Abstract
Antibody-producing plasma cells are a major source of protective immunity in vertebrates, including salmon. During the spawning phase, salmon undergo drastic, hormonally driven changes in their physiology, including elevated levels of cortisol, which are known to suppress the immune system. As adult fish need to survive their long journey to the spawning grounds, we hypothesized that humoral immunity, in the form of IgM-secreting plasma cells, remains functional until post-spawning. This was investigated by measuring changes in membrane and secreted immunoglobulin heavy chain mu and Pax5 transcripts in spleen and kidney from migrating sockeye salmon, using real-time qPCR. As an additional measurement, the abundance of developing B, mature B, and plasma cells was determined in spawning fish, using flow cytometry. Immune tissue samples were collected from fish from the Kenai River drainage and Main Bay, Prince William Sound. Our results reveal that spawning fish express high levels of secreted heavy chain mu transcripts in their spleen and anterior kidney throughout the spawning journey. Furthermore, we show that IgM-secreting PCs (HCmu++/Pax5−) remain abundant in anterior kidney and spleen of post-spawning sockeye salmon, with a concomitant loss in developing B cells (HCmu−/Pax5+). This suggests that successful spawners retain their PCs throughout the spawning journey and post-spawning.
INTRODUCTION
Anadromous species of salmon, including the sockeye salmon (Oncorhynchus nerka: O.nerka), die after spawning. During their journey from rearing site to the ocean, and again, upon their return from the ocean to the spawning site 2 to 5 years later, salmonids make high levels of cortisol, which have immunosuppressive effects. However, the salmon ‘s ability to survive the spawning journey, despite increasing pathogen load, suggests that their immune system is not completely suppressed.
Fish within the Oncorhynchus genus, including the most commonly studied rainbow trout (Oncorhynchus mykiss; O.mykiss) and the less studied sockeye salmon (O.nerka), possess T and B-lymphocytes similar to those in mammalian species, but lack bone marrow and lymph nodes. Hematopoiesis takes place in the anterior kidney, which is functionally analogous to mammalian bone marrow (Hansen and Zapata 1998, Zwollo et al, 2010). We, and others have proposed that once B cells have matured in the anterior kidney, such cells migrate to the spleen (and possibly the posterior kidney) and are primarily activated there (Zwollo et al 2005, 2008, 2010, Bromage et al 2004). The spleen appears to be the main site where activated B cells are induced to proliferate (the plasmablast stage), followed by differentiation into transitional (T-PC) and/or mature (M-PC) plasma cells (Barr et al, 2011). There is evidence that a subset of these immunoglobulin-secreting PCs home to the anterior kidney where they become long-lived plasma cells (LLPCs), as occurs in the bone marrow of mammalian species, constitutively secreting antibody independent of the presence of antigen for months, or possibly, years (Bromage et al, 2004, Kaattari et al, 2005, Ye et al, 2011, Ma, et al. 2013). Recently, we hypothesized that LLPCs play crucial roles in immune protection in spawning salmon (Zwollo, 2012).
To begin investigating patterns of humoral immune protection during back-migration to natal grounds, we measured expression of immunoglobulin heavy chain mu (HCmu) and Pax5 transcripts in immune tissues from spawning sockeye salmon, Expression patterns were investigated at three different locations during the spawning journey. Furthermore, abundance of B cell subsets was determined by flow cytometric analysis. Our data show that HCmu is not strongly expressed until the mature B cell stage, while its expression increases dramatically during terminal B cell differentiation. The transcription factor Pax5 is first expressed during early B cell development, remains expressed through the plasmablast and T-PC stages, but is completely downregulated in mature plasma cells (M-PC) (Zwollo et al, 2010, Barr et al, 2011). Here we show that HCmu++/Pax5-, PCs remain abundant in anterior kidney and spleen of post-spawning sockeye salmon, with a concomitant loss in HCmu−/Pax5+, developing B cells. Hence, successful spawners retain their PCs throughout the spawning journey and post-spawning.
MATERIALS AND METHODS
Tissue Collection and Storage
Outbred adult rainbow trout (20-30 cm; The Virginia Institute of Marine Science/The College of William and Mary Trout Facility) were used as controls. Immune tissues from wild adult sockeye salmon (O. nerka) were collected from sites in South Central Alaska, including Main Bay, Prince William Sound (Latitude/Longitude: 60.31170/-148.05300; hatchery fish; collected June 2011, 2012), Mouth of the Kenai River (Latitude/Longitude: 60.32580/-151.15560; wild fish; collected July 2011, 2012), confluence of the Moose and Kenai Rivers, named here Mouth of the Moose River (Latitude/Longitude: 60.32070/-150.45230; wild fish; collected July 2012), and Quartz Creek-Upper Kenai River, (Latitude/Longitude: 60.29380/-129.41450; wild fish; August 2011, 2012) as shown in Figure 1. Juvenile sockeye salmon (6-10 cm; collected April/May 2011, 2012) were raised at the Trail Lake Hatchery (TLH), near Moose Pass. Live fish at Mouth of the Kenai, Mouth of Moose River, and Main Bay were donated by recreational fishermen, and processed on site. For all fish, euthanasia was applied immediately by cerebral concussion. Tissues from pre-and post-spawning fish at Quartz Creek were collected and processed on site immediately, as above. TLH fish were processed at the hatchery site, and euthanized using MS-222. Tissue samples were collected in accordance to the rules and regulations of the Alaska Department of Fish and Game and fish resource permits (ADFG2011-277, ADFG2012-258) were obtained prior to collection. Approximately equal numbers of males and females were collected at each site, except for the Mouth of the Kenai, at which site more males (5) than females (2) were collected. For RNA experiments, 6-10 cubic mm tissue from spleen, anterior kidney or posterior kidney were collected into 750 μL of RNAlater reagent (Applied Biosystems Inc.) and stored overnight at 4C, then frozen at −20C until use. For flow cytometric analysis, freshly isolated cell suspensions were immediately fixed in paraformaldehyde on site, as described below.
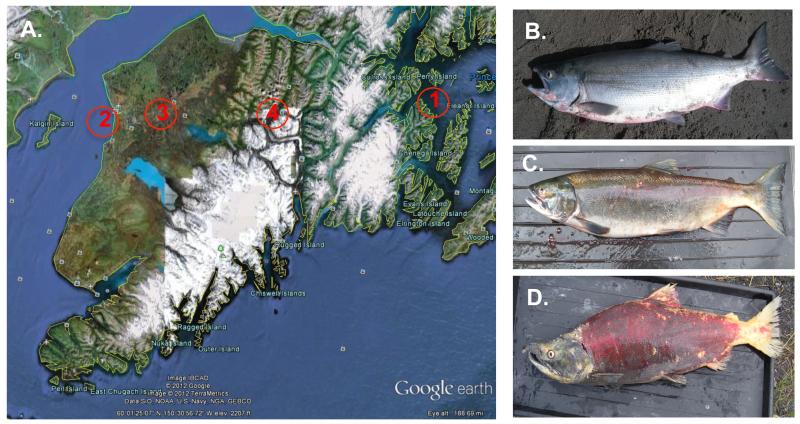
Figure 1.
Sampling sites (A) and phenotypic changes in sockeye salmon during the spawning journey (B-D). A. Locations of sampling sites at the Kenai Peninsula and Prince William Sound; red numbers: 1: Main Bay, Prince William Sound; 2: Mouth of the Kenai River; 3: Confluence of Kenai and Mouth of Moose River; 4: Quartz Creek. B-D. Photographs illustrating dramatic phenotypic changes in sockeye salmon during the spawning journey. B. As fish enter the Kenai River. C. At spawning site, Quartz Creek, pre-spawning fish, and D. At spawning site Quartz Creek, post-spawning.
RNA Extraction
RNA was extracted using MagMAX-96 for Microarrays Kit (Ambion/Applied Biosystems, Inc.) and MagMAX 24Express instrument (Applied Biosystems, Inc). Each sample (6-10 cubic mm piece of tissue) was deposited in 1 mL of TRI Reagent in a Lysing Matrix D beads tube (Molecular Probes, Inc.) and then pulverized in BeadRuptor instrument (OMNI International): one 20 sec pulse at speed 5.5. Next, 50 μL of bromo-chloropropane (BCP) was added followed by a 15 second vortex. Samples were then centrifuged at 10,500rpm, 4C, 10 min and 100 ul of a clear upper layer was placed on MagMAX sample tray. All other MagMAX-96 kit components were placed on the tray according to the manufacturer protocol and RNA was isolated with the AM1389spin program on the MagMAX 24Express instrument. RNA concentration was determined using the NanoDrop™ ND-1000 full-spectrum (220nm-750nm) spectrophotometer (Thermo Fisher Scientific).
cDNA and Real-Time PCR
1μg of total RNA was used to make 20μL of cDNA using the iScript kit (BioRad). Quantitative real-time PCR was performed using a PerfeCta SYBR Green SuperMix, ROX PCR kit (Quanta Biosciences). Each sample was amplified in triplicate. For the detection of HCmu transcripts, a reaction mix containing 12.5 μL SYBR Green, 9.5 μL RNase free water, 1 μL Forward Primer (3 μM), 1 μL Reverse Primer 3 μM (Table 1), and 1 μL of 1:4 diluted cDNA (12.5ng) was used. For Pax5 detection, 1.50 μL cDNA (75ng) was used. Trout α- tubulin (Tubulin 60; Table 1) was used as an endogenous control for HCmu, and Tubulin 56 (Table I), for Pax5. Primers for Pax5 and HCmu are indicated in Table I. The reactions were performed on an Applied Biosystems StepOne Real-Time PCR instrument. Negative controls were included in each plate set-up where cDNA template was substituted by equal amount of nuclease free water. For secreted and membrane HCmu PCR, cDNA was denatured at 95.0C for 10min. After this, samples were cycled as follows: 95C for 15sec and 60C for 1min for 40 cycles in a two-step PCR reaction. For Pax5 detection, cDNA was denatured at 95C for 10min, then samples were cycled as follows: 95C for 15sec and 56C for 1min for 40 cycles in a two-step PCR reaction, using the Pax5 primers in Table I. Melt curve analysis was performed for both cycling conditions to verify the specificity of primers and lack of primer-dimer artifacts. Specificity of the primer pairs was further confirmed by sequencing real-time PCR fragments using capillary electrophoresis on ABI 3130 Avant Genetic Analyzer. Sequences were analyzed on ABI sequencing software version 5.1. An NCBI Blast tool was used to align obtained sequences with the published Oncorhynchus sp. sequence data.
Table 1.
PCR-Primer Information
| Primer Name | Primer Target | Primer Sequence |
|---|---|---|
| qtHCmu.F | Secreted and Membrane IgM (60C annealing) |
5′-GCG CTG TAG ATC ACA TGG AA-3′ |
| qt.HCmu.sec.R | Secreted IgM (60C annealing) |
5′-GCA AGT CAG GGT CAC CGT AT-3′ |
| qtHCmu.mem.R | Membrane IgM (60C annealing) |
5′-TTT CAC CTT GAT GGC AGT TG-3 |
| Ttub60.F | alpha-Tubulin (60C Annealing) |
5′-CGT CCC CAG GTG TCC ACT-3′ |
| Ttub60.R | alpha-Tubulin (60C Annealing) |
5′-GTA GGT GGG GCG CTC AAT-3′ |
| tPax5E2.F | Pax5 (56C annealing) | 5′-CAC GGA TGT GTC AGC AAG AT-3′ |
| tPax5/E3-4.R | Pax5 (56C annealing) | 5′-CCT GAT GAT CCT GTT GAT AGA-3′ |
| Ttub56.F | alpha-Tubulin (56C annealing) |
5′-TTC ACC TCC CTG CTG ATG-3′ |
| Ttub56.R | alpha-Tubulin (56C annealing) |
5′-CCA CCA TGA AGG CAC AGT-3′ |
Analysis of Quantitative Real-Time PCR
CT values were uploaded into DataAssist Software (ABI). Expression levels were calculated as fold-change relative to the reference sample (N=5; averaged O. mykiss CT) for the O. nerka samples. Expression of individual genes from each sample was normalized to relative expression of trout α-tubulin within the same experiment. The fold change, or amount of target, was calculated according to the Fold Change = 2−ΔΔCT equation (Livak and Schmittgen, 2001). Samples with fold-change values that were more than 3-fold different from the average value, were excluded, which excluded <1% of the samples.
Antibodies
The polyclonal rabbit anti-Pax5 antibody (ED-1) has been described previously [Zwollo et al, 1998]. The mouse-anti-trout IgM (I-14) monoclonal antibody recognizes both membrane and secreted forms of Oncorhynchus HCmu [DeLuca et al, 1983]. For flow cytometric analyses, antibodies were conjugated to Alexa Fluor 555 or Alexa Fluor 647 using protein-labeling kits according to manufacturer’s instructions (Molecular Probes). Isotype control antibodies included rabbit IgG (Molecular probes) or mouse IgG (eBiosciences) conjugated to Alexa 555 or Alexa 647. Antibody aliquots were stored in 1% BSA at −20C.
Fixation, permeabilization, staining, and flow cytometry
Tissues from anterior kidney, posterior kidney and spleen tissue were collected in RPMI-1640 and cell suspensions made. Cells were then washed in PBS plus 0.02% sodium azide (PBS-SA) and fixed and permeabilized as described previously [Zwollo et al, 2008]. The next day, cells were refixed in 1% paraformaldehyde for 10 minutes, and stored in fetal bovine serum containing 10% dimethyl sulfoxide (DMSO) at −80C until analysis. For flow cytometric analysis, fixed and permeabilized cells were stained at a concentration of 107 cells/ml with final antibody concentration of 0.5-2 μg/50,000 cells/50 μl, and analyzed as described previously (Zwollo et al, 2008). 20,000-30,000 events were acquired per sample using a BD FACSArray (BD Biosciences). Duplicate samples were analyzed for each experiment. Experiments were repeated a minimum of three times. Contour graphs were generated using WinMDI 2-8 (J.Trotter 1993-1998) software, and are shown as log algorithms with intervals of 50%. Means and standard errors were calculated for each experiment.
RESULTS
First, we investigated possible changes in membrane and secreted HCmu transcripts of adult sockeye salmon during the spawning journey, using qPCR. Three immune tissues were analyzed, namely anterior kidney, spleen, and posterior kidney. As reference sample, we used the average value of 5 independent O.mykiss samples for each tissue. Juvenile, pre-smolting O.nerka were used as (non-spawning) controls. Figure 1B-D illustrates the severe phenotypic changes that fish undergo during their spawning journey as they enter Mouth of the Kenai (Figure 1B), to pre-spawning at Quartz Creek (Figure 1C), to post-spawning at the same site (Figure 1D).
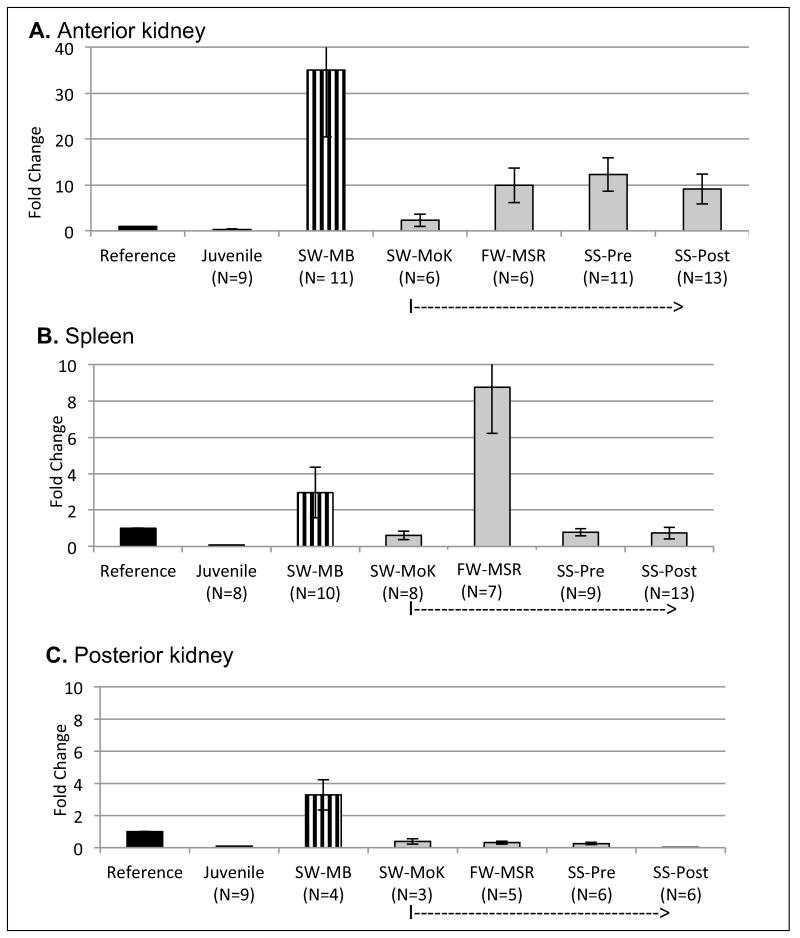
Figure 2 shows the result of qPCR analysis using the secreted HCmu primers. The anterior kidney is the main site for B lymphopoiesis in fish, but also houses IgM-secreting (LL)PCs. Of all groups, juvenile O. nerka expressed the lowest levels of secreted HCmu in anterior kidney (Figure 2A). A site-dependent increase in secreted HCmu transcripts was observed in migrating fish from the Kenai run, being lowest in saltwater pre-spawning fish (mouth of the Kenai; SW-MoK), increasing in freshwater pre-spawning fish (mouth of Moose River; FW-MSR), and in pre-spawning fish at Quartz Creek (SS-Pre). Pre-spawning fish at Quartz Creek had the highest average levels of secreted HCmu transcripts, with a drop in post-spawned fish at the same site (SS-Post; Figure 2A). Interestingly, pre-spawning fish from a different run, collected at Main Bay in Prince William Sound (SW-MB; Figure 1A), had the highest levels of secreted HCmu in their anterior kidney of all groups.
Figure 2.
Results from qPCR to determine relative RNA levels of secreted HCmu, indicated in fold-change on the Y-axis relative to the reference sample. Mean fold-change values with SE. N values indicated underneath each group on X-axis. Reference sample (in black) was mature, non-spawning O. mykiss. All other groups are O. nerka. SW-MB: salt-water, pre-spawning, at Main Bay (in vertical stripe). Juvenile (white): pre-smolts from Trail Lake Hatchery. Kenai run: in grey, note a dotted arrow on X-axis. SW-MoK: salt-water, pre-spawning, at Mouth of the Kenai; FW-MSR: fresh-water, pre-spawning, at confluence of Mouth of the Kenai and Moose River; SS-Pre: fresh-water, pre-spawning, at Quartz Creek; SS-Post: fresh-water, post-spawning, at Quartz Creek. A. Anterior kidney. B. Spleen. C. Posterior Kidney.
Secreted immunoglobulin patterns in spleens of Kenai-run fish had a different pattern (Figure 2B): levels did not change significantly in spawning fish during their fresh water phase, or compared to the reference sample, with the exception of fish at the freshwater site FW-MSR. Again, juveniles had the lowest levels of secreted HCmu of all groups. In posterior kidney, levels of secreted HCmu were lower than reference samples, in fish arriving at the Mouth of the Kenai, and levels decreased even further as fish moved upstream in fresh water (Figure 2C). Spleens of SW-MB fish again had high levels of secreted HCmu.
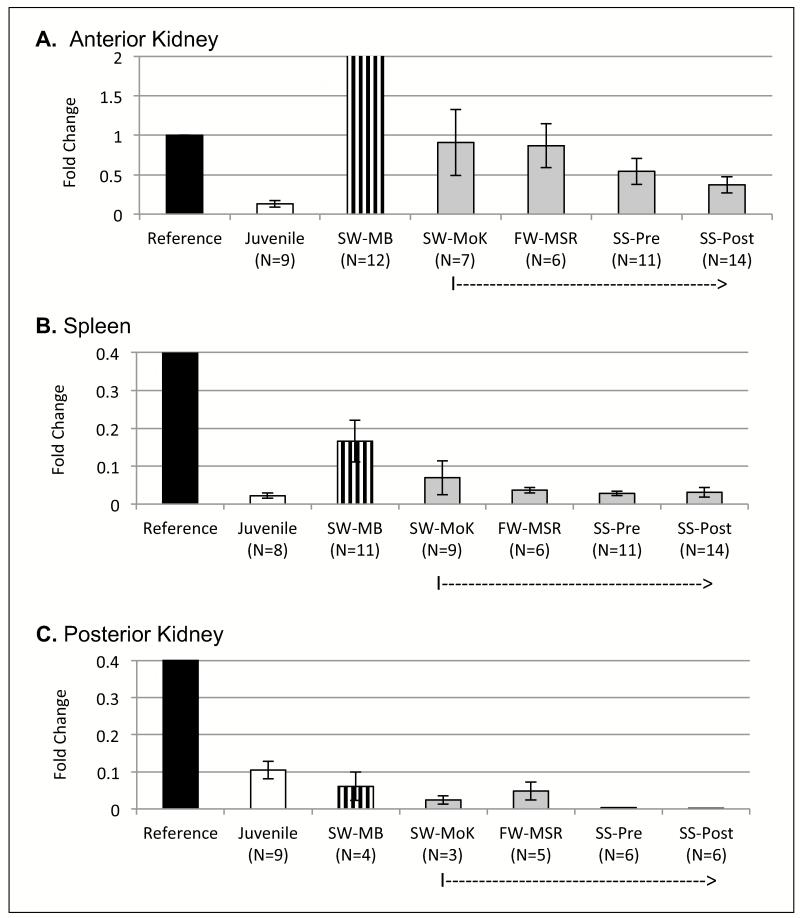
Next, the levels of membrane HCmu were determined (Figure 3). Juveniles expressed the lowest levels of membrane HCmu transcripts of all groups, for both spleen and anterior kidney, but not posterior kidney (Figure 3A-3C). For Kenai fish, there was a gradual reduction in levels of membrane IgM transcripts as fish moved from the Mouth of the Kenai towards their spawning site at Quartz Creek (from SW-MoK to SS-post) for all three tissues (Figures 3A-3C). Of note, juvenile fish made higher levels of membrane HCmu in the posterior kidney than adult, spawning fish did. Membrane HCmu levels in posterior kidneys of SW-MB fish were similar to those from Kenai fish.
Figure 3.
Results from qPCR to determine relative RNA levels of membrane HCmu, indicated in fold-change on the Y-axis relative to the reference sample. Mean fold-change values with SE. N values indicated underneath each group on X-axis. Reference sample (in black) was mature, non-spawning O. mykiss. All other groups are O. nerka. SW-MB: salt-water, pre-spawning, at Main Bay (in vertical stripe). Juvenile (white): pre-smolts from Trail Lake Hatchery. Kenai run: in grey, note a dotted arrow on X-axis. SW-MoK: salt-water, pre-spawning, at Mouth of the Kenai; FW-MSR: fresh-water, pre-spawning, at confluence of Mouth of the Kenai and Moose River; SS-Pre: fresh-water, pre-spawning, at Quartz Creek; SS-Post: fresh-water, post-spawning, at Quartz Creek. A. Anterior kidney. B. Spleen. C. Posterior Kidney.
Comparison of male and female qPCR patterns did not show significant differences in levels of secreted or membrane Hcmu in the spleens of spawning fish, with the exception of fish at their spawning site (Quartz Creek). In spleens of such fish, the secreted HCmu levels of males were higher than in females, both in pre-spawning and post-spawned fish (pre-spawning males: 2.3 fold [SE +/− 1.1] higher; post-spawned males: 4.6 times higher (SE +/− 1.7)]. Furthermore, both pre- and post-spawned males had higher levels of membrane HCmu transcripts in their spleens than females (5.4-fold [+/− 1.0] and 3.1-fold [+/− 1.8] respectively). For anterior kidney, the same pattern was present: with sex differences detected only for fish at the spawning site at Quartz Creek, and males expressing significantly higher levels of secreted Hcmu transcripts compared to females (9.8 [SE +/− 5.5] and 25.3 [SE +/− 9.6] -fold respectively). In the anterior kidney, levels of membrane HCmu were also higher in males collected at Quartz Creek, compared to females collected at this site, both in pre-spawners (5.8 (+/− 1.3) and post-spawners (2.6 (+/− 1.2). No gender-specific differences were detectable in posterior kidney at any site.
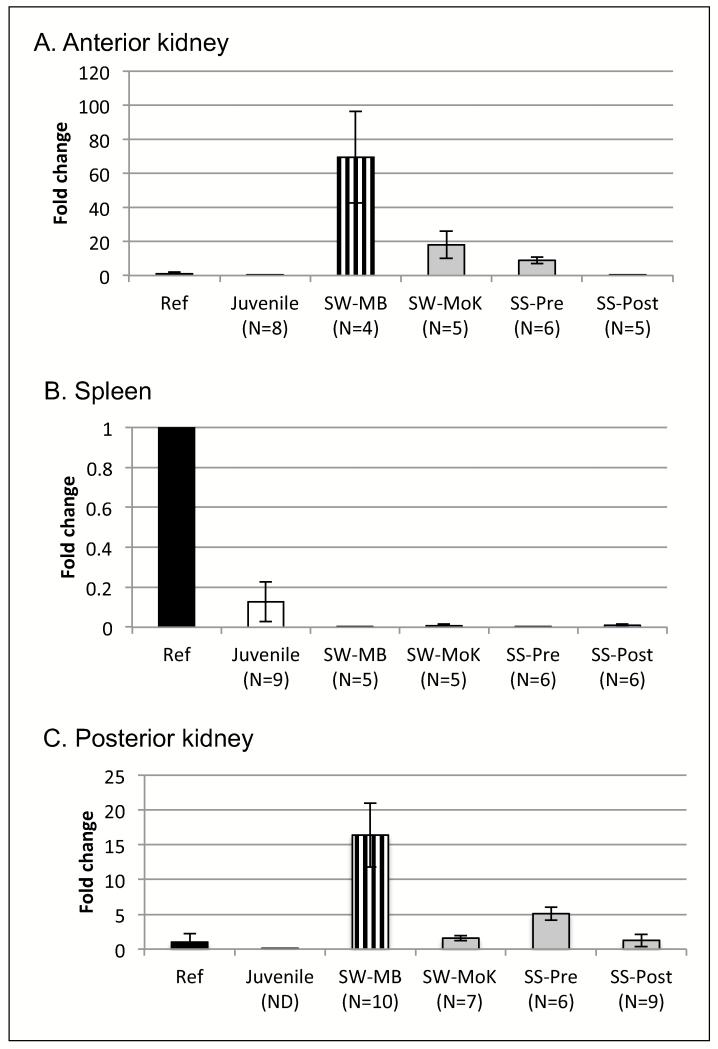
To further investigate changes in B cell populations during spawning, expression of Pax5 RNA was determined. This transcription factor is first expressed during the late pro-B cell stage and remains expressed throughout the plasmablast and T-PC stages, but is absent from M-PCs (Zwollo et al. 2008, 2010, 2011). Compared to reference and juvenile samples, Pax5 levels were much higher in anterior kidney of pre-spawning fish, including SW-MB, and SW-MoK fish (and to a lower extend, in SS-Pre), but not post-spawners (SS-Post; Figure 4A). In contrast, spleens of spawning fish had strongly reduced levels of Pax5 compared to reference or juveniles (Figure 4B). Pax5 levels in posterior kidney showed a pattern most similar to that of anterior kidney, with increased levels of Pax5 in pre-spawning, but not post-spawned fish (Figure 4C).
Figure 4.
Results from qPCR to determine relative RNA levels of Pax5, indicated in fold-change on the Y-axis relative to the reference sample. Mean fold-change values with SE. N values indicated underneath each group on X-axis. Reference sample (in black) was mature, non-spawning O. mykiss. All other groups are O. nerka. SW-MB: salt-water, pre-spawning, at Main Bay (in vertical stripe). Juvenile (white): pre-smolts from Trail Lake Hatchery. Kenai run: in grey, note a dotted arrow on X-axis. SW-MoK: salt-water, pre-spawning, at Mouth of the Kenai; FW-MSR: fresh-water, pre-spawning, at confluence of Mouth of the Kenai and Moose River; SS-Pre: fresh-water, pre-spawning, at Quartz Creek; SS-Post: fresh-water, post-spawning, at Quartz Creek. A. Anterior kidney. B. Spleen. C. Posterior Kidney.
Previously, we found a strong correlation between Pax5 expression and membrane HCmu expression in spleens of non-spawning adult rainbow trout (Zwollo et al 2008). Such correlations are normally lacking in anterior kidney, because of the abundance of developing B cells in this primary immune organ (including cells which express Pax5, but not HCmu). Hence, the degree of correlation between Pax5 and membrane HCmu expression can be used as an indicator of immune state of the anterior kidney, reflecting changes in developing (Pax5+/HCmu−), mature (Pax5+/HCmu+), and/or terminally differentiated (HCmu++/Pax5−) cells.
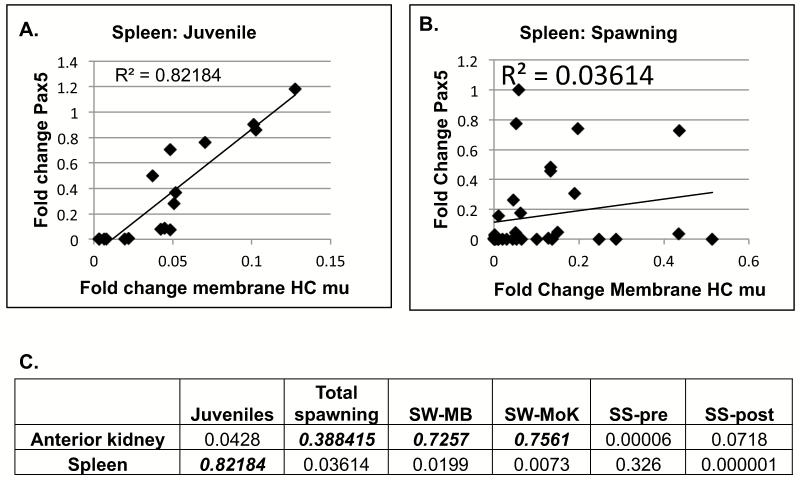
The Scatter Plot in Figure 5A shows the expected correlation between membrane HCmu and Pax5 transcript levels in juvenile spleens (R2 of 0.821). This correlation, as shown in Figure 5B, is absent in samples from spawning fish (combined SW-MB, SW-MoK, SS-Pre and SS-Post splenic samples). Figure 5C compares the correlation strength between Pax5 and HCmu at different collection sites, in anterior kidney and spleen. Spleens from spawning salmon, independent of the collection site, had all lost their Pax5/HCmu correlation. However, the anterior kidney of SW fish appeared to transiently gain such a “spleen-like” correlation (SW-MB and SW-MoK; Figure 5C), suggesting a loss of developing B cells and/or an increase in (im)mature B cells at this stage.
Figure 5.
Scatter plots to determine level of correlation between membrane HCmu (fold change) and Pax5 (fold change) transcripts. A. Strong correlation in juvenile O. nerka spleens. N=18. B. No correlation in spleens of spawning O. nerka. N=26. C. Summary of correlations for anterior kidney and spleen samples from different sites. Numbers represent R2 values. Numbers in bold italics indicate that a correlation between HCmu and Pax5 expression is present.
Since qPCR revealed changes in the levels of secreted HCmu, membrane HCmu, and Pax5 during the spawning journey, we would predict that the distribution of B cell subsets (or B cell signatures) of immune tissues would also change during the spawning journey. Based on the qPCR patterns, we expected to see relatively high percentages of plasma cells (M-PC) in anterior kidney samples from both pre- and post-spawning fish, while juveniles were expected to have the lowest percentages of M-PC. Two-color flow cytometry was performed on a subset of anterior kidney and spleen tissue samples. Fixed and permeabilized cells from 5 groups were used: juveniles, saltwater pre-spawners (SW-MB), pre-spawners at spawning site (SS-Pre), post-spawners (SS-Post), and the reference group (adult O. mykiss). Fluorescently labeled anti-Pax5 and anti-HCmu antibodies were used to identify the various B cell subsets. In anterior kidney tissue, developing (pro/pre) B cells are Pax5+/HCmu−, and (im)mature B cells have the phenotype Pax5+/HCmu+ (Zwollo et al, 2010). IgM+ mature B cells in spleen are HCmu+/Pax5+, while plasmablasts and T-PCs have the phenotype Pax5+/Hcmu++, and M-PCs are Pax5−/HCmu++ (Barr et al, 2011). It should be noted that the earliest stages of B cell development, including CLPs and early pro-B cells, will not stain with the Pax5 antibody (Zwollo et al. 2010), hence these subsets were not analyzed in this experiment. Figure 6 illustrates contour graphs of B cell signatures for each group, and Table II lists the average percentages (+/− SE) of cells for each B cell subset. Equal numbers of males and females were tested.
Figure 6.
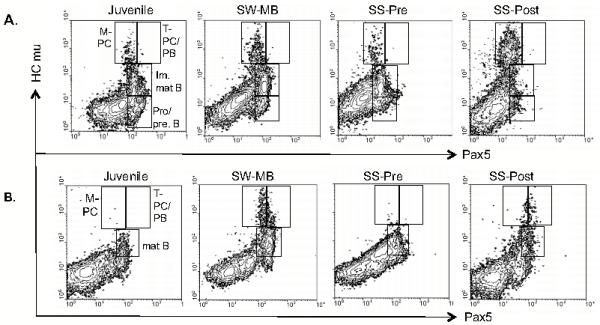
Contour graphs from flow cytometric analyses in fixed and permeabilized sockeye salmon immune tissues. Shown are representative contour graphs for each collection site and for each study group. N=4 for each group except for juveniles, for which N=7. Y-axis: anti-HCmu labeled with Alexa555. X-axis: anti-Pax5, paired domain-specific antibody labeled with Alexa647. A. Anterior kidney; M-PC: HCmu++/Pax5−; plasmablasts/T-PCs: HCmu++/Pax5+; Pro/pre-B: HCmu−/Pax5+; (im)mature B: HCmu+/Pax5+. B. Spleen, as A except mature B: HCmu+/Pax5+.
TABLE II.
Mean percentage of immune cells of each phenotype, with standard error in parentheses.
| ANTERIOR KIDNEY | |||||
|---|---|---|---|---|---|
|
Pro/pre-B (HCmu−/Pax5+) |
(Im)mat B (HCmu+/Pax5+) |
Plasmablast/T-PC (HCmu++/Pax5+) |
M-PC (HCmu++/Pax5−) |
Total ASC (mu++) |
|
| Adult OM | 9.23 (3.22) | 11.7 (1.18) | 0.52 (0.18) | 0.75 (0.28) | 1.27 |
| Juvenile | 13.9 (1.39) | 6.5 (0.85) | 0.47 (0.06) | 0.18 (0.02) | 0.65 |
| SW-MB | 8.7 (1.25) | 5.6 (0.89) | 0.91 (0.03) | 0.9 (0.11) | 1.81 |
| SS-Pre | 3.36 (1.22) | 0.91 (0.51) | 0.03 (0.02) | 0.29 (0.09) | 0.32 |
| SS-Post | 1.1 ( 0.4) | 1.5 (0.50) | 0.83 (0.20) | 2.4 (0.50) | 3.23 |
| SPLEEN | ||||
|---|---|---|---|---|
|
Mu+ mature B (HCmu+/Pax5+) |
Plasmablast/T-PC (HCmu++/Pax5+) |
M-PC (HCmu++/Pax5−) |
Total ASC (mu++) |
|
| Adult OM | 40.3 (3.90) | 1.05 (0.18) | 0.11 (0.03) | 1.16 |
| Juvenile | 3.97 (0.48) | 0.12 (0.02) | 0.02 (0.01) | 0.14 |
| SW-MB | 7.8 (1.80) | 2.28 (0.52) | 0.07 (0.06) | 2.35 |
| SS-Pre | 8.8 (1.40) | 0.27 (0.03) | 0.04 (0.01) | 0.31 |
| SS-Post | 1.3 (0.43) | 0.71 (0.23) | 0.73 (0.12) | 1.44 |
Unexpectedly, the post-spawning group (SS-Post) had the highest percentage of M-PCs in their anterior kidney (Figure 6A and Table II): at least 8 times more than pre-spawning fish at the same site. Additionally, the total percentage of ASCs (including plasmablasts, T-PCs, and M-PCs) was 10-fold higher in post-spawners compared to pre-spawners at the same site. The spleens of post-spawners also harbored dramatic increases, particularly for M-PCs (>18-fold increase), with a more moderate increase in total ASCs (<5-fold). In agreement with the qPCR results using secreted HCmu-specific primers, fish collected from Main Bay (SW-MB) also had relatively high percentages of M-PC and T-plasma cells/plasmablasts in anterior kidney and spleen, although not as high as the percentages seen in the post-spawners (SS-Post; Figure 6B and Table II). These patterns appeared independent of the gender of the fish.
In agreement with the qPCR results using membrane HCmu-specific primers and Pax5, flow cytometric analysis confirmed a loss of developing B cell populations (both HCmu−/Pax5+ pro/pre-B, and HCmu+/Pax5+ (im)mature B cells) in the anterior kidney, which became more severe later in the spawning journey (Figure 6A, Table II). Similarly, a dramatic loss of mature B cells (HCmu+/Pax5+) was detected in spleens of post-spawning fish (Figure 6B, Table II).
DISCUSSION
This study aimed to determine the shifting patterns of developing B, mature B and IgM-secreting plasma cells during the spawning journey of adult sockeye salmon, to better understand the connection between humoral immunity and pre-spawning survival in fish. It is clear from the data shown here that spawning sockeye salmon do NOT lose their ability to generate and secrete IgM during their migratory journey. In fact, a gradual increase in secreted HCmu transcript levels was observed as spawning fish moved from saltwater (Mouth of the Kenai) to a spawning site (Quartz Creek) and this correlated with a sharp increase in the percentage of ASCs for fish at these sites. Interestingly, data from post-spawned fish indicated that, while the average level of secreted HCmu transcripts in anterior kidney had dropped at this stage, the percentage of M-PCs dramatically increased. There were several important differences between what was measured in each approach; qPCR measured average levels of secreted HCmu transcripts in each tissue, while flow cytometry measured the percentage of cells that were actively secreting the end-product of these HCmu transcripts, the HCmu polypeptide that forms IgM. One explanation for this difference is that HCmu RNA is more susceptible to degradation in tissues of post-spawned fish, compared to pre-spawners. Nevertheless, both approaches gave important information on the humoral immune state of a spawning fish, but only flow cytometric analysis could reveal the significant increase (or survival) of a minor population of the total cells (<3% of lymphoid cells), PCs, in post-spawned fish.
It is unknown how PCs resist cortisol-induced cell death at terminal life-stages of spawning fish, and how PCs survive even in post-spawned fish. Little is known about cortisol resistance in fish PCs, but human LLPCs appear to be highly resistant to glucocorticoid-induced apoptosis [Hoyer et al, 2008]. We speculate that the selective “retention” of PCs in anterior kidney and spleen of spawning fish is the result of glucocorticoid resistance, similar to what has been observed in human (Igarashi et al, 2005) and fish lymphocytes (Yada et al, 2008). This makes cells less responsive to chronic exposure to cortisol, and consequently, increases resistance to cortisol-induced apoptosis, thereby protecting important humoral memory in spawning fish.
Using only the qPCR data, it would appear that IgM-secretion in spleens of spawning fish did not change much during the spawning journey, with the exception of the (transient) increase in fish from the FW-MSR site. However, as for anterior kidney, flow cytometric analysis of spleen tissues revealed that the percentage of M-PCs was much higher in post-spawners than in pre-spawners from the same site. Our results illustrate the complex dynamics of B cell activation in the spleen: while qPCR analysis can determine the average level of secreted HCmu transcripts in a tissue, it cannot distinguish between multiple, terminally differentiating B cell populations (activated B, plasmablasts, T-PC, and M-PC), each expressing a different amount of secreted HCmu (Barr et al, 2011). Hence, while HCmu RNA levels are a good indicator of an active humoral immune state in a tissue, flow cytometry analysis is the approach of choice to detect the percentage of IgM-secreting cells in that tissue.
Our results are in agreement with previous observations by others. Using ELISPOT and ELISA assays, it has been shown that spawning carp retained high frequencies of IgM-secreting cells and serum IgM, in blood, spleen and anterior kidney, in spite of high cortisol levels [Saha et al, 2002]. Others have suggested that in vitro-induced B cell proliferation and antibody production are reduced in spawning fish [Tripp et al, 1987], but do not give any information on the percentages and the nature of ASCs in immune tissues.
Our combined data also show that during the spawning journey, levels of membrane-HCmu RNA and the percentage of membrane-bound, IgM+ (im)mature B cells are gradually reduced in the anterior kidney, and more dramatically, in the spleen. Consequently, by the time fish reach their spawning ground, they have very few resting mature B cells left, and must heavily rely on their existing ASCs for protection from infections. The great majority of ASCs appear to be M-PCs, but additionally, some plasmablasts/T-PCs (Pax5+/HCmu++) are present, which may have been generated from mature B cells in response to pathogens, perhaps earlier in the spawning journey. The observed loss of splenic mature B cells also correlated with reduced Pax5 RNA levels in the spleen of spawning fish. This may be the result of activation of most of the remaining mature B (or memory B?) cells, and subsequent downregulation of Pax5, as B cells terminally differentiate into plasma cells. Our data are in agreement with other studies, which have shown that generation of new B lymphocytes in fish is inhibited in the presence of high cortisol, while LPS-activated (but not resting) fish B lymphocytes are induced to undergo apoptosis [Slater et al, 1997, Maule et al, 1990, Saha et al, 2003, and Weyts et al, 1998.].
The high levels of Pax5 found in the anterior kidney of pre-spawning, saltwater fish were unexpected, and suggests a boost in B lymphopoiesis prior to, or upon fish entering fresh water. However, increased cortisol levels during the freshwater period likely inhibit further B lymphopoiesis in the final stages of the spawning journey. The low percentage of developing B cells in anterior kidneys of fish at their spawning site (SS-Pre) supports such a cortisol-induced loss. Virtually nothing is known about cortisol-induced changes in early developing B cells in fish. However, in humans, cortisol has an inhibiting effect on B lymphopoiesis. [Igarashi et al, 2005]. Human pre-B cells are highly sensitive to glucocorticoid, which induces apoptosis; concomitantly, Pax5 protein and transcript levels decrease when pre-B cells are cultured in glucocorticoid-containing medium [Rahman et al, 2001]. However, this topic needs further study.
Interestingly, pre-spawning fish collected at Main Bay (SW-MB) had strikingly high levels of membrane HCmu, secreted HCmu and Pax5 transcripts, as well as relatively high percentages of plasmablasts/T-PCs and plasma cells. This may suggest that the genetic stock of Main Bay sockeye salmon generates and/or retains more B cells in their immune organs, and/or has a higher capacity for humoral (antibody) response and PC formation compared to Kenai sockeye. This may have important consequences in terms of their capacity to generate (LL)PCs, and will need more extensive analysis.
Juvenile salmon had the lowest percentage of M-PCs of all groups, in both their anterior kidney and spleen, and this was supported by qPCR data using secreted HCmu-specific primers. This suggests that juveniles are still building their (LL)PC repertoire at this stage, the pre-smolt stage. Of interest in this regard is our observation that mature smolts (collected at TLH in July from the same stock) had on the average more than twice as many M-PCs as pre-smolts (collected at TLH in april/may) suggesting that accumulation of PCs in anterior kidney increases during the late pre-smolt stage (Zwollo, unpublished data). Juveniles also expressed significantly lower levels of membrane HCmu and Pax5 in their spleen, compared to reference fish, suggesting they have relatively low percentages of mature B cells at this stage. Although juveniles possessed fewer (im)mature B and ASCs than adult fish, they did have higher levels of developing B cells (Pax5+/HCmu−) in their anterior kidney, indicating the potential for generation of mature B, PCs, and LLPCs as the fish mature.
Although sample size in this study was small, our data may suggest that sockeye salmon males that have reached their spawning ground express higher levels of both secreted and membrane HCmu transcripts, compared to females; hence, sexually mature males may keep generating ASCs longer than females. It can be speculated that this is caused by cortisol differences between the sexes: a study on spawning wild sockeye salmon (Sandblom et al, 2009) showed that females approaching their spawning ground had significantly higher levels of cortisol than males. Considering the immunosuppressive effects of cortisol on development and survival of B cells, this may cause spawning females to lose their humoral immunocompetence earlier than males. Clearly, this will require additional research.
We recently proposed the “immunological imprinting hypothesis” (Zwollo 2012). This hypothesis proposes that LLPCs play crucial roles during the spawning stage of anadromous salmon, as they protect fish from pre-spawning mortality through continuous secretion of antibodies with specificity to the pathogen footprint of its natal site. Here, we provide evidence that IgM-secreting M-PC cells survive in anterior kidney and spleens of spawning salmon, while developing B and mature B cells are lost. Most of the relative increases in M-PCs cells are likely a direct result of loss of other immune cells, and as such, represent the retention of T-PCs. However, new IgM-secreting cells maybe produced during the journey, as suggested by the high frequency of (HCmu++/Pax5+) plasmablast/T-PCs in post-spawned fish, and the continued presence of PCs in the spleen. The question of relative contribution of plasmablast, T-PC versus M-PC, to immune protection in spawning fish, will need to be addressed in future studies.
Surviving post-spawners are, per definition, “successful” fish: they possess essential, favorable markers necessary for both survival of the long spawning journey and spawning itself. Hence, we speculate that the observed high levels of secreted HCmu transcripts and the high percentages of IgM-secreting PCs in post-spawned fish, reflect an acute necessity to retain a sufficient level of PCs. Future studies, now in progress in our laboratory, aim to determine if, and to which extend, such PCs represent natal-pathogen-specific LLPCs. If so, they would be expected to promote pre-spawning survival from natal-stream specific infections in the absence of B lymphopoiesis, and as such, increase the likelihood of successful production of offspring.
Highlights.
-
>
spawning sockeye salmon express high levels of secreted heavy chain mu transcripts in their spleen and anterior kidney
-
>
IgM-secreting plasma cells remain abundant in anterior kidney and spleen of post-spawning sockeye salmon
-
>
HCmu−/Pax5+ developing B cells are lost during spawning.
Acknowledgements
We thank Dr. Steve Kaattari for critical reading of the manuscript. The authors are also grateful for donation of juvenile sockeye salmon, help and support from Tom Prochazka and others at the Trail Lake Hatchery. We also wish to thank Drs. Oya Yazgan, Hui-Ching Kuo, Carey Bagdassarian, Alan Tigert, and Chris Pallister for help with collection of samples, Gulf of Alaska Keeper for donation of shipboarding time and help with sample collections, and the ADFG for important advise and information for the project. This research was supported by National Institute of Health R15 award A1070249-02, three Undergraduate Research Scholarships from the Howard Hughes Medical Institute (AB, TC, and JS) and a Vera Barkley Associate Professor Award from The College of William and Mary.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barr M, Mott K, Zwollo P. Defining terminally differentiating B cell populations using the transcription factor XbpI. Fish and Shellfish Immunology. 2011;31:727–735. doi: 10.1016/j.fsi.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromage ES, Kaattari IM, Zwollo P, Kaattari SL. Plasmablast and plasma cell production and distribution in trout immune tissues. J Immunol. 2004;173:7317–23. doi: 10.4049/jimmunol.173.12.7317. [DOI] [PubMed] [Google Scholar]
- DeLuca D, Wilson M, Warr GW. Lymphocyte heterogeneity in the trout, Salmo gairdneri, defined with monoclonal antibodies to IgM. Eur. J. Immunol. 1983;13:546–51. doi: 10.1002/eji.1830130706. [DOI] [PubMed] [Google Scholar]
- Hansen JD, Zapata AG. Lymphocyte development in fish and amphibians. Immunol Rev. 1998;166:199–220. doi: 10.1111/j.1600-065x.1998.tb01264.x. [DOI] [PubMed] [Google Scholar]
- Hoyer BF, Mumtaz IM, Yoshida T, Hiepe F, Radbruch A. How to cope with pathogenic long-lived plasma cells in autoimmune diseases. Ann Rheum Dis. 2008;67(Suppl 3):iii87–9. doi: 10.1136/ard.2008.098418. [DOI] [PubMed] [Google Scholar]
- Igarashi H, Medina KL, Yokota T, Rossi MI, Sakaguchi N, Comp PC, Kincade PW. Early lymphoid progenitors in mouse and man are highly sensitive to glucocorticoids. Int Immunol. 2005;17:501–11. doi: 10.1093/intimm/dxh230. [DOI] [PubMed] [Google Scholar]
- Kaattari Analysis of long-lived plasma cell production and regulation. Aquaculture. 2005;246:1–9. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. N Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma C, Ye J, Kaattari SL. Differential compartmentalization of memory B cells versus plasma cells in salmonid fish. Eur. J. Immunol. 2013 doi: 10.1002/eji.201242570. Epub ahead of print. DOI: 10.1002./eji.201242570. [DOI] [PubMed] [Google Scholar]
- Maule AG, Schreck CB. Glucocorticoid receptors in leukocytes and gill of juvenile coho salmon (Oncorhynchus kisutch) Gen Comp Endocrinol. 1990;77:448–55. doi: 10.1016/0016-6480(90)90236-f. [DOI] [PubMed] [Google Scholar]
- Miller KM, Li S, Kaukinen KH, Ginther N, Hammill E, Curtis JM, Patterson DA, Sierocinski T, Donnison L, Pavlidis P, Hinch SG, Hruska KA, Cooke SJ, English KK, Farrell AP. Genomic signatures predict migration and spawning failure in wild Canadian salmon. Science. 2011;331:214–7. doi: 10.1126/science.1196901. [DOI] [PubMed] [Google Scholar]
- Rahman M, Hirabayashi Y, Ishii T, Watanabe M, Maolin L, Sasaki T. Prednisolone sodium succinate down-regulates BSAP/Pax5 and causes a growth arrest in the Nalm6 pre-B cell line. Tohoku J Exp Med. 2001;193:237–44. doi: 10.1620/tjem.193.237. [DOI] [PubMed] [Google Scholar]
- Saha NR, Usami T, Suzuki Y. Seasonal changes in the immune activities of common carp (Cypinus carpio) Fish Physiology and Biochemistry. 2002;26:379–87. [Google Scholar]
- Saha NR, Usami T, Suzuki Y. A double staining flow cytometric assay for the detection of steroid induced apoptotic leucocytes in common carp (Cyprinus carpio) Dev Comp Immunol. 2003;27:351–63. doi: 10.1016/s0145-305x(02)00116-7. [DOI] [PubMed] [Google Scholar]
- Sandblom E, Clark TD, Hinch SG, Farrell AP. Sex-specific differences in cardiac control and hematology of sockeye salmon (O.nerka) approaching their spawning ground. Am. J. Physiol. Reg. Comp. Physiol. 2009;297:R1136–R1143. doi: 10.1152/ajpregu.00363.2009. [DOI] [PubMed] [Google Scholar]
- Slater CH, Schreck CB. Physiological levels of testosterone kill salmonid leukocytes in vitro. Gen Comp Endocrinol. 1997;106:113–9. doi: 10.1006/gcen.1996.6858. [DOI] [PubMed] [Google Scholar]
- Tripp RA, Maule AG, Schreck CB, Kaattari SL. Cortisol mediated suppression of salmonid lymphocyte responses in vitro. Dev Comp Immunol. 1987;11:565–76. doi: 10.1016/0145-305x(87)90045-0. [DOI] [PubMed] [Google Scholar]
- Weyts FA, Flik G, Rombout JH, Verburg-van Kemenade BM. Cortisol induces apoptosis in activated B cells, not in other lymphoid cells of the common carp, Cyprinus carpio L. Dev Comp Immunol. 1998;22:551–62. doi: 10.1016/s0145-305x(98)00033-0. [DOI] [PubMed] [Google Scholar]
- Ye J, Kaattari I, Kaattari S. Plasmablasts And Plasma Cells; Reconsidering Teleost Immune System Organization. Dev. Comp. Immunol. 2011;35:1273–81. doi: 10.1016/j.dci.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Yada T, Hyodo S, Schreck CB. Gen. Comp. Endocrinol. 2008;156:622–627. doi: 10.1016/j.ygcen.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Zwollo P, Rao S, Wallin JJ, Gackstetter ER, Koshland ME. J. Biol. Chem. 1998;273:18647–18655. doi: 10.1074/jbc.273.29.18647. [DOI] [PubMed] [Google Scholar]
- Zwollo P, Cole S, Bromage E, Kaattari S. B cell heterogeneity in the teleost kidney: evidence for a maturation gradient from anterior to posterior kidney. J Immunol. 2005;174:6608–16. doi: 10.4049/jimmunol.174.11.6608. [DOI] [PubMed] [Google Scholar]
- Zwollo P, Haines A, Rosato P, Gumulak-Smith J. Molecular and cellular analysis of B-cell populations in the rainbow trout using Pax5 and immunoglobulin markers. Dev Comp Immunol. 2008;32:1482–96. doi: 10.1016/j.dci.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwollo P, Mott K, Barr M. Comparative analyses of B cell populations in trout kidney and mouse bone marrow: establishing “B cell signatures”. Dev Comp Immunol. 2010;34:1291–9. doi: 10.1016/j.dci.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwollo P. Dissecting teleost B cell differentiation using transcription factors. Dev Comp Immunol. 2011;35:898–905. doi: 10.1016/j.dci.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwollo P. Perspective: Why salmon return to their natal stream; the immunological imprinting hypothesis. Dev. Comp. Immunol. 2012;38:27–29. doi: 10.1016/j.dci.2012.03.011. [DOI] [PubMed] [Google Scholar]