acid isolated from olive leaves is a natural agonist and the generation of Tgr5−/− mice (Keitel and Haussinger, 2012; Porez et al., 2012b; Thomas et al., 2009).
TGR5 is highly expressed in non-parenchymal cells of the liver, that include the gallbladder epithelial cells, cholangiocytes (epithelial cells of the bile duct), sinusoidal epithelium, Küpffer cells as well as immune cells (Keitel and Haussinger, 2011). However, TGR5 is also expressed in many other tissues including brown adipose tissue, intestinal enteroendocrine cells that secrete the incretin hormone glucagon-like peptide-1 (GLP-1), enteric neurons and the brain (Keitel et al., 2010; Keitel and Haussinger, 2012; Vassileva et al., 2006). Activation of TGR5 results in receptor internalization, increased cAMP levels and activation of protein kinase A leading to increased phosphorylation of target proteins, including the cAMP response element binding protein (CREBP) transcription factor (Nguyen and Bouscarel, 2008; Pols et al., 2011b; Porez et al., 2012b). The results of such activation are broad and cell-specific and include anti-inflammatory effects in macrophages, increased energy expenditure in brown adipose tissue, improved glucose metabolism and insulin sensitivity, gall bladder relaxation and increased intestinal motility. The role of TGR5 in the brain is likely independent of bile acids. The finding that TGR5 also responds to nanomolar concentrations of the neurosteroid 5β-pregnane-3α-ol-20-one (Keitel et al., 2010) raises the possibility that additional endogenous agonists may co-exist in the circulation.
i) TGR5 and the Immune System
A role for TGR5 in the immune system was discovered when it was shown that activation of TGR5 on isolated macrophages or in mice, suppressed the release of pro-inflammatory cytokines, such as interleukin-1α and interleukin-6, in response to LPS (Kawamata et al., 2003; Keitel et al., 2008; Wang et al., 2011). The mechanism involves a TGR5-dependent decrease in the phosphorylation of an inhibitor of nuclear factor κB (NFκB), thus preventing the nuclear translocation NFκB-p65 and reducing the transcription of pro-inflammatory cytokines by NFκB (Pols et al., 2011a; Wang et al., 2011). The physiological importance of the anti-inflammatory effects of macrophage TGR5 was further elucidated following transplantation of bone marrow from wild type or Tgr5−/− mice into recipient Ldlr−/− and Ldlr−/−Tgr5−/− mice (Pols et al., 2011a). Treatment of the transplanted mice with a TGR5 agonist attenuated both the levels of pro-inflammatory cytokines and atherosclerosis in Ldlr−/−, but not in Ldlr−/−Tgr5−/− mice. These studies identify an important physiological role for macrophage TGR5 in suppressing cytokine expression and reducing disease in mice (Pols et al., 2011a). Whether this finding can be exploited in humans remains unknown as clinical trials with TGR5 agonists have yet to be reported.
ii) TGR5 and Energy Metabolism
Early evidence for TGR5 influencing energy metabolism came from feeding rodents a high fat diet supplemented with CA. The supra-physiological bile acid levels achieved in blood resulted in decreased obesity and insulin resistance (Watanabe et al., 2006). A mechanism for the decreased obesity was postulated to involve bile-acid dependent activation of TGR5 in brown adipose tissue leading to an increase in energy expenditure. TGR5, via increased cAMP, activated type 2 iodothyronine deiodinase (DIO2) leading to increase levels of active thyroid hormone (Watanabe et al., 2006). These effects could be mimicked by treatment of mice with INT-777, a TGR5-specific agonist, but not with the FXR agonist GW4064 (Thomas et al., 2009), thus identifying TGR5 as the essential receptor. The significance of TGR5 as an important regulator of energy expenditure is supported by the findings that female Tgr5−/− mice fed a high fat diet are more obese than controls (Maruyama et al., 2006) and that administration of a TGR5 agonist to wild-type mice reduced obesity and glucose levels in response to a high fat diet (Sato et al., 2007).
iii) TGR5 and Glucose Metabolism
Numerous studies have shown that activation of TGR5 in high fat-fed wild type or TGR5-transgenic mice lowers plasma glucose, enhances glucose tolerance and increases insulin sensitivity. Despite the finding that basal plasma glucose levels are normal in chow-fed Tgr5−/− mice, these data identify a role for TGR5 in regulating carbohydrate metabolism, (Sato et al., 2007; Thomas et al., 2009). Current evidence indicates that the mechanism involved in the control of glucose homeostasis is indirect and requires activation of TGR5 on intestinal enteroendocrine L-cells leading to secretion of GLP-1 into the circulation. It is hypothesized that GLP-1 then binds to the GLP-1 receptor in the liver leading to the improved insulin sensitivity (Katsuma et al., 2005; Keitel and Haussinger, 2012; Thomas et al., 2009). Important confirmation of this pathway would be gained from studies utilizing mice that lack TGR5 in enteroendocrine L-cells. At the current time, it is unclear whether TGR5 agonists might provide an alternative treatment for type 2 diabetes.
iv) Bile Acid Sequestrants, TGR5 and Glycemic Control
Bile acid sequestrants are approved for treatment of patients with hypercholesterolemia. These resins bind bile acids in the intestinal lumen and consequently impair bile acid resorption, thus interrupting the enterohepatic circulation of bile acids. The result is increased bile acid excretion, reduced plasma LDL levels, and, in type 2 diabetic patients, decreased plasma glucose and increase insulin sensitivity (Staels et al., 2010). A mechanism to explain the improved control of plasma glucose levels was recently identified when it was shown that treatment of mice with a bile acid sequestrant led to increased secretion of preproglucagon, the precursor of GLP-1, from the colon of wild-type but not Tgr5−/− mice (Harach et al., 2012). It was proposed that the increased levels of bile acids in the colon after bile-acid sequestrant treatment were sufficient to activate colonic TGR5 and enhance plasma GLP-1 levels (Harach et al., 2012). If a similar mechanism exists in humans, it may explain at least in part, the hypoglycemic effects observed when type 2 diabetics are treated with bile acid sequestrants.
v) TGR5, Gall Bladder Filling and Fluid Secretion
TGR5 protein is present on the surface of cholangiocytes, on the epithelial cells lining the gall bladder, on smooth muscle cells and on intracellular recycling endosomes (Keitel et al., 2009; Keitel and Haussinger, 2012). TGR5 expressed on these cells in the biliary tree controls fluid and chloride secretion by the epithelial cells, as well as post-prandial relaxation of the gallbladder prior to refilling with bile.
One mechanism was revealed when treatment of human gallbladder tissue or murine gallbladder epithelial cells with TGR5 agonists was shown to activate the cystic fibrosis transmembrane conductance regulator (CFTR) and increase chloride secretion (Keitel et al., 2009). Based on these, and a number of other studies, Keitel and Haussinger recently proposed that TGR5 may function as a bile acid sensor that links bile acid concentration in the biliary tree to bile acid absorption and fluid secretion (Keitel and Haussinger, 2012).
The finding that Tgr5−/− mice have smaller gallbladders than wild type mice, especially after feeding a cholic acid-enriched diet, suggested a role for this G-protein coupled receptor in gall bladder filling (Li et al., 2011). Consistent with this model, treatment of wild-type, but not Tgr5−/− mice with a TGR5 agonist caused relaxation of biliary smooth muscles and gallbladder filling (Lavoie et al., 2010; Li et al., 2011). Thus, there are two bile acid-dependent pathways that promote gallbladder relaxation following contraction and release of bile into the duodenum in response to CCK. One pathway involves bile acid activation of TGR5 expressed on the surface of smooth muscle cells of the gall bladder, and the second involves FGF15/19-dependent activation of FGFR4/β-klotho on the gall bladder following release of the hormone from enterocytes.
A broader role for TGR5 in bile acid metabolism is suggested by the finding that Tgr5−/− mice have a reduced bile acid pool size, despite normal bile acid synthesis. These mice are also resistant to gallstone formation following administration of a lithogenic diet (Vassileva et al 2006). However, the mechanisms leading to these changes have yet to be identified.
vi) TGR5 and Intestinal Motility
Luminal concentrations of bile salts are known to affect intestinal motility (Hofmann, 2010). Based on recent studies with Tgr5−/− mice, TGR5 transgenic mice and TGR5 agonists, it appears that normal peristalsis and transit time in the colon are dependent upon activation of TGR5 on enterochromaffin cells and inhibitory colonic neurons. Thus, transit time is decreased and constipation increased in Tgr5−/− mice (Alemi et al., 2013). The generation of tissue-specific TGR5 knockout mice that lack TGR5 in enterochromaffin cells and inhibitory neurons would provide a powerful model to further substantiate this hypothesis.
Clinical Applications: Alteration of Bile Acid Homeostasis and FXR- or TGR5-specific Agonists
For over 3000 years, bear bile has been used in traditional Chinese medicine to treat various ailments. Unfortunately, in most cases the bile is obtained from live bears housed under appalling conditions. Sadly, bear farming and distribution of their bile continues, despite being illegal in many countries, including the USA.
In Western medicine, oral administration of pure, chemically synthesized CDCA or UDCA has been used for only four decades. One of the earliest uses in modern clinical medicine involved chronic oral administration of bile acids (originally CDCA but subsequently UDCA) over months or years to patients with cholesterol gallstones (Table 2) (Iser and Sali, 1981). The change in the bile acid pool slowly solubilized the cholesterol gallstones in the gall bladder or ducts. However, the efficacy of UDCA is limited and this approach has been largely replaced by surgery. Oral administration of UDCA or nor-UDCA are also approved treatments to improve liver function in patients with primary biliary cirrhosis or sclerosing cholangitis, respectively (Table 2). UDCA has also been successfully used to prevent the occurrence of fatal veno-occlusive disease (VOD), a type of graft vs. host reaction, in recipients of bone marrow transplants (Ruutu et al., 2002).
Table 2. Bile Acids and FXR agonists: Current Clinical Status.
Clinically approved treatments that involve bile acid sequestrants, bile acids or specific FXR agonists are listed, together with their target disease and real or potential beneficial effect. Where noted, FXR agonists are in phase I-III clinical trials. LDL, low density lipoprotein; LDLR, LDL receptor.
| Compound | Clinical Status | Target | Target Disease | Effect |
|---|---|---|---|---|
| Bile Acid (BA) Sequestrant | Approved | Increased BA excretion | Hyperlipidemia | Increased hepatic LDLR Decreased plasma LDL |
| Type 2 Diabetes | Increased insulin sensitivity | |||
|
| ||||
| Cholic Acid | Approved | Provide bile acids (BAs) for lipid absorption | Inborn errors of BA metabolism that inhibit BA synthesis | Increased lipid absorption Improved liver function |
| CDCA | Approved | Replacement BA | Cerebrotendinous xanthomatosis | Reduce endogenous BA synthesis |
|
| ||||
| UDCA | Approved | Gallstone dissolution | Gallstones | Solubilize cholesterol gallstones |
|
| ||||
| UDCA | Approved | Liver | Primary biliary cirrhosis (PBC) | Improved liver function |
|
| ||||
| INT-747 (6-ECDCA; Obeticholic Acid/OCA) | Phase III (2011) | FXR | Type 2 Diabetes | Improved insulin sensitivity |
|
| ||||
| OCA | Phase I (2011) | FXR | Non-alcoholic steatohepatitis (NAFLD) | Improved insulin sensitivity Improved liver function |
|
| ||||
| OCA | Phase III (2011) | FXR | PBC | Improved liver function |
|
| ||||
| PX-102 | Phase I (2011) | FXR | Metabolic Syndrome NAFLD | Improved insulin sensitivity Improved liver function |
|
| ||||
| nor-UDCA | Phase I (2011) | Liver | Sclerosing cholangitis | Improved liver function |
Inborn errors of 9 of the bile acid synthetic genes have been identified, resulting in impaired bile acid synthesis and accumulation of intermediates (Heubi et al., 2007). The clinical symptoms of such inborn errors are heterogeneous. Patients may present with cholestasis, malabsorption of lipids and vitamins, fibrosis, neurological disease or hypercholesterolemia, and if untreated, progressive liver disease may result in high morbidity rates (Heubi et al., 2007). Treatment of some disorders by oral administration of CA or CDCA improves lipid adsorption, prevents liver injury, and inhibits the synthesis of toxic bile acid intermediates, thus providing an alternative to liver transplantation (Table 2) (Heubi et al., 2007).
A number of clinical trials (phase I-III) have been undertaken using the synthetic FXR agonists INT-747 (6-ECDCA; obeticholic acid), FXR-450 or PX-102 (Table 2) (Porez et al., 2012b). Although no detrimental effects were reported, larger scale clinical trials will be necessary to assess whether FXR-specific agonists will prove useful in the treatment of Type 2 diabetes, metabolic syndrome, non-alcoholic steatohepatitis or primary biliary cirrhosis. To our knowledge, no clinical trials have been completed with synthetic agonists for TGR5.
Summary and Future Perspectives
The proposal that bile acids might activate three nuclear receptors and a G-protein-coupled receptor and bring about broad changes in metabolism would have met with considerable skepticism before 1999. However, a recent study in which treatment of primary rodent hepatocytes with bile salts led to activation of ERK1/2, in a sphingosine-1-phosphate receptor-dependent pathway (Studer et al., 2012), suggests that other bile acid receptors remain to be identified. In this latter case, the generation and analysis of mice that lack this, or other potential bile acid receptors, will be particularly important.
The understanding of the role bile acids play in human health and disease will likely benefit from recent advances in technology that have enabled genome-wide association studies (GWAS) of disorders of bile acid metabolism. Numerous GWAS have been carried out for bile acid disorders, including Biliary Artesia (Garcia-Barcelo et al., 2010), gallstones (Buch et al., 2007), gall bladder cancer (Cha et al., 2012) and primary biliary cirrhosis (Liu et al., 2010; Mells et al., 2011; Nakamura et al., 2012). Some associations have identified genes known to affect the disease; for example, gallstones were associated with variations at the ABCG5/G8 locus, the transporter involved in biliary cholesterol efflux (Buch et al., 2007). However, many loci containing genes not known to be associated with bile acid metabolism have been identified in several of these studies, including loci containing inflammatory genes identified in the GWAS for primary biliary cirrhosis (Liu et al., 2010; Mells et al., 2011). Genes involved in bile acid metabolism have also been associated with other traits such as the association of Cyp7a1 with total and LDL cholesterol (Teslovich et al., 2010). The expansion of these studies will likely open up many new avenues for discovery of novel genes involved in bile acid metabolism.
The finding that initial phase I–III clinical trials to test the efficacy of FXR agonists have been successful is reason for optimism. However, the development of clinically useful agonists for FXR and TGR5 appears to be particularly challenging given that these bile acid receptors are present in many tissues where they affect numerous genes and distinct metabolic pathways. In this regard, the development of tissue selective agonists for FXR or TGR5 may prove to be important as their use may limit unwanted side effects. Finally, when we consider the numerous novel physiological functions of bile acids that have been identified in the last 13 years it seems highly likely that many more remain to be discovered.
Acknowledgments
This work was supported in part by United States Public Health Service, grants HL30568 (to P.A.E.) and the Laubish fund at UCLA (P.A.E.). T.Q. de A.V. (11POST7240070) and E.J.T. (11POST7300060) were partially supported by postdoctoral fellowships from the American Heart Association Western States Affiliate. We thank Drs Jake Lusis and Simon Beavan for useful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alemi F, Poole DP, Chiu J, Schoonjans K, Cattaruzza F, Grider JR, Bunnett NW, Corvera CU. The receptor TGR5 mediates the prokinetic actions of intestinal bile acids and is required for normal defecation in mice. Gastroenterology. 2013;144:145–154. doi: 10.1053/j.gastro.2012.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KE, Kok E, Javitt NB. Bile acid synthesis in man: metabolism of 7 -hydroxycholesterol- 14 C and 26-hydroxycholesterol- 3 H. J Clin Invest. 1972;51:112–117. doi: 10.1172/JCI106780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelin B, Bjorkhem I, Einarsson K, Ewerth S. Hepatic uptake of bile acids in man. Fasting and postprandial concentrations of individual bile acids in portal venous and systemic blood serum. J Clin Invest. 1982;70:724–731. doi: 10.1172/JCI110668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelin B, Larsson TE, Rudling M. Circulating fibroblast growth factors as metabolic regulators - A critical appraisal. Cell Metab. 2012;16:693–705. doi: 10.1016/j.cmet.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Bennett BJ, Vallim TQdA, Wang Z, Shih D, Meng Y, Gregory J, Allayee H, Lee R, Graham M, Crooke R, et al. Genetic and Dietary Regulation of Trimethylamine-N-Oxide, a Metabolite Associated with Atherosclerosis. Cell Metab. 2013;17:1–12. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkhem I, Leoni V, Meaney S. Genetic connections between neurological disorders and cholesterol metabolism. J Lipid Res. 2010;51:2489–2503. doi: 10.1194/jlr.R006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L, Travaglione S, Falzano L, Gauthier NC, Popoff MR, Lemichez E, Fiorentini C, Fabbri A. Rac GTPase instructs nuclear factor-kappaB activation by conveying the SCF complex and IkBalpha to the ruffling membranes. Mol Biol Cell. 2004;15:1124–1133. doi: 10.1091/mbc.E03-05-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch S, Schafmayer C, Volzke H, Becker C, Franke A, von Eller-Eberstein H, Kluck C, Bassmann I, Brosch M, Lammert F, et al. A genome-wide association scan identifies the hepatic cholesterol transporter ABCG8 as a susceptibility factor for human gallstone disease. Nat Genet. 2007;39:995–999. doi: 10.1038/ng2101. [DOI] [PubMed] [Google Scholar]
- Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat Rev Mol Cell Biol. 2012;13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariou B, van Harmelen K, Duran-Sandoval D, van Dijk TH, Grefhorst A, Abdelkarim M, Caron S, Torpier G, Fruchart JC, Gonzalez FJ, et al. The farnesoid X receptor modulates adiposity and peripheral insulin sensitivity in mice. J Biol Chem. 2006;281:11039–11049. doi: 10.1074/jbc.M510258200. [DOI] [PubMed] [Google Scholar]
- Cha PC, Zembutsu H, Takahashi A, Kubo M, Kamatani N, Nakamura Y. A genome-wide association study identifies SNP in DCC is associated with gallbladder cancer in the Japanese population. J Hum Genet. 2012;57:235–237. doi: 10.1038/jhg.2012.9. [DOI] [PubMed] [Google Scholar]
- Chandra R, Liddle RA. Cholecystokinin. Curr Opin Endocrinol Diabetes Obes. 2007;14:63–67. doi: 10.1097/MED.0b013e3280122850. [DOI] [PubMed] [Google Scholar]
- Cheng J, Shah YM, Gonzalez FJ. Pregnane X receptor as a target for treatment of inflammatory bowel disorders. Trends Pharmacol Sci. 2012;33:323–330. doi: 10.1016/j.tips.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, Moschetta A, Bookout AL, Peng L, Umetani M, Holmstrom SR, Suino-Powell K, Xu HE, Richardson JA, Gerard RD, et al. Identification of a hormonal basis for gallbladder filling. Nat Med. 2006;12:1253–1255. doi: 10.1038/nm1501. [DOI] [PubMed] [Google Scholar]
- Chong HK, Infante AM, Seo YK, Jeon TI, Zhang Y, Edwards PA, Xie X, Osborne TF. Genome-wide interrogation of hepatic FXR reveals an asymmetric IR-1 motif and synergy with LRH-1. Nucleic Acids Res. 2010;38:6007–6017. doi: 10.1093/nar/gkq397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson PA. Bile Secretion and the Enterohepatic Circulation. Sleisenger and Fordtran’s gastrointestinal and liver disease: pathophysiology/diagnosis/management. 2010:1075–1088. [Google Scholar]
- Dawson PA, Haywood J, Craddock AL, Wilson M, Tietjen M, Kluckman K, Maeda N, Parks JS. Targeted deletion of the ileal bile acid transporter eliminates enterohepatic cycling of bile acids in mice. J Biol Chem. 2003;278:33920–33927. doi: 10.1074/jbc.M306370200. [DOI] [PubMed] [Google Scholar]
- Dawson PA, Hubbert ML, Rao A. Getting the mOST from OST: Role of organic solute transporter, OSTalpha-OSTbeta, in bile acid and steroid metabolism. Biochim Biophys Acta. 2010;1801:994–1004. doi: 10.1016/j.bbalip.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denson LA, Sturm E, Echevarria W, Zimmerman TL, Makishima M, Mangelsdorf DJ, Karpen SJ. The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp. Gastroenterology. 2001;121:140–147. doi: 10.1053/gast.2001.25503. [DOI] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duane WC, Javitt NB. 27-hydroxycholesterol: production rates in normal human subjects. J Lipid Res. 1999;40:1194–1199. [PubMed] [Google Scholar]
- Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687–693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- Galman C, Angelin B, Rudling M. Pronounced variation in bile acid synthesis in humans is related to gender, hypertriglyceridaemia and circulating levels of fibroblast growth factor 19. J Intern Med. 2011;270:580–588. doi: 10.1111/j.1365-2796.2011.02466.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Barcelo MM, Yeung MY, Miao XP, Tang CS, Cheng G, So MT, Ngan ES, Lui VC, Chen Y, Liu XL, et al. Genome-wide association study identifies a susceptibility locus for biliary atresia on 10q24.2. Hum Mol Genet. 2010;19:2917–2925. doi: 10.1093/hmg/ddq196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf GA, Yu L, Li WP, Gerard RD, Tuma PL, Cohen JC, Hobbs H. ABCG5 and ABCG8 are obligate heterodimers for protein trafficking and biliary cholesterol excretion. J Biol Chem. 2003;278:48275–48282. doi: 10.1074/jbc.M310223200. [DOI] [PubMed] [Google Scholar]
- Harach T, Pols TW, Nomura M, Maida A, Watanabe M, Auwerx J, Schoonjans K. TGR5 potentiates GLP-1 secretion in response to anionic exchange resins. Sci Rep. 2012;2:430. doi: 10.1038/srep00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heubi JE, Setchell KD, Bove KE. Inborn errors of bile acid metabolism. Seminars in Liver Disease. 2007;27:282–294. doi: 10.1055/s-2007-985073. [DOI] [PubMed] [Google Scholar]
- Higashiyama H, Kinoshita M, Asano S. Immunolocalization of farnesoid X receptor (FXR) in mouse tissues using tissue microarray. Acta histochem. 2008;110:86–93. doi: 10.1016/j.acthis.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Hofmann AF. Comprehensive Physiology. John Wiley & Sons, Inc; 2010. Enterohepatic Circulation of Bile Acids; pp. 567–596. [Google Scholar]
- Holt JA, Luo G, Billin AN, Bisi J, McNeill YY, Kozarsky KF, Donahee M, Wang DY, Mansfield TA, Kliewer SA, et al. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17:1581–1591. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Iser JH, Sali A. Chenodeoxycholic acid: a review of its pharmacological properties and therapeutic use. Drugs. 1981;21:90–119. doi: 10.2165/00003495-198121020-00002. [DOI] [PubMed] [Google Scholar]
- Ishibashi S, Schwarz M, Frykman PK, Herz J, Russell DW. Disruption of cholesterol 7alpha-hydroxylase gene in mice. I Postnatal lethality reversed by bile acid and vitamin supplementation. J Biol Chem. 1996;271:18017–18023. doi: 10.1074/jbc.271.30.18017. [DOI] [PubMed] [Google Scholar]
- Ito S, Fujimori T, Furuya A, Satoh J, Nabeshima Y. Impaired negative feedback suppression of bile acid synthesis in mice lacking betaKlotho. J Clin Invest. 2005;115:2202–2208. doi: 10.1172/JCI23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA. Physiology of FGF15/19. Adv Exp Med Biol. 2012;728:171–182. doi: 10.1007/978-1-4614-0887-1_11. [DOI] [PubMed] [Google Scholar]
- Kalaany NY, Mangelsdorf DJ. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol. 2006;68:159–191. doi: 10.1146/annurev.physiol.68.033104.152158. [DOI] [PubMed] [Google Scholar]
- Katsuma S, Hirasawa A, Tsujimoto G. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem Biophys Res Commun. 2005;329:386–390. doi: 10.1016/j.bbrc.2005.01.139. [DOI] [PubMed] [Google Scholar]
- Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- Keitel V, Cupisti K, Ullmer C, Knoefel WT, Kubitz R, Haussinger D. The membrane-bound bile acid receptor TGR5 is localized in the epithelium of human gallbladders. Hepatology. 2009;50:861–870. doi: 10.1002/hep.23032. [DOI] [PubMed] [Google Scholar]
- Keitel V, Donner M, Winandy S, Kubitz R, Haussinger D. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem Biophys Res Commun. 2008;372:78–84. doi: 10.1016/j.bbrc.2008.04.171. [DOI] [PubMed] [Google Scholar]
- Keitel V, Gorg B, Bidmon HJ, Zemtsova I, Spomer L, Zilles K, Haussinger D. The bile acid receptor TGR5 (Gpbar-1) acts as a neurosteroid receptor in brain. Glia. 2010;58:1794–1805. doi: 10.1002/glia.21049. [DOI] [PubMed] [Google Scholar]
- Keitel V, Haussinger D. TGR5 in the biliary tree. Dig Dis. 2011;29:45–47. doi: 10.1159/000324127. [DOI] [PubMed] [Google Scholar]
- Keitel V, Haussinger D. Perspective: TGR5 (Gpbar-1) in liver physiology and disease. Clin Res Hepatol Gastroenterol. 2012;36:412–419. doi: 10.1016/j.clinre.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Kerr TA, Saeki S, Schneider M, Schaefer K, Berdy S, Redder T, Shan B, Russell DW, Schwarz M. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev Cell. 2002;2:713–720. doi: 10.1016/s1534-5807(02)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kir S, Beddow SA, Samuel VT, Miller P, Previs SF, Suino-Powell K, Xu HE, Shulman GI, Kliewer SA, Mangelsdorf DJ. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science. 2011;331:1621–1624. doi: 10.1126/science.1198363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong B, Wang L, Chiang JY, Zhang Y, Klaassen CD, Guo GL. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology. 2012;56:1034–1043. doi: 10.1002/hep.25740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B, Balemba OB, Godfrey C, Watson CA, Vassileva G, Corvera CU, Nelson MT, Mawe GM. Hydrophobic bile salts inhibit gallbladder smooth muscle function via stimulation of GPBAR1 receptors and activation of KATP channels. J Physiol. 2010;588:3295–3305. doi: 10.1113/jphysiol.2010.192146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FY, de Aguiar Vallim TQ, Chong HK, Zhang Y, Liu Y, Jones SA, Osborne TF, Edwards PA. Activation of the farnesoid X receptor provides protection against acetaminophen-induced hepatic toxicity. Mol Endocrinol. 2010;24:1626–1636. doi: 10.1210/me.2010-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Zhang Y, Lee FY, Nelson SF, Gonzalez FJ, Edwards PA. FXR regulates organic solute transporters alpha and beta in the adrenal gland, kidney, and intestine. J Lipid Res. 2006;47:201–214. doi: 10.1194/jlr.M500417-JLR200. [DOI] [PubMed] [Google Scholar]
- Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- Li T, Holmstrom SR, Kir S, Umetani M, Schmidt DR, Kliewer SA, Mangelsdorf DJ. The G protein-coupled bile acid receptor, TGR5, stimulates gallbladder filling. Mol Endocrinol. 2011;25:1066–1071. doi: 10.1210/me.2010-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Invernizzi P, Lu Y, Kosoy R, Lu Y, Bianchi I, Podda M, Xu C, Xie G, Macciardi F, et al. Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nat Genet. 2010;42:658–660. doi: 10.1038/ng.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Binz J, Numerick MJ, Dennis S, Luo G, Desai B, MacKenzie KI, Mansfield TA, Kliewer SA, Goodwin B, et al. Hepatoprotection by the farnesoid X receptor agonist GW4064 in rat models of intra- and extrahepatic cholestasis. J Clin Invest. 2003;112:1678–1687. doi: 10.1172/JCI18945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundasen T, Galman C, Angelin B, Rudling M. Circulating intestinal fibroblast growth factor 19 has a pronounced diurnal variation and modulates hepatic bile acid synthesis in man. J Intern Med. 2006;260:530–536. doi: 10.1111/j.1365-2796.2006.01731.x. [DOI] [PubMed] [Google Scholar]
- Ma K, Saha PK, Chan L, Moore DD. Farnesoid X receptor is essential for normal glucose homeostasis. J Clin Invest. 2006;116:1102–1109. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, Sugiyama E, Itadani H, Tanaka K. Identification of membrane-type receptor for bile acids (M-BAR) Biochem Biophys Res Commun. 2002;298:714–719. doi: 10.1016/s0006-291x(02)02550-0. [DOI] [PubMed] [Google Scholar]
- Maruyama T, Tanaka K, Suzuki J, Miyoshi H, Harada N, Nakamura T, Miyamoto Y, Kanatani A, Tamai Y. Targeted disruption of G protein-coupled bile acid receptor 1 (Gpbar1/M-Bar) in mice. J Endocrinol. 2006;191:197–205. doi: 10.1677/joe.1.06546. [DOI] [PubMed] [Google Scholar]
- Matsubara T, Li F, Gonzalez FJ. Mol Cell Endocrinol. 2012. FXR signaling in the enterohepatic system. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mells GF, Floyd JA, Morley KI, Cordell HJ, Franklin CS, Shin SY, Heneghan MA, Neuberger JM, Donaldson PT, Day DB, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2011;43:329–332. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Nishida N, Kawashima M, Aiba Y, Tanaka A, Yasunami M, Nakamura H, Komori A, Nakamuta M, Zeniya M, et al. Genome-wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am J Hum Genet. 2012;91:721–728. doi: 10.1016/j.ajhg.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A, Bouscarel B. Bile acids and signal transduction: role in glucose homeostasis. Cell Signal. 2008;20:2180–2197. doi: 10.1016/j.cellsig.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Nicolaou M, Andress EJ, Zolnerciks JK, Dixon PH, Williamson C, Linton KJ. Canalicular ABC transporters and liver disease. J Pathol. 2012;226:300–315. doi: 10.1002/path.3019. [DOI] [PubMed] [Google Scholar]
- Oelkers P, Kirby LC, Heubi JE, Dawson PA. Primary bile acid malabsorption caused by mutations in the ileal sodium-dependent bile acid transporter gene (SLC10A2) J Clin Invest. 1997;99:1880–1887. doi: 10.1172/JCI119355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai R, French D, Ma N, Hotzel K, Plise E, Salphati L, Setchell KD, Ware J, Lauriault V, Schutt L, et al. Antibody-mediated inhibition of fibroblast growth factor 19 results in increased bile acids synthesis and ileal malabsorption of bile acids in cynomolgus monkeys. Toxicol Sci. 2012;126:446–456. doi: 10.1093/toxsci/kfs011. [DOI] [PubMed] [Google Scholar]
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, et al. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- Pols TW, Nomura M, Harach T, Lo Sasso G, Oosterveer MH, Thomas C, Rizzo G, Gioiello A, Adorini L, Pellicciari R, et al. TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab. 2011a;14:747–757. doi: 10.1016/j.cmet.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols TW, Noriega LG, Nomura M, Auwerx J, Schoonjans K. The bile acid membrane receptor TGR5: a valuable metabolic target. Dig Dis. 2011b;29:37–44. doi: 10.1159/000324126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porez G, Prawitt J, Gross B, Staels B. Bile acid receptors as targets for the treatment of dyslipidemia and cardiovascular disease: Thematic Review Series: New Lipid and Lipoprotein Targets for the Treatment of Cardiometabolic Diseases. J Lipid Res. 2012a;53:1723–1737. doi: 10.1194/jlr.R024794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porez G, Prawitt J, Gross B, Staels B. Bile acid receptors as targets for the treatment of dyslipidemia and cardiovascular disease: Thematic Review Series: New Lipid and Lipoprotein Targets for the Treatment of Cardiometabolic Diseases. J Lipid Res. 2012b;53:1723–1737. doi: 10.1194/jlr.R024794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, Boney-Montoya J, Choi M, He T, Sunny NE, Satapati S, Suino-Powell K, Xu HE, Gerard RD, Finck BN, et al. FGF15/19 regulates hepatic glucose metabolism by inhibiting the CREB-PGC-1alpha pathway. Cell Metab. 2011;13:729–738. doi: 10.1016/j.cmet.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, Kliewer SA, Mangelsdorf DJ. Endocrine fibroblast growth factors 15/19 and 21: from feast to famine. Genes Dev. 2012;26:312–324. doi: 10.1101/gad.184788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A, Haywood J, Craddock AL, Belinsky MG, Kruh GD, Dawson PA. The organic solute transporter alpha-beta, Ostalpha-Ostbeta, is essential for intestinal bile acid transport and homeostasis. Proc Natl Acad Sci USA. 2008;105:3891–3896. doi: 10.1073/pnas.0712328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- Russell DW. Fifty years of advances in bile acid synthesis and metabolism. J Lipid Res. 2009;50(Suppl):S120–125. doi: 10.1194/jlr.R800026-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruutu T, Eriksson B, Remes K, Juvonen E, Volin L, Remberger M, Parkkali T, Hagglund H, Ringden O. Ursodeoxycholic acid for the prevention of hepatic complications in allogeneic stem cell transplantation. Blood. 2002;100:1977–1983. doi: 10.1182/blood-2001-12-0159. [DOI] [PubMed] [Google Scholar]
- Sato H, Genet C, Strehle A, Thomas C, Lobstein A, Wagner A, Mioskowski C, Auwerx J, Saladin R. Anti-hyperglycemic activity of a TGR5 agonist isolated from Olea europaea. Biochem Biophys Res Commun. 2007;362:793–798. doi: 10.1016/j.bbrc.2007.06.130. [DOI] [PubMed] [Google Scholar]
- Sato H, Macchiarulo A, Thomas C, Gioiello A, Une M, Hofmann AF, Saladin R, Schoonjans K, Pellicciari R, Auwerx J. Novel potent and selective bile acid derivatives as TGR5 agonists: biological screening, structure-activity relationships, and molecular modeling studies. J Med Chem. 2008;51:1831–1841. doi: 10.1021/jm7015864. [DOI] [PubMed] [Google Scholar]
- Schaap FG. Role of fibroblast growth factor 19 in the control of glucose homeostasis. Curr Opin Clin Nutr Metab Care. 2012;15:386–391. doi: 10.1097/MCO.0b013e3283547171. [DOI] [PubMed] [Google Scholar]
- Schaap FG, van der Gaag NA, Gouma DJ, Jansen PL. High expression of the bile salt-homeostatic hormone fibroblast growth factor 19 in the liver of patients with extrahepatic cholestasis. Hepatology. 2009;49:1228–1235. doi: 10.1002/hep.22771. [DOI] [PubMed] [Google Scholar]
- Schmidt DR, Holmstrom SR, Fon Tacer K, Bookout AL, Kliewer SA, Mangelsdorf DJ. Regulation of bile acid synthesis by fat-soluble vitamins A and D. J Biol Chem. 2010;285:14486–14494. doi: 10.1074/jbc.M110.116004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz M, Lund EG, Setchell KD, Kayden HJ, Zerwekh JE, Bjorkhem I, Herz J, Russell DW. Disruption of cholesterol 7alpha-hydroxylase gene in mice. II Bile acid deficiency is overcome by induction of oxysterol 7alpha-hydroxylase. J Biol Chem. 1996;271:18024–18031. doi: 10.1074/jbc.271.30.18024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz M, Russell DW, Dietschy JM, Turley SD. Alternate pathways of bile acid synthesis in the cholesterol 7alpha-hydroxylase knockout mouse are not upregulated by either cholesterol or cholestyramine feeding. J Lipid Res. 2001;42:1594–1603. [PubMed] [Google Scholar]
- Selwyn F, Klaassen CD. Characterization of Bile Acid homeostasis in Germ-free Mice. FASEB J. 2012;26:1155.1. [Google Scholar]
- Seol W, Choi HS, Moore DD. Isolation of proteins that interact specifically with the retinoid X receptor: two novel orphan receptors. Mol Endocrinol. 1995;9:72–85. doi: 10.1210/mend.9.1.7760852. [DOI] [PubMed] [Google Scholar]
- Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- Sonoda J, Pei L, Evans RM. Nuclear receptors: decoding metabolic disease. FEBS Lett. 2008;582:2–9. doi: 10.1016/j.febslet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staels B, Handelsman Y, Fonseca V. Bile Acid Sequestrants for Lipid and Glucose Control. Current Diabetes Reports. 2010;10:70–77. doi: 10.1007/s11892-009-0087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci USA. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer E, Zhou X, Zhao R, Wang Y, Takabe K, Nagahashi M, Pandak WM, Dent P, Spiegel S, Shi R, et al. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology. 2012;55:267–276. doi: 10.1002/hep.24681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung JY, Costerton JW, Shaffer EA. Defense system in the biliary tract against bacterial infection. Dig Dis Sci. 1992;37:689–696. doi: 10.1007/BF01296423. [DOI] [PubMed] [Google Scholar]
- Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, Nicholson JK, Holmes E. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AM, Hart SN, Kong B, Fang J, Zhong XB, Guo GL. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology. 2010;51:1410–1419. doi: 10.1002/hep.23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- Trottier J, Bialek A, Caron P, Straka RJ, Milkiewicz P, Barbier O. Profiling circulating and urinary bile acids in patients with biliary obstruction before and after biliary stenting. PLoS One. 2011;6:e22094. doi: 10.1371/journal.pone.0022094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassileva G, Golovko A, Markowitz L, Abbondanzo SJ, Zeng M, Yang S, Hoos L, Tetzloff G, Levitan D, Murgolo NJ, et al. Targeted deletion of Gpbar1 protects mice from cholesterol gallstone formation. Biochem J. 2006;398:423–430. doi: 10.1042/BJ20060537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DQ, Cohen DE, Carey MC. Biliary lipids and cholesterol gallstone disease. J Lipid Res. 2009a;50(Suppl):S406–411. doi: 10.1194/jlr.R800075-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- Wang L, Lee YK, Bundman D, Han Y, Thevananther S, Kim CS, Chua SS, Wei P, Heyman RA, Karin M, et al. Redundant pathways for negative feedback regulation of bile acid production. Dev Cell. 2002;2:721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- Wang XX, Jiang T, Shen Y, Adorini L, Pruzanski M, Gonzalez FJ, Scherzer P, Lewis L, Miyazaki-Anzai S, Levi M. The farnesoid X receptor modulates renal lipid metabolism and diet-induced renal inflammation, fibrosis, and proteinuria. Am J Physiol Renal Physiol. 2009b;297:F1587–1596. doi: 10.1152/ajprenal.00404.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YD, Chen WD, Yu D, Forman BM, Huang W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor kappa light-chain enhancer of activated B cells (NF-kappaB) in mice. Hepatology. 2011;54:1421–1432. doi: 10.1002/hep.24525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, Moore DD, Auwerx J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland HO. Nobel Lectures, Chemistry 1922–1941. Amsterdam: 1966. Nobel Prize Lecture (1928): The Chemistry of the Bile Acids. [Google Scholar]
- Yu C, Wang F, Kan M, Jin C, Jones RB, Weinstein M, Deng CX, McKeehan WL. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J Biol Chem. 2000;275:15482–15489. doi: 10.1074/jbc.275.20.15482. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Edwards PA. FXR signaling in metabolic disease. FEBS Lett. 2008;582:10–18. doi: 10.1016/j.febslet.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hagedorn CH, Wang L. Role of nuclear receptor SHP in metabolism and cancer. Biochim Biophys Acta. 2011a;1812:893–908. doi: 10.1016/j.bbadis.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hong JY, Rockwell CE, Copple BL, Jaeschke H, Klaassen CD. Effect of bile duct ligation on bile acid composition in mouse serum and liver. Liver Int. 2012;32:58–69. doi: 10.1111/j.1478-3231.2011.02662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci USA. 2006;103:1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YK, Guo GL, Klaassen CD. Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PLoS One. 2011b;6:e16683. doi: 10.1371/journal.pone.0016683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollner G, Trauner M. Molecular mechanisms of cholestasis. Wiener Medizinische Wochenschrift. 2006;156:380–385. doi: 10.1007/s10354-006-0312-7. [DOI] [PubMed] [Google Scholar]