Abstract
Over the past 20 years our understanding of the basic mechanisms of gene regulation has vastly expanded due to the unexpected roles of small regulatory RNAs, in particular, microRNAs (miRNAs). miRNAs add another layer of complexity to the regulation of effector molecules for nearly every physiological process, making them excellent candidate molecules as therapeutic targets, biomarkers, and disease predictors. Hormonal contributions to mature miRNA expression, biosynthetic processing, and downstream functions have only just begun to be investigated. Elucidating the physiological consequences of miRNA sexual dimorphism, and their associated regulatory processes, may be key towards understanding both normal and pathological processes in the brain. This short review provides a basic overview of miRNA biosynthesis, their role in normal brain development, and potential links to neurological diseases. We conclude with a brief discussion of the current knowledge of sex-specific miRNA processes in both the brain and the heart to conceptually integrate the relevance of miRNAs with the overarching theme (“sex differences in health and disease: brain and heart connections”) of this special topics issue.
Introduction
MicroRNAs (miRNAs) have emerged over the last decade as powerful regulatory agents for virtually all fundamental biological processes, and therefore functional dysregulation of miRNAs has been implicated as a causative factor in a variety of pathogenic conditions. Belonging to a larger class of non-coding regulatory RNA molecules, these tiny ~22-nucleotide (nt) RNA species participate in the cell as highly effective gene silencers through the post-transcriptional regulation of their target gene mRNA transcripts [1–3]. Pioneering work in the laboratories of Victor Ambros [4–6], Gary Ruvkun [7], and David Baulcombe [8, 9] first identified these small regulatory RNAs in C. elegans and various plant species, but evolutionary conserved miRNAs have since been described in all animal species studied to date, including humans [10, 11] (see Table 1 for current nomenclature conventions). While our understanding of the intracellular processing of miRNAs and the basics of miRNA:target gene interactions has increased significantly in the past few years, many questions remain about how miRNAs are regulated, especially with respect to age and hormones.
Table 1.
Nomenclature guidelines for miRNAs. Reference [44] and www.mirbase.org
| ANNOTATION | CRITERIA |
|---|---|
| First 3 lower case letters | Denotes species (i.e. hsa = human, rno = rat, dme = drosophila) |
| mir/miR | Lower case “r” indicates pri- or pre- forms. Upper case “R” indicates mature sequence |
| Number | Number is assigned based on date of sequence validation (i.e. lower numbers were discovered first) |
| Lower case letter after number | Denotes closely related sequences (i.e. miR-20a, miR-20b) |
| -3p/-5p | Indicates the arm of the precursor hairpin from which the mature sequence was derived. |
Note – the ‘minor’ miRNA product (usually transcribed from the -3p end and expressed at lower concentrations in the cell) was previously designated with an asterisk (*). This annotation will no longer be recognized beginning with miRBase17 and will be replaced with the -3p/-5p annotation.
miRNA Biosynthesis
The majority of miRNAs in humans and higher vertebrates have promoter sequences that are autonomously situated between other genes and are often found in clusters, with multiple miRNA promoter sequences lying in close proximity to one another [11]. The canonical miRNA biosynthetic pathway in mammals begins with the generation of a primary transcript (pri-miRNA) encoded from these promoters and initiated by RNA Polymerase II, similar to the biosynthetic pathways for protein coding messenger RNA (mRNA) [12, 13]. First strand pri-mRNA transcription results in a relatively long (i.e. 100 – 1000 bp) double-stranded molecule with a classical hairpin loop that results from its high degree of self-complementarity. The subsequent processing of the pri-miRNA is accomplished through successive cleavage events orchestrated by two RNase III enzymes: the nuclear enzyme drosha and the cytoplasmic enzyme dicer. Drosha forms a heterodimer with DiGeorge syndrome Critical Region 8 (DGCR8), a RNA binding protein that positions the catalytic domain of drosha near the base of the pri-miRNA hairpin loop [14, 15]. Drosha cleaves the pri-miRNA hairpin yielding a ~70 nt stem-loop precursor miRNA (pre-miRNA) that is exported out of the nucleus in a RAN-GTP dependent manner by the cotransporter Exportin 5 [16, 17]. Dicer completes the biosynthetic process by cleaving pre-miRNA precursors into the canonical ~22 nt duplex miRNA.
Interestingly, a small fraction of miRNA coding sequences, termed “mirtrons”, have their precursor sequences encoded within the intronic region of another gene, allowing them to bypass drosha processing. In those cases, transcription of the pre-miRNA is dependent on the upstream regulation of its “host” gene. Mirtrons are more commonly found in lower organisms that have an abundance of genes containing short introns (i.e. similar in size to that of a pre-miRNA), such as insects and nematodes [18, 19]. One explanation for this evolutionary switch in miRNA processing might be that higher organisms evolved the more complex two-step processing pathway (drosha-dicer) to better regulate miRNA activity, and their respective target genes, within more highly specialized cell types, such as neurons. Alternatively, the small percentage of evolutionarily retained mirtrons in higher organisms may provide an alternate pathway for mature miRNA synthesis and another level of regulation during complex early developmental processes. This scenario would facilitate the immediate miRNA-mediated post-transcriptional modulation of embryonic stem cell target genes that determine cell fate specification and differentiation pathways, such as Hox genes.
While the biochemical events required for processing of the pri-miRNA and pre-miRNA sequences are becoming more transparent, the precise molecular mechanisms regulating transformation of the miRNA duplex to a single-stranded effector remain unclear. Most studies suggest that one strand of the duplex, termed the “guide” strand, is preferentially selected for incorporation into the RNA-induced silencing complex (RISC), while the opposite “passenger” strand (formerly designated as miR*) is typically degraded [20–22]. The RISC complex is a ribonucleoprotein aggregate that contains two primary functional units: the mature guide strand miRNA and a member of the Argonaute (Ago) family of proteins. Because the guide and passenger strands are inverse complements it is reasonable to predict that their target genes would be vastly different, making the strand selection process a critical regulatory step for directing the functional consequences of miRNA-mediated gene repression. However, the mechanisms dictating the process of strand selection remain unresolved. One model suggests that the miRNA is incorporated into the Ago protein as a duplex; the duplex is then unwound based on the thermodynamic stability of the first 1–4 nucleotides at the 5′ end; the least thermodynamically stable strand is retained in the Ago protein as the “guide”; and finally, the passenger strand is discarded [23, 24]. One important assumption for this model is that it requires active ATP-dependent loading of the miRNA duplex into the Ago protein. An alternative model posits that the miRNA duplex is unwound by specialized RNA helicases prior to guide strand incorporation into the Ago protein and then the single-stranded effector can be spontaneously incorporated in an ATP-independent manner [25]. Evidence for this model is based largely on theoretical predictions about the inherent properties of RNA helicases and the thermodynamic properties governing the miRNA asymmetrical strand selection. Although several putative RNA helicases have been proposed to support this model, none have yet been shown to be sufficient and required at a specific step in the RISC assembly process.
miRNA-mediated gene silencing
A highly conserved protein from the Argonaute superfamily is at the core of all RISC complexes. Single-stranded miRNAs associated with a particular Ago protein direct the ribonucleoprotein RISC complex to a specified target gene, accomplished through miRNA recognition of a partial complementary binding sequence on the target mRNA 3′UTR. There are four identified Ago protein subfamilies in humans (AGO1-4), but AGO2 is the only one that has demonstrated endonuclease (“slicer”) activity [26] thereby, defining it as a critical component in vertebrate RNA silencing complexes (see [27] for an excellent review of Ago proteins). Alternatively AGO1, 3, and 4 induce translational repression of their target genes in the absence of mRNA cleavage, which raises the fundamental question of what mechanisms confer miRNA specificity for a given Ago protein. Indeed, the highly conserved structure of the Ago protein domains suggest that a great deal of overlap exists among the members of the Ago superfamily, allowing for multiple small RNA molecules, including siRNAs, to bind more than one Ago protein subtype. In plants, some evidence suggests that the length of the mature miRNA guide strand (21 nt – 22 nt) and the nucleotide identity at the 5′ end are factors determining specificity for Ago proteins. Nonetheless translational repression, as opposed to mRNA cleavage, is now widely recognized as the primary mechanism by which miRNAs achieve gene silencing in animals.
Sex differences in miRNA silencing mechanisms remain a relatively unexplored area of research. Although several studies have described the effects of sex steroid hormones, primarily 17β-estradiol (E2), but also testosterone (T), on steady-state mature miRNA expression and some of the processing enzymes, such as drosha and dicer, none have examined whether protein constituents of the RISC complex or mechanisms of miRNA silencing action are regulated by sex steroid hormones.
miRNAs in neurodevelopment
Whole-genome sequencing data from a variety of species have underscored the importance of post-transcriptional and post-translational modifications needed to achieve extensive phenotypic diversity. miRNAs have the ability to fine-tune the downstream physiological effects of gene transcription by regulating the effectors (i.e. proteins) of those genes. Central nervous system (CNS) development, in particular, requires a precise temporal orchestration of events that is uniquely suited for the fine-tuning attributes of miRNAs. The importance of miRNAs in embryonic development was originally demonstrated using transgenic animal models that manipulated miRNA biosynthetic processing enzymes, such as dicer and DGCR8. There is a single gene that encodes dicer in C. elegans, mice, and humans and depletion of dicer results in severe developmental consequences. Studies in dicer-mutant zebrafish showed they had disrupted embryonic morphogenesis and neural differentiation [28]. More specifically, the brains lacked ventricles, neuronal positioning was disrupted suggesting migration defects, and many neurons had defasciculated axons [28]. Strikingly, injections of miRNAs from the miR-430 family (miR-430a/b/c) reversed many of the brain morphogenic defects that resulted from dicer deletion in the zebrafish, revealing a direct connection between mature miRNAs and dicer during development. The partial rescue of neuronal defects in this study also provided some of the first evidence for tissue-specific effects of miRNAs [28, 29]. Global dicer deletion in mice is embryonic lethal [30] prompting the generation of tissue-specific conditional dicer-null mouse models. In the developing neocortex, the absence of dicer resulted in a smaller cortex, improper cortical layering, increased apoptosis, as well as an overall reduction in neural progenitor cells and oligodendrocytes [31–33]. Dicer is also a critical enzyme for mediating the effective maturation and maintenance of olfactory, purkinje, dopaminergic and forebrain neurons [34–37]. One important consideration is that dicer, an RNase III enzyme, mediates the biosynthetic processing of all small regulatory RNAs, including siRNA, and could possibly be an important regulator of other, as yet unidentified, physiological processes. Therefore, caution must be exercised when drawing conclusions about the direct cause of the phenotypic manifestations observed in dicer-null animals. Deletion of DGCR8 in murine embryonic stem cells also results in a phenotype that reflects a loss of miRNA function and disrupted miRNA biosynthesis including excess accumulation of primary miRNA transcripts, lack of mature miRNA expression, abnormal expression of differentiation markers, and early arrested development. Although lethal in early embryonic development, the magnitude of neuronal defects observed in DCGR8-null mice is not as striking as those in dicer-null mice [38], suggesting that other compensatory mechanisms can override the phenotypic consequences of DGCR8 deletion.
The discrete biological functions of individual miRNAs during neuronal development are less well understood. Lsy-6 was the first specific miRNA recognized to have a role in nervous system development in vivo, where it was shown to regulate left/right asymmetrical patterning of the taste receptor neurons in C. elegans [39]. Two other miRNAs, miR-9 and miR-10, are highly expressed in the brain and have been shown to play important roles in the brain development of many species including humans, rodents, zebrafish and drosophila, demonstrating a high degree of evolutionary conservation among these miRNAs. Specifically, miR-9 promotes migration and proliferation in human neural progenitor cells by targeting stathmin, a gene required for microtuble assembly [40] and peripheral nervous system sensory organ development in drosophila [41]. Further, miR-9 is significantly reduced in the presenilin-1 null mouse model, which exhibits severe CNS developmental defects, during specific stages of development compared with wild-type mice [42]. Another important miRNA during development is miR-10, which targets members of the HOX gene family; a highly conserved group of transcription factors that coordinate anterior-posterior body axis alignment in zebrafish and other species during development [43].
In addition to posttranscriptional regulation of mRNA, alternative pre-mRNA splicing is another well-described mechanism for increasing phenotypic diversity with a limited pool of protein-coding genes. Recent studies have shown that miRNAs actively participate in the regulation of alternative splicing events in neurons by targeting genes that are involved in pre-mRNA splicing processes. For instance, the neuronal-specific miRNA, miR-124, was shown to directly target the RNA-binding protein PTBP1, which reduces neuron-specific alternative splicing [44]. Taken together, these studies revealed that specific miRNAs and their processing enzymes are critical for proper gene expression and brain function throughout development, however once the brain is fully formed, constant monitoring of ongoing cellular process is yet another critical function for miRNAs in the brain.
miRNAs in neuronal cell maintenance
miRNAs vital role in neuronal maintenance processes position them as attractive targets for therapeutic strategies to combat a variety of neurodegenerative conditions. Ongoing cellular maintenance processes are critical facilitators of neuronal plasticity, which results from dynamic changes in dendritic spine density. Specific miRNAs, such as miR-29a/b and miR-134, have been identified as participants in this process by regulating components of actin polymerization that are required for dendritic spine formation and remodeling. In hippocampal neurons, miR-29a/b targeted expression of the protein ARPc3, which is a subunit of the ARP2/3 actin nucleation complex [45]. Additionally, miR-134 decreased dendritic spine size by targeting a synaptic kinase, LIMPK1, that enhances actin polymerization in hippocampus neurons [46]. Further evidence for miRNA participation in ongoing neuronal maintenance has been elucidated in studies investigating miR-124, which is highly enriched in neuronal tissues. In both chick and xenopus, miR-124 was not required for neuronal determination [47, 48]. Rather, Sanuki and colleagues showed that miR-124 promoted neuronal maturation and axonal growth in embryonic and adult mice demonstrating its ongoing role in CNS maintenance [49].
miRNAs beyond early development
The role of miRNAs beyond early developmental periods is just beginning to be investigated. Mature miRNA expression is age-dependent and accordingly, miRNAs regulate both early developmental gene expression changes as well as those that occur throughout the lifespan in various species [50–53]. A deep sequencing study recently highlighted 75 miRNAs that were differentially expressed in the brain with age, 71 of which were downregulated, including miR-124 and miR-34a [53]. As mentioned above, miR-124 is highly expressed in differentiating and mature neurons and its possible primary function is to suppress non-neuronal genes and promote the expression of neuronal-specific phenotypes in adult neurons, as demonstrated by the induction of a neuronal-like phenotype in HeLa cells following overexpression of miR-124 [54, 55]. miRNAs that are important for continuing brain development are not restricted to rodents and zebrafish. A predictive bioinformatics analysis has recently suggested that miRNAs are responsible for differences in gene expression from birth to adult in the primate hippocampus. Importantly, the miRNAs of interest in this study were also expressed in the human hippocampus, yet some miRNAs were specific to the non-human primate [56]. A recent comparison of human, chimpanzee and macaque using Affymetrix microarrays found that gene expression patterns diverged at a faster evolutionary pace in humans compared to macaques and chimpanzees, suggesting that miRNAs might have significantly contributed to the evolution of advanced forebrain architecture in humans [57] and that miRNA-mediated gene regulation could have been the driving force behind the evolutionary development in the human brain [57].
miRNAs in the aging brain
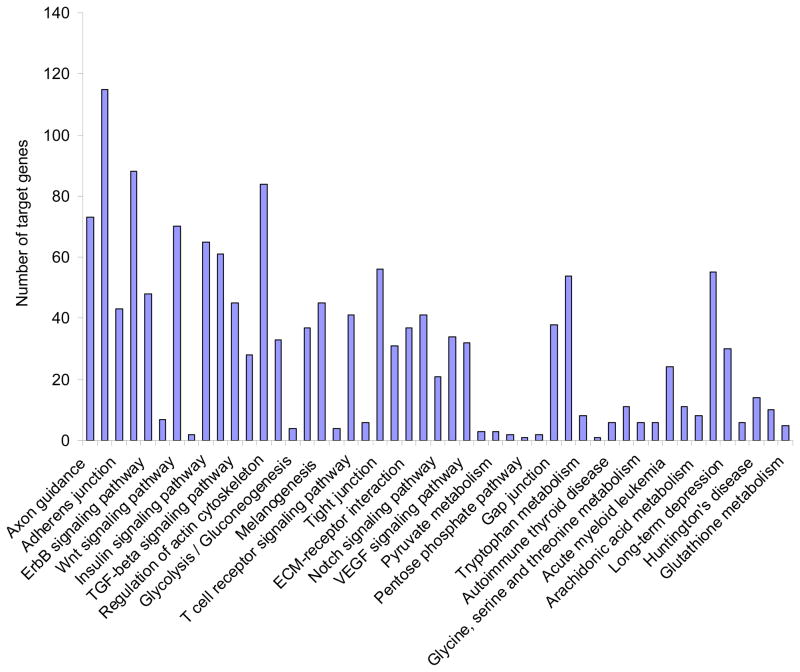
Gene expression profiles of aging human and primate brains have shown that steady state mRNA levels are altered with age particularly for those genes involved with regulating metabolism, synaptic activity, dendritic growth, and cell survival [58–61]. Overall, there is a global decrease in gene expression in the aging brain [58, 59, 61], which can result in a cellular environment that promotes disease. The exact cause of the observed age-related global decrease in gene expression has not been determined, though reports suggest that altered transcription factor expression and/or oxidative damage to gene promoter regions may be contributing factors [62]. Additionally, overall changes in gene expression could potentially be attributed to global changes in RNA processing, for which, regulatory RNAs have come to the forefront as important regulators of age-related gene expression and neurological disease [53, 63–65]. miRNAs were first discovered as important regulators of embryonic development in C. elegans, and as aging often recapitulates development, it follows that one of the first pieces of evidence that miRNAs are crucial for normal aging was also described in C. elegans [51, 66]. However, more recent mammalian studies have provided greater insight into the role of miRNAs in brain aging. In mice, global miRNA expression is increased with age, corresponding precisely with overall decreases in mRNA expression [53, 63]. Interestingly, some of the miRNAs downregulated with age have been shown to target genes involved with oxidative phosphorylation [63]. Further, microarray data obtained from studies in our laboratory revealed that 21 miRNAs were differentially regulated by age alone in the hippocampus of female rats (Table 2, unpublished data). Target pathway prediction programs, such as DIANA-mirpath [67], indicated that these 21 miRNAs could regulate genes involved in multiple cellular signaling pathways, but especially those important for regulating neuronal plasticity (Fig. 1). Human and primate studies have revealed additional levels of complexity for miRNA interaction in the aging brain. Somel and colleagues used macaque and human prefrontal cortex brain samples to track changes in miRNA, mRNA, and protein expression over the lifetime [65]. Their study showed that miRNA expression is highly dynamic during developmental and aging processes, reflecting inverse expression patterns for both mRNA and protein however, the underlying mechanisms behind these age-related changes in miRNA/mRNA expression remain to be elucidated. One hypothesis is that a combination of synergistic factors including adaptive responses to environmental stressors, oxidative DNA damage, altered hormone/growth factor levels, and perhaps circadian rhythms, can all contribute to changing gene expression profiles with age [65]. A significant limitation from the available miRNA/mRNA expression datasets is that they almost all used prefrontal cortex as basis for the study, which may not accurately represent how aging occurs in the other brain regions. Indeed, each brain region has a unique expression profile and associated physiological function. Another important consideration is that cognitive aging, and associated functional decline, are not equivalent for men and women. For example, specific neurological diseases, such as Alzheimer Disease (AD), depression, and anxiety, all present with a bias towards females, indicating that sex hormones and/or chromosomal contributions also play a role in the molecular processes governing these conditions [68–70].
Table 2. miRNAs signifcantly altered by age alone in the rat female hippocampus.
Female Fisher 344 rats at 3 and 18 mo. old were ovariectomized. Brains were removed 7 days post-ovariectomy, hippocampus microdissected, total RNA isolated, and processed for mature miRNA expression using a rat microRNA microarray platform (Sanger 18.0 miRBase probes, LC Sciences).
| miRNA | p-value |
|---|---|
| miR-218a* | 1.95E-03 |
| miR-495* | 3.23E-03 |
| miR-26a* | 3.26E-03 |
| miR-423-3p | 2.08E-02 |
| miR-382 | 2.12E-02 |
| miR-376b-3p | 2.88E-02 |
| miR-379 | 2.95E-02 |
| let-7d-3p | 3.03E-02 |
| miR-125b-5p* | 3.46E-02 |
| miR-137 | 3.97E-02 |
| miR-539 | 4.15E-02 |
| miR-7a* | 5.92E-02 |
| miR-21 | 5.82E-03 |
| miR-219-2-3p | 7.52E-03 |
| miR-150 | 1.87E-02 |
| miR-329 | 2.56E-02 |
| miR-181b* | 4.04E-02 |
| miR-29a* | 4.28E-02 |
| miR-323 | 5.08E-02 |
| miR-34c | 6.25E-02 |
| miR-132 | 6.92E-02 |
An * indicates miRNAs that were also significantly altered by E2 treatment.
Fig. 1. Cellular pathway analysis of predicted miRNA target genes.
Putative miRNA target gene cellular pathways altered by age were generated using the software program DIANA-miRpath [67]. Mature miRNA expression levels were analyzed from the hippocampus of female Fisher 344 rats at 3 and 18 mo. of age. All animals were ovariectomized 7 days prior to tissue collection and total RNA isolation. miRNA expression levels were analyzed using a rat microRNA microarray platform (LC Biosciences, Sanger 18.0 miRbase probes). Loyola University Institutional Animal Care and Use Committee approved all animal experiments.
miRNAs in neurodegenerative disorders
miRNA expression profiles have been reported for a variety of neurodegenerative disorders, however the best studied to date is AD. The precise role of miRNAs in the pathogenic progression of AD is not clear, although mature miRNAs are differentially expressed in diseased brains [71–75]. Specific miRNAs that are commonly dysregulated in many AD patients include miR-7, miR-9, miR-26, miR-29, miR-34, miR-125a/b, miR-181, and miR-495. Interestingly, all of these miRNAs have also been shown to be regulated in the brain by age alone ([65], Table 1), making their contribution to age-related pathological conditions difficult to discern. Several of these miRNAs are also predicted, or have been shown to directly target, β-site APP-cleaving enzyme 1 (BACE1) and amyloid precursor protein (APP). These proteins are critical in the accumulation of β-amyloid plaques, one hallmark of AD pathology, thought to be a critical component of neuronal death and cognitive impairments. Recent studies of sporadic AD patients have shown that there are increased BACE1 protein levels and enzymatic activity in the brain [71, 76], yet no corresponding increase in BACE1 mRNA, suggesting the potential for miRNA-mediated post-transcriptional regulation of BACE1 [71]. Two predicted regulators of BACE1 are miR-29a/b and miR-9, both of which have reduced expression levels in AD brains [71]. miR-9 expression is also reduced in Huntington’s disease, which has similar pathologies to AD [77], suggesting that certain miRNAs might have important roles in specific cellular pathways that have a high propensity for generating diseased states when dysregulated. Due to the correlative nature of the available datasets from AD patients, it is not possible to determine if miRNAs are causing AD, or if their altered expression profiles are merely another consequence of AD pathogenesis. A thorough understanding of how miRNAs contribute to the normal aging process is required to determine if they play a causative role in AD or any other neurodegenerative disorders.
Empirical determination of miRNA targets is crucial towards understanding their impact and specific role in the progression of neurological diseases, however this has proved to be one of the key challenges in the miRNA field. One of the best experimental strategies currently available to identify miRNA gene targets is High-throughput Sequencing of RNA isolated by Crosslinking Immunoprecipitation (HITS-CLIP). HITS-CLIP is a novel technique that isolates mRNA bound to the RISC complex by UV crosslinking and immunoprecipitation [78]. Once the mRNA is isolated, RNA sequencing can be used to determine what transcripts are bound to the RISC complex. Elucidation of the exact miRNA target genes can then be determined by examining the 3′ UTR of the isolated mRNA transcripts for seed sequence matches. Using a HITS-CLIP approach on AD brain samples would reveal all of the mRNAs that are being repressed by the RISC complex, thereby providing valuable insight into the role of differentially expressed miRNAs in AD affected brains.
Another major obstacle for treating AD patients is the lack of available early diagnostic markers. Diagnosis at the onset of AD would enable more targeted and rigorous treatment possibilities with the ultimate goal of slowing disease progression. Notably, miRNAs have recently been proposed for use as potential biomarkers for AD, as well as other diseases, such as various cancers, since many of the miRNAs dysregulated in AD and aging are also expressed in the plasma of cancer patients [79, 80]. Importantly, these miRNAs are enclosed in exosomes that protect them from degradation, making them a potentially reliable biomarker for many diseases.
miRNAs in mental illness
Mental health disorders such as depression, anxiety, and schizophrenia are often highly correlated with advanced age [81–83]. Collectively, mental health disorders are characterized by disrupted neuronal communication, and typically involve the dysfunction of many different brain regions, making these disorders extraordinarily complex to diagnose and treat. Communication between neurons requires tight temporal regulation of multiple genes, which can be finely controlled by miRNAs. In support of this idea, dendrites and synapses express mature miRNAs as well as the components required for miRNA biogenesis and repression, indicating that miRNAs are likely involved in the regulation of neuronal communication [46, 84–86]. miRNA expression is decreased overall in patients diagnosed with depression, whereas miRNA expression is generally increased in those with schizophrenia, suggesting that small RNAs may have a role in these diseases. Much like in AD, it is not yet known if altered expression patterns of miRNAs in schizophrenic patients are responsible for inducing the pathology or are a resulting consequence. The individual miRNAs that are differentially expressed in these patients are predicted to target genes involved in mediating synaptic plasticity [87, 88]. In particular, abnormal expression of miR-132 and miR-137 were positively associated with schizophrenia [88, 89] and previous studies have demonstrated a vital role for miR-132 in promoting dendritic growth, dendritic spine formation, and synaptic integration [88, 90–92]. Studies have also shown that members of the miR-34 family are dysregulated in depression and anxiety disorder [93, 94]. For instance, miR-34c repressed corticotropin-releasing factor receptor type 1 (CRFR1) after acute and chronic stress, resulting in neurons that were less responsive to corticotropin-releasing factor (CRF) [93]. Desensitization and/or hyper-reactivity of the hypothalamo-pituitary-adrenal (HPA) axis, of which CRF is a key neuropeptide, has been proposed as an important contributor to the development of affective disorders, such as depression and anxiety [95]. miR-34 is also an age-regulated miRNA, highlighting a potential molecular mechanism for how the aging process contributes to the manifestation of these disorders [65], however no study to date has directly examined the role of age-regulated miRNAs in the context of mental health disorders. Taken together, these studies suggest that specific miRNAs may be a common underlying factor in the etiology of mental illness and the study of their target genes can reveal the molecular pathways involved.
Hormone regulation of miRNAs
miRNAs are estimated to control the translation of more than a third of all protein-coding genes [96], many of which are implicated in sex-biased diseases. Manifestations of such sex-specific morphological and pathophysiological phenotypes are often rooted in differences in circulating gonadal steroid hormones, such as androgens and estrogens, although chromosomal differences can also be contributing factors [97]. Hormonal regulation of steady-state mature miRNA expression was first reported in Drosophila melanogaster [98, 99]. In this species, the hormones ecdysone (Ecd) and juvenile hormone (JH) exert opposing actions on the progression from larval-to-pupal, and pupal-to-adult, stages of development. Sempere and colleagues [98] found that two miRNAs, lin-4 and let-7, mediated the expression of genes (lin-14, lin-28, and lin-41, respectively) required for facilitating the successful transitions between these developmental stages. Moreover, the steady-state expression of lin-4 and let-7 were dependent on increased levels of Ecd, which peak just prior to the larval-pupal transition [98]. A follow up study identified 3 additional miRs (mir-100, mir-125, and mir-34) in D. melanogaster that were responsive to Ecd and/or JH [99], raising the possibility that hormones might also influence miRNAs in vertebrate and mammalian species.
Ecdysone is a steroid hormone and, in addition to its role in regulating molting, it is the primary sex hormone produced in D. melanogaster [100]. The Ecd receptor is a member of the highly conserved superfamily of steroid receptors, which collectively function as ligand-activated transcription factors in both vertebrate and invertebrate animals. In mammals, T and its primary androgenic metabolite, 5α-dihydrotestosterone (DHT), exert their actions through high affinity androgen receptors (AR), while E2, the estrogenic metabolite of T, exerts its action through estrogen receptors (ER). Classically these receptors directly alter gene transcription by binding to cognate DNA response elements in the nucleus [101] therefore, these classical mechanisms of steroid hormone action are termed “genomic”. Alternatively, non-genomic effectors of hormone:receptor complexes include multiple kinases and phospholipases that can activate intracellular second messenger cascades.
Coincident with reports of Ecd regulating miRNAs in D. melanogaster, Calin and colleagues [102] observed decreased expression of miR-15 and miR-16 in cases of chronic lymphocytic leukemia. Their study suggested that miRNAs might play an important role in cancer pathogenesis and prompted several subsequent miRNA expression profiling studies in other types of cancer. Convergence of the evidence for miRNAs putative role in cancer, along with the evidence suggesting hormones could regulate them, made breast cancer an attractive model system to explore whether there was a definitive mechanistic link between hormones and miRNAs. Indeed, our current understanding of hormonal regulation of miRNAs in mammals has been most thoroughly derived from studies using breast cancer cell lines as model systems and microarray technology as endpoint detection strategies. These broad-based screening approaches have yielded an enormous amount of data, yet there have been few in-depth studies investigating the roles of individual miRNAs and their respective biological functions. Unfortunately, even among studies that used the same well-characterized MCF-7 breast cancer cell line as a model, many of these efforts have also yielded contradictory and ambiguous results, possibly due to variations in E2 treatment paradigms and differing microarray platform technologies [103].
Collectively, early studies demonstrated that miRNA expression profiles could potentially be used as markers to accurately predict the phenotype of a breast cancer tumor [104, 105]. For instance, 29 miRNAs were significantly different (either up or downregulated) between normal and tumorigenic tissue samples [104] and 43 were specifically upregulated in ER-positive compared to ER-negative tumors [105]. Interestingly, miR30s (miR-30a-5p, 30b, 30c, 30d) were significantly downregulated in both ERα- and progesterone receptor- (PR) negative breast cancer tumors [104]. Breast tumor phenotypes that do not express hormone receptors (mainly ERα) typically do not respond as well to conventional therapies and predict a poorer prognosis compared with ER-positive tumors. The significant decrease of mature miRNAs from the miR-30 family that is observed in hormone receptor-negative tumors could indicate that hormones normally regulate these miRNAs, and/or that they target a specific subset of oncogenes that are preferentially expressed in hormone receptor-negative cell types. Importantly, ERβ, which acts in opposition to many ERα-mediated effects in breast cancer tumors, inhibits pri-miRNA synthesis of miR-30a [106], highlighting another mechanism for ERβ-mediated antagonism of ERα signaling pathways. Notably, this was one of the first reports showing a direct effect of ERβ on miRNA regulation in any system.
In contrast to the number of studies describing hormonal regulation of miRNAs in classically steroid-responsive tissues, such as breast [103], ovary [107, 108], uterus [109–112], prostate [113], and testis [108], there are a paucity of studies investigating the effects of hormones on miRNAs in the brain of any species. In the teleost fish, Hippoglossus hippoglossus (i.e. Atlantic halibut) miRNA expression in the brain varied according to both age and sex, but few of the observed sexually dimorphic miRNAs were validated using qRT-PCR [114]. Specifically noted was a significant increase in miR-451, and a decrease in miR-9, in 3yr-old females compared with males; however, because the potential mRNA targets are still unknown the biological significance of this sex difference is not readily apparent. Further, Morgan and Bale [115] recently demonstrated that 7 miRNAs were significantly different between male and female rat brains at postnatal day (PND) 0. Remarkably, a single injection of an aromatase inhibitor, which prevents T-to-E2 conversion, was sufficient to induce a female-like miRNA profile in the PND 0 male brains, suggesting that some miRNAs in the rat brain preferentially target genes that regulate sexual differentiation [115]. One limitation of both of these studies was that the entire brain was used in the assay samples, which perhaps diluted the magnitude of results that might have been observed from any brain-region specific effects. A more targeted approach analyzing adult mouse hippocampus, cerebellum, and cortex showed that there were significant sex differences in all brain regions, with the greatest number of sexually dimorphic miRNAs being expressed in the hippocampus [116]. Consistent with those findings, studies from our laboratory demonstrated that specific miRNAs in the hippocampus, paraventricular nucleus, and central amygdala were E2-responsive, with the largest effects observed in the hippocampus (unpublished data). Importantly, not only were these miRNAs E2- responsive, but a few were also differentially regulated by E2 dependent on age. Further analysis using algorithmic target prediction programs (i.e. Targetscan [117, 118], microRNA.org [119], and MicroCosm [120]) revealed that these miRNAs potentially target genes important for mediating learning and memory, the stress response, and maintaining synaptic connections, all of which are physiological processes with well-documented sexually dimorphic phenotypes. Although these miRNAs have not yet been analyzed in males, we can infer that they would have sexually dimorphic expression patterns due to the inherent differences in circulating E2 levels between males and females.
Sex differences in miRNA expression in the brain would be especially relevant to examine in the context of sexually dimorphic neurological diseases, such as stroke, AD, and schizophrenia. To date, several studies have examined the miRNA expression profiles in post-mortem brains from patients afflicted with AD and/or schizophrenia, but only one differentiated the data by sex [121]. They found that miR-30b, which is estrogen responsive, was expressed at significantly lower levels in female schizophrenic patients compared to males, however a putative target for miR-30b in those patients was not determined [121]. Another study examined sequence variations in mature or precursor miRNAs in 193 male schizophrenic patients compared with age-matched controls [122]. They hypothesized that schizophrenia might be associated with point mutations in miRNAs leading to altered or ineffective downstream gene targeting. Importantly, they limited their analysis to miRNAs encoded on the X-chromosome, due mainly to the propensity for males to develop schizophrenia and their associated decreased fertility rates. Their results showed that eight variants in miRNA genes, all of which they described as “ultra-rare”, were present in schizophrenic patients, but only one was found in the controls [122]. Overall, this study provided evidence for chromosome-based sex differences in miRNAs that are potentially independent of hormonal influence. A similar study examined the influence of miRNAs on the posttranscriptional regulation of X-linked inhibitor of apoptosis (XIAP), a caspase inhibitor that is thought to contribute to the sex differences observed following cerebral ischemia [123]. Importantly, the baseline and stroke-induced sex differences in XIAP were independent of ovarian steroid hormones, suggesting the observed sex effects were due solely to differences in chromosomal contribution. Siegel and colleagues verified that miR-23a bound directly to the 3′UTR of XIAP and miR-23a knock-down increased XIAP mRNA expression levels in HeLa cells [123]. Moreover, miR-23a sex differences in the brain were absent at baseline, yet following cerebral ischemia there was a significant increase in miR-23a expression in females [123]. Taken together, these data indicated that the sex-specific miR-23a regulation of XIAP was crucial for the hormone-independent neuroprotective effects seen in the female ischemic brain.
Data from breast cancer model systems laid a valuable foundation for understanding how the hormonal control of miRNA activity acts as an underlying mechanism to regulate mammalian gene expression patterns in normal physiological processes. Similar to the lack of studies investigating the hormonal regulation of miRNAs in the brain, there has, to our knowledge, only been one report describing a miRNA sex differences in the heart [124]. Connexins are critical proteins that contribute to the formation of gap junctions in the heart, facilitating cell-cell communication pathways. Abnormal expression of connexin 43, a contributing factor to sudden cardiac death, may be important in women, for which cardiac arrhythmia is more common compared to men. In one study, female adult rat ventricular myocytes had greater Cx43 mRNA and protein expression at baseline compared with male rats, suggesting the potential for increased gap junction formation and/or function [124]. Further, female ventricular myocytes had a greater increase in total, as well as phosphorylated, Cx43 protein expression compared with males following stimulation with phenylephrine (PE, an α-adrenergic receptor agonist). These results provided a basis for the sex differences observed in cardiac arrhythmias and raised the possibility that miRNAs could target Cx43 in a sexually dimorphic manner. Indeed, the authors revealed that miR-1, a muscle-specific miRNA previously reported as having a role in intensifying arrhythmic phenotypes [125], was differentially expressed in males and females after PE treatment [124]. Apart from this single study, no other work to date has been done to identify sex-specific miRNA regulation of cardiac function.
Conclusions
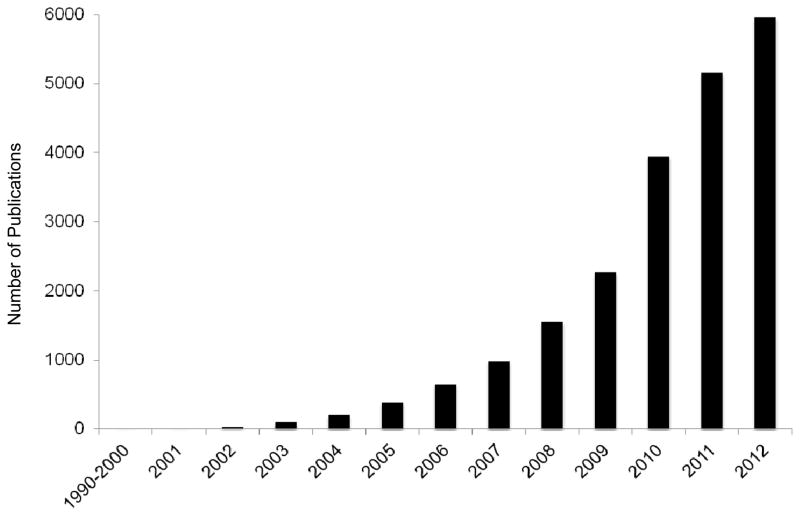
The field of miRNAs is still very young, just over a decade since their discovery, and the amount of knowledge we have gained in that short period of time has expanded exponentially. A recent PubMed search using the term “microRNA” revealed nearly 20,000 published reports, with the majority of those occurring in just the last few years (Fig. 2). Conversely, adding the terms “hormones” and “brain” yielded a mere 45 publications, most of which do not describe any empirical testing of how hormones affect miRNAs in the brain. Generally it is assumed that the fundamental cellular processes underlying all posttranscriptional and posttranslational events are equivalent for both sexes. With regard to general miRNA processing, there have not been any studies designed to determine whether the RISC complex is assembled and functions the same way in both sexes, whether there are sex differences in polyribosome occupancy of miRNAs, whether there are temporal differences in miRNA turnover between sexes, or potential sexual dimorphism in miRNA biosynthetic processing. The lack of such studies represents a significant gap in our knowledge base and underscores an important direction for future efforts. Imperative to this is the need for direct hormonal manipulations in animal models in order to dissect the relative contributions of individual hormones to miRNA-mediated physiological processes. In this review we cite studies detailing the contribution of miRNAs to neuronal development, cellular maintenance, and the etiology neurological diseases. Importantly, sex steroid hormones play an essential role in maintaining a fine balance between homeostatic and pathological outcomes for all of these processes. From sexual differentiation in the developing brain to the presentation of sex disparities in mental health and AD, it is clear that both hormones and miRNAs are central regulators of these processes, and the overlap between the regulatory processes governing each is vastly understudied.
Fig. 2. Number of miRNA publications listed in PUBMED.
The number of total publication listed in PUBMED database according to year of publication. Search term used was “microRNA”. Total publications include all types of articles.
Footnotes
This article is published as part of the special issue on “sex differences in health and disease: brain and heart connections”
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Q, Paroo Z. Biochemical principles of small RNA pathways. Annu Rev Biochem. 2010;79:295–319. doi: 10.1146/annurev.biochem.052208.151733. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 6.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216(2):671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 7.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 8.Baulcombe DC. RNA as a target and an initiator of post-transcriptional gene silencing in transgenic plants. Plant Mol Biol. 1996;32(1–2):79–88. doi: 10.1007/BF00039378. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286(5441):950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 10.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 11.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9(2):175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21(17):4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 16.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 17.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28(2):328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448(7149):83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404(6775):293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 21.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297(5589):2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 22.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431(7006):343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 23.Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci. 2010;35(7):368–376. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 24.Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol. 2010;17(1):17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambrus AM, Frolov MV. The diverse roles of RNA helicases in RNAi. Cell Cycle. 2009;8(21):3500–3505. doi: 10.4161/cc.8.21.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15(2):185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Mallory A, Vaucheret H. Form, function, and regulation of ARGONAUTE proteins. Plant Cell. 2010;22(12):3879–3889. doi: 10.1105/tpc.110.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giraldez AJ. MicroRNAs Regulate Brain Morphogenesis in Zebrafish. Science. 2005;308(5723):833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 29.Plasterk RHA. Micro RNAs in Animal Development. Cell. 2006;124(5):877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 30.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nature Genetics. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 31.McLoughlin HS, Fineberg SK, Ghosh LL, Tecedor L, Davidson BL. Dicer is required for proliferation, viability, migration and differentiation in corticoneurogenesis. Neuroscience. 2012;223:285–295. doi: 10.1016/j.neuroscience.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawase-Koga Y, Otaegi G, Sun T. Different timings of dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Developmental Dynamics. 2009;238(11):2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135(23):3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi PS, Zakhary L, Choi WY, Caron S, Alvarez-Saavedra E, Miska EA, McManus M, Harfe B, Giraldez AJ, Horvitz RH, Schier AF, Dulac C. Members of the miRNA-200 Family Regulate Olfactory Neurogenesis. Neuron. 2008;57(1):41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA Feedback Circuit in Midbrain Dopamine Neurons. Science. 2007;317(5842):1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaefer A, O’Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. Journal of Experimental Medicine. 2007;204(7):1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, Ullian EM. Conditional Loss of Dicer Disrupts Cellular and Tissue Morphogenesis in the Cortex and Hippocampus. Journal of Neuroscience. 2008;28(17):4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babiarz JE, Hsu R, Melton C, Thomas M, Ullian EM, Blelloch R. A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus Dicer1 knockouts and small RNA sequencing. Rna. 2011;17(8):1489–1501. doi: 10.1261/rna.2442211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426(6968):845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 40.Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, Gao FB. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell stem cell. 2010;6(4):323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes & Development. 2006;20(20):2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krichevsky AM. A microRNA array reveals extensive regulation of microRNAs during brain development. Rna. 2003;9(10):1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woltering JM, Durston AJ. MiR-10 represses HoxB1a and HoxB3a in zebrafish. PLoS ONE. 2008;3(1):e1396. doi: 10.1371/journal.pone.0001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 Promotes Neuronal Differentiation by Triggering Brain-Specific Alternative Pre-mRNA Splicing. Molecular Cell. 2007;27(3):435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lippi G, Steinert JR, Marczylo EL, D’Oro S, Fiore R, Forsythe ID, Schratt G, Zoli M, Nicotera P, Young KW. Targeting of the Arpc3 actin nucleation factor by miR-29a/b regulates dendritic spine morphology. The Journal of Cell Biology. 2011;194(6):889–904. doi: 10.1083/jcb.201103006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439(7074):283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 47.Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21(5):531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu R, Liu K, Liu Y, Mo W, Flynt AS, Patton JG, Kar A, Wu JY, He R. The role of miR-124a in early development of the Xenopus eye. Mechanisms of development. 2009;126(10):804–816. doi: 10.1016/j.mod.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanuki R, Onishi A, Koike C, Muramatsu R, Watanabe S, Muranishi Y, Irie S, Uneo S, Koyasu T, Matsui R, Cherasse Y, Urade Y, Watanabe D, Kondo M, Yamashita T, Furukawa T. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nature Neuroscience. 2011;14(9):1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- 50.Somel M, Guo S, Fu N, Yan Z, Hu HY, Xu Y, Yuan Y, Ning Z, Hu Y, Menzel C, Hu H, Lachmann M, Zeng R, Chen W, Khaitovich P. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Research. 2010;20(9):1207–1218. doi: 10.1101/gr.106849.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kato M, Chen X, Inukai S, Zhao H, Slack FJ. Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. Rna. 2011;17(10):1804–1820. doi: 10.1261/rna.2714411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kapsimali M, Kloosterman WP, de Bruijn E, Rosa F, Plasterk RHA, Wilson SW. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biology. 2007;8(8):R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inukai S, de Lencastre A, Turner M, Slack F. Novel microRNAs differentially expressed during aging in the mouse brain. PLoS ONE. 2012;7(7):e40028. doi: 10.1371/journal.pone.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baroukh NN, Van Obberghen E. Function of microRNA-375 and microRNA-124a in pancreas and brain. FEBS Journal. 2009;276(22):6509–6521. doi: 10.1111/j.1742-4658.2009.07353.x. [DOI] [PubMed] [Google Scholar]
- 55.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nature Neuroscience. 2009;12(4):399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Favre G, Banta Lavenex P, Lavenex P. miRNA regulation of gene expression: a predictive bioinformatics analysis in the postnatally developing monkey hippocampus. PLoS One. 2012;7(8):e43435. doi: 10.1371/journal.pone.0043435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Somel M, Liu X, Tang L, Yan Z, Hu H, Guo S, Jiang X, Zhang X, Xu G, Xie G, Li N, Hu Y, Chen W, Paabo S, Khaitovich P. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS biology. 2011;9(12):e1001214. doi: 10.1371/journal.pbio.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Erraji-Benchekroun L, Underwood MD, Arango V, Galfalvy H, Pavlidis P, Smyrniotopoulos P, Mann JJ, Sibille E. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol Psychiatry. 2005;57(5):549–558. doi: 10.1016/j.biopsych.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 59.Kumar A, Gibbs J, Beilina A, Dillman A, Kumaran R, Trabzuni D, Ryten M, Walker R, Smith C, Traynor B, Hardy J, Singleton A, Cookson M. Age-associated changes in gene expression in human brain and isolated neurons. Neurobiology of aging. 2012 doi: 10.1016/j.neurobiolaging.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee C, Weindruch R, Prolla T. Gene-expression profile of the ageing brain in mice. Nature genetics. 2000;25(3):294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 61.Loerch P, Lu T, Dakin K, Vann J, Isaacs A, Geula C, Wang J, Pan Y, Gabuzda D, Li C, Prolla T, Yankner B. Evolution of the aging brain transcriptome and synaptic regulation. PloS one. 2008;3(10) doi: 10.1371/journal.pone.0003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429(6994):883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 63.Li N, Bates D, An J, Terry D, Wang E. Up-regulation of key microRNAs, and inverse down-regulation of their predicted oxidative phosphorylation target genes, during aging in mouse brain. Neurobiology of aging. 2011;32(5):944–955. doi: 10.1016/j.neurobiolaging.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 64.Noren Hooten N, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman A, Evans M. icroRNA expression patterns reveal differential expression of target genes with age. PloS one. 2010;5(5) doi: 10.1371/journal.pone.0010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Somel M, Guo S, Fu N, Yan Z, Hu H, Xu Y, Yuan Y, Ning Z, Hu Y, Menzel C, Hu H, Lachmann M, Zeng R, Chen W, Khaitovich P. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome research. 2010;20(9):1207–1218. doi: 10.1101/gr.106849.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lehrbach NJ, Castro C, Murfitt KJ, Abreu-Goodger C, Griffin JL, Miska EA. Post-developmental microRNA expression is required for normal physiology, and regulates aging in parallel to insulin/IGF-1 signaling in C. elegans. RNA. 2012;18(12):2220–2235. doi: 10.1261/rna.035402.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vlachos IS, Kostoulas N, Vergoulis T, Georgakilas G, Reczko M, Maragkakis M, Paraskevopoulou MD, Prionidis K, Dalamagas T, Hatzigeorgiou AG. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic acids research. 2012;40(Web Server issue):W498–504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brookmeyer R, Gray S, Kawas C. Projections of Alzheimer’s disease in the United States and the public health impact of delaying disease onset. Am J Public Health. 1998;88(9):1337–1342. doi: 10.2105/ajph.88.9.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McPherson S, Back C, Buckwalter JG, Cummings JL. Gender-related cognitive deficits in Alzheimer’s disease. Int Psychogeriatr. 1999;11(2):117–122. doi: 10.1017/s1041610299005670. [DOI] [PubMed] [Google Scholar]
- 70.Weissman MM, Bland RC, Canino GJ, Faravelli C, Greenwald S, Hwu HG, Joyce PR, Karam EG, Lee CK, Lellouch J, Lepine JP, Newman SC, Rubio-Stipec M, Wells JE, Wickramaratne PJ, Wittchen H, Yeh EK. Cross-national epidemiology of major depression and bipolar disorder. JAMA. 1996;276(4):293–299. [PubMed] [Google Scholar]
- 71.Hebert S, Horre K, Nicolai L, Papadopoulou A, Mandemakers W, Silahtaroglu A, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(17):6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shioya M, Obayashi S, Tabunoki H, Arima K, Saito Y, Ishida T, Satoh J. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathology and applied neurobiology. 2010;36(4):320–330. doi: 10.1111/j.1365-2990.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- 73.Wang WX, Huang Q, Hu Y, Stromberg AJ, Nelson PT. Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 2011;121(2):193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang X, Liu P, Zhu H, Xu Y, Ma C, Dai X, Huang L, Liu Y, Zhang L, Qin C. miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer’s disease, inhibits bcl2 translation. Brain research bulletin. 2009;80(4–5):268–273. doi: 10.1016/j.brainresbull.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 75.Zhu HC, Wang LM, Wang M, Song B, Tan S, Teng JF, Duan DX. MicroRNA-195 downregulates Alzheimer’s disease amyloid-β production by targeting BACE1. Brain research bulletin. 2012;88(6):596–601. doi: 10.1016/j.brainresbull.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 76.Holsinger R, McLean C, Beyreuther K, Masters C, Evin G. Increased expression of the amyloid precursor beta-secretase in Alzheimer’s disease. Annals of neurology. 2002;51(6):783–786. doi: 10.1002/ana.10208. [DOI] [PubMed] [Google Scholar]
- 77.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28(53):14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chi S, Zang J, Mele A, Darnell R. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460(7254):479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Geekiyanage H, Jicha GA, Nelson PT, Chan C. Blood serum miRNA: non-invasive biomarkers for Alzheimer’s disease. Exp Neurol. 2012;235(2):491–496. doi: 10.1016/j.expneurol.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sheinerman KS, V, Tsivinsky G, Crawford F, Mullan MJ, Abdullah L, Umansky SR. Plasma microRNA biomarkers for detection of mild cognitive impairment. Aging (Albany NY) 2012;4(9):590–605. doi: 10.18632/aging.100486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Byers AL, Yaffe K, Covinsky KE, Friedman MB, Bruce ML. High occurrence of mood and anxiety disorders among older adults: The National Comorbidity Survey Replication. Arch Gen Psychiatry. 2010;67(5):489–496. doi: 10.1001/archgenpsychiatry.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Colantuoni C, Hyde T, Mitkus S, Joseph A, Sartorius L, Aguirre C, Creswell J, Johnson E, Deep-Soboslay A, Herman M, Lipska B, Weinberger D, Kleinman J. Age-related changes in the expression of schizophrenia susceptibility genes in the human prefrontal cortex. Brain structure & function. 2008;213(1–2):255–271. doi: 10.1007/s00429-008-0181-5. [DOI] [PubMed] [Google Scholar]
- 83.Steffens DC, Fisher GG, Langa KM, Potter GG, Plassman BL. Prevalence of depression among older Americans: the Aging, Demographics and Memory Study. Int Psychogeriatr. 2009;21(5):879–888. doi: 10.1017/S1041610209990044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lugli G, Larson J, Demars MP, Smalheiser NR. Primary microRNA precursor transcripts are localized at post-synaptic densities in adult mouse forebrain. J Neurochem. 2012;123(4):459–466. doi: 10.1111/j.1471-4159.2012.07921.x. [DOI] [PubMed] [Google Scholar]
- 85.Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem. 2005;94(4):896–905. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- 86.Lugli G, V, Torvik I, Larson J, Smalheiser NR. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem. 2008;106(2):650–661. doi: 10.1111/j.1471-4159.2008.05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haramati S, Navon I, Issler O, Ezra-Nevo G, Gil S, Zwang R, Hornstein E, Chen A. MicroRNA as repressors of stress-induced anxiety: the case of amygdalar miR-34. J Neurosci. 2011;31(40):14191–14203. doi: 10.1523/JNEUROSCI.1673-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, Willoughby D, Kenny PJ, Elsworth JD, Lawrence MS, Roth RH, Edbauer D, Kleiman RJ, Wahlestedt C. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci U S A. 2012;109(8):3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Green M, Cairns M, Wu J, Dragovic M, Jablensky A, Tooney P, Scott R, Carr V. Genome-wide supported variant MIR137 and severe negative symptoms predict membership of an impaired cognitive subtype of schizophrenia. Molecular psychiatry. 2012 doi: 10.1038/mp.2012.84. [DOI] [PubMed] [Google Scholar]
- 90.Magill S, Cambronne X, Luikart B, Lioy D, Leighton B, Westbrook G, Mandel G, Goodman R. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(47):20382–20387. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mellios N, Sugihara H, Castro J, Banerjee A, Le C, Kumar A, Crawford B, Strathmann J, Tropea D, Levine S, Edbauer D, Sur M. miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nature neuroscience. 2011;14(10):1240–1242. doi: 10.1038/nn.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pathania M, Torres-Reveron J, Yan L, Kimura T, Lin T, Gordon V, Teng Z-Q, Zhao X, Fulga T, Van Vactor D, Bordey A. miR-132 enhances dendritic morphogenesis, spine density, synaptic integration, and survival of newborn olfactory bulb neurons. PloS one. 2012;7(5) doi: 10.1371/journal.pone.0038174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Earls LR, Fricke RG, Yu J, Berry RB, Baldwin LT, Zakharenko SS. Age-dependent microRNA control of synaptic plasticity in 22q11 deletion syndrome and schizophrenia. J Neurosci. 2012;32(41):14132–14144. doi: 10.1523/JNEUROSCI.1312-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smalheiser NR, Lugli G, Rizavi HS, Torvik VI, Turecki G, Dwivedi Y. MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS One. 2012;7(3):e33201. doi: 10.1371/journal.pone.0033201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vreeburg SA, Hoogendijk WJ, van Pelt J, Derijk RH, Verhagen JC, van Dyck R, Smit JH, Zitman FG, Penninx BW. Major depressive disorder and hypothalamic-pituitary-adrenal axis activity: results from a large cohort study. Arch Gen Psychiatry. 2009;66(6):617–626. doi: 10.1001/archgenpsychiatry.2009.50. [DOI] [PubMed] [Google Scholar]
- 96.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 97.Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nat Rev Genet. 2008;9(12):911–922. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V. The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev Biol. 2002;244(1):170–179. doi: 10.1006/dbio.2002.0594. [DOI] [PubMed] [Google Scholar]
- 99.Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev Biol. 2003;259(1):9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 100.Garen A, Kauvar L, Lepesant JA. Roles of ecdysone in Drosophila development. Proc Natl Acad Sci U S A. 1977;74(11):5099–5103. doi: 10.1073/pnas.74.11.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Beato M. Gene regulation by steroid hormones. Cell. 1989;56(3):335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 102.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Klinge CM. miRNAs and estrogen action. Trends Endocrinol Metab. 2012;23(5):223–233. doi: 10.1016/j.tem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 105.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Paris O, Ferraro L, Grober OM, Ravo M, De Filippo MR, Giurato G, Nassa G, Tarallo R, Cantarella C, Rizzo F, Di Benedetto A, Mottolese M, Benes V, Ambrosino C, Nola E, Weisz A. Direct regulation of microRNA biogenesis and expression by estrogen receptor beta in hormone-responsive breast cancer. Oncogene. 2012;31(38):4196–4206. doi: 10.1038/onc.2011.583. [DOI] [PubMed] [Google Scholar]
- 107.Fiedler SD, Carletti MZ, Hong X, Christenson LK. Hormonal regulation of MicroRNA expression in periovulatory mouse mural granulosa cells. Biol Reprod. 2008;79(6):1030–1037. doi: 10.1095/biolreprod.108.069690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mishima T, Takizawa T, Luo SS, Ishibashi O, Kawahigashi Y, Mizuguchi Y, Ishikawa T, Mori M, Kanda T, Goto T. MicroRNA (miRNA) cloning analysis reveals sex differences in miRNA expression profiles between adult mouse testis and ovary. Reproduction. 2008;136(6):811–822. doi: 10.1530/REP-08-0349. [DOI] [PubMed] [Google Scholar]
- 109.Burney RO, Hamilton AE, Aghajanova L, Vo KC, Nezhat CN, Lessey BA, Giudice LC. MicroRNA expression profiling of eutopic secretory endometrium in women with versus without endometriosis. Mol Hum Reprod. 2009;15(10):625–631. doi: 10.1093/molehr/gap068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nothnick WB, Healy C. Estrogen induces distinct patterns of microRNA expression within the mouse uterus. Reprod Sci. 2010;17(11):987–994. doi: 10.1177/1933719110377472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pan Q, Luo X, Chegini N. Differential expression of microRNAs in myometrium and leiomyomas and regulation by ovarian steroids. J Cell Mol Med. 2008;12(1):227–240. doi: 10.1111/j.1582-4934.2007.00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 112.Pan Q, Luo X, Toloubeydokhti T, Chegini N. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol Hum Reprod. 2007;13(11):797–806. doi: 10.1093/molehr/gam063. [DOI] [PubMed] [Google Scholar]
- 113.Narayanan R, Jiang J, Gusev Y, Jones A, Kearbey JD, Miller DD, Schmittgen TD, Dalton JT. MicroRNAs are mediators of androgen action in prostate and muscle. PLoS One. 2010;5(10):e13637. doi: 10.1371/journal.pone.0013637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bizuayehu TT, Babiak J, Norberg B, Fernandes JM, Johansen SD, Babiak I. Sex-biased miRNA expression in Atlantic halibut (Hippoglossus hippoglossus) brain and gonads. Sex Dev. 2012;6(5):257–266. doi: 10.1159/000341378. [DOI] [PubMed] [Google Scholar]
- 115.Morgan CP, Bale TL. Early prenatal stress epigenetically programs dysmasculinization in second-generation offspring via the paternal lineage. J Neurosci. 2011;31(33):11748–11755. doi: 10.1523/JNEUROSCI.1887-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Koturbash I, Zemp F, Kolb B, Kovalchuk O. Sex-specific radiation-induced microRNAome responses in the hippocampus, cerebellum and frontal cortex in a mouse model. Mutat Res. 2011;722(2):114–118. doi: 10.1016/j.mrgentox.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 117.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(Database issue):D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mellios N, Galdzicka M, Ginns E, Baker SP, Rogaev E, Xu J, Akbarian S. Gender-specific reduction of estrogen-sensitive small RNA, miR-30b, in subjects with schizophrenia. Schizophrenia bulletin. 2012;38(3):433–443. doi: 10.1093/schbul/sbq091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Feng J, Sun G, Yan J, Noltner K, Li W, Buzin CH, Longmate J, Heston LL, Rossi J, Sommer SS. Evidence for X-chromosomal schizophrenia associated with microRNA alterations. PloS one. 2009;4(7):e6121. doi: 10.1371/journal.pone.0006121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Siegel C, Li J, Liu F, Benashski SE, McCullough LD. miR-23a regulation of X-linked inhibitor of apoptosis (XIAP) contributes to sex differences in the response to cerebral ischemia. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(28):11662–11667. doi: 10.1073/pnas.1102635108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stauffer BL, Sobus RD, Sucharov CC. Sex differences in cardiomyocyte connexin43 expression. Journal of cardiovascular pharmacology. 2011;58(1):32–39. doi: 10.1097/FJC.0b013e31821b70b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, Chen G, Wang Z. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nature medicine. 2007;13(4):486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]