Abstract
DNA methylation is important for gene regulation and is vulnerable to early-life exposure to environmental contaminants. We found that direct waterborne benzo[a]pyrene (BaP) exposure at 24 μg/L from 2.5 to 96 hours post fertilization (hpf) to zebrafish embryos significantly decreased global cytosine methylation by 44.8% and promoter methylation in vasa by 17%. Consequently, vasa expression was significantly increased by 33%. In contrast, BaP exposure at environmentally relevant concentrations did not change CpG island methylation or gene expression in cancer genes such as ras-association domain family member 1 (rassf1), telomerase reverse transcriptase (tert), c-jun, and c-myca. Similarly, BaP did not change gene expression of DNA methyltransferase 1 (dnmt1) and glycine N-methyltransferase (gnmt). While total DNMT activity was not affected, GNMT enzyme activity was moderately increased. In summary, BaP is an epigenetic modifier for global and gene specific DNA methylation status in zebrafish larvae.
Keywords: Benzo[a]pyrene, DNA methylation, zebrafish, vasa
1. Introduction
Many drugs and environmental chemicals are being identified as epigenetic modifiers that can cause epimutations, particularly by affecting DNA methylation patterns (Bollati and Baccarelli 2010). In vitro studies have shown that benzo[a]pyrene (BaP), a carcinogen and developmental and reproductive toxicant, is also an epigenetic modifier. In the early 1980s, a few studies used BaP and its mutagenic metabolite anti-7β,8α-dihydroxy-9α,10α-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene (BPDE) to investigate the modulation of DNA methylation in vitro. They found that modification of DNA with BPDE resulted in impairment of DNA methyltransferase (DNMT) activity (Pfeifer et al. 1984; Wilson and Jones 1983; Wilson and Jones 1984; Wojciechowski and Meehan 1984). Furthermore, treatment of murine cell lines with BaP decreased the DNA 5-methylcytosine content (Wilson and Jones 1983). BPDE preferentially binds to methylated DNA (Augoustides-Savvopoulou et al. 2003; Denissenko et al. 1996; Hu et al. 2003). Two more recent studies described BaP-induced hypo- and hypermethylation events in in vitro cell line models (Sadikovic et al. 2004; Sadikovic and Rodenhiser 2006). Despite no alteration in mRNA expression of dnmt1, dnmt3a, or dnmt13b by BPDE, there was an increased expression of DNMT1 protein and promoter hypermethylation of several genes of a panel of 30 genes analyzed in immortalized bronchial epithelial cells (Damiani et al. 2008). However, when non-transformed cells were treated with BPDE in vitro, no significant change in methylation status was noted (Tommasi et al. 2010).
All the above studies were performed in cell lines in vitro with high concentrations of BaP or BPDE. Studies in human cohorts exposed to PAHs are suggesting further relationships between exposure and potential altered methylation status of offspring (Breton et al. 2009; Herbstman et al. 2012; Joubert et al. 2012; Suter et al. 2011). In the study described here, an in vivo model, zebrafish, was used to investigate BaP effects on DNA methylation during early development. We exposed zebrafish embryos to environmentally relevant concentrations of waterborne BaP and measured both global and gene specific DNA methylation of five developmentally important genes, namely vasa, ras-association domain family member 1 (rassf1), telomerase reverse transcriptase (tert), c-jun and c-myca. In addition, gene expression and enzyme activity of two methylation related enzymes, DNMT1 and glycine N-methyltransferase (GNMT), were measured. BaP significantly reduced DNA methylation globally and gene-specifically in vasa promoter but not the other target genes. GNMT, but not DNMT, activity was increased by BaP exposure.
2. Methods
2.1 Zebrafish care
AB line wild-type zebrafish were purchased from Zebrafish International Resource Center (ZFIN, Eugene, OR) and raised according to the IACUC approved conditions. Fish were kept in Aquatic Habitats ZF0601 Zebrafish Stand-Alone System (Aquatic Habitats, Apopka, FL). Culture water characteristics were: pH of 7.0-7.5; salinity of 60 parts per million (ppm, Instant Ocean, Cincinnati, OH); and temperature of 24-30°C. The light-dark cycle was 14:10. Adult fish were fed twice daily with both tropical flake fish food (TetraMin, Tetra Werke, Germany) and live brine shrimp (Salt Creek Inc., Salt Lake City, UT). Sexually mature fish were selected as breeders and their eggs were collected for the studies described below.
2.2 Zebrafish embryo BaP exposure
Fertilized eggs were cleaned and disinfected with 0.4 ppm methylene blue for 1 minute and then randomly sorted into four treatment groups (3-6 replicates per group), namely control dimethylsulfoxide (DMSO, 1 μl/mL), 0.24, 2.4 and 24 μg/L BaP (stock solution 23.7 ± 0.9 μg/mL in DMSO; final DMSO concentration was 1 μl/mL in all treatment groups). BaP concentration in the stock was confirmed by gas chromatography (Agilent 6890) coupled with mass spectrometry (Agilent 5973N) in selected ion monitoring mode for ions 252 and 253. Thirty fertilized eggs were pooled randomly and raised in 10 ml of zebrafish water in glass vials. Exposures for each experimental treatment began at approximately 2.5 hpf. Water was changed and eggs were re-dosed daily. Survival rate and hatching efficiency were measured daily until 72 hpf. Embryos or larvae were collected at different time-points (embryos would typically hatch at 48-72 hpf).
For DNA extraction, larvae were put in 100% ethanol, snap frozen in liquid nitrogen and kept in a −80°C freezer. For RNA extraction, larvae were stored in RNAlater at −80°C immediately. For GNMT and DNMT enzyme activity assays, larvae were frozen at −80°C in 1 ml HEGD buffer (HEPES 10 mM, EDTA 1.5 mM, glycerol 10% v/v, DTT 1 mM, PMSF 0.5 mM and pH 8.0).
2.3 Global DNA methylation measurement
Genomic DNA was isolated from zebrafish larvae by using DNeasy Blood & Tissue Easy Kit (Qiagen, Valencia, CA) according to manufacturer’s protocol. DNA samples were treated with RNase A to remove RNA contaminant. The Methylamp™ Global DNA Methylation Quantification Kit (Epigentek Group, Farmingdale, NY) was used to quantitate the percentage of methylated cytosine in the genomic DNA. The methylated cytosine amount was in proportion to the OD intensity at 450 nm measured with a HTS 7000 Bioassay Reader (Perkin Elmer, Waltham, MA). The cytosine methylation percentage was calculated by using the formula: cytosine methylation % = (OD (sample-negative control) / 36.5%) / (OD (positive control-negative control) × 10); 36.5% is the GC content in zebrafish genome (Han and Zhao 2008); and 10 was the dilution factor for the positive control. Each sample was measured in duplicate.
2.4 Sodium bisulfite sequencing
Genomic DNA was treated with sodium metabisulfite, as described (Raizis et al. 1995). Desulphonation, purification, and recovery of DNA were done with EZ Bisulfite DNA Clean-up Kit (ZYMO Research). Bisulfite specific primers were designed with Methyl Primer Express v1.0 (Applied Biosystems, Foster City, CA) or Methprimer (Table 1). Target genes were amplified from bisulfite converted DNA by PCR with ZymoTaq™ PreMix (Hot start DNA taq polymerase, ZYMO Research). PCR products were ligated into pGEM® T Easy Vector System (Promega, Madison, WI) and transformed into DH5α E.coli competent cells (Invitrogen, Grand Island, NY). Each treatment had three biological samples and at least eight white colonies from each sample were selected for sequencing. Plasmid DNA was isolated in 96-well format using a modified alkaline-lysis method and then sequenced in the ABI 3730xl sequencer using BigDye v3.1 Terminator/Buffer Ready Rxn Cycle Sequencing kit (Applied Biosystems). Sequence data was analyzed with DNAstar (SeqMan, Madison, WI). CpG methylation percentage was calculated as (total number of methylated CpG) / (number of CpG sites in each gene × number of colonies sequenced).
Table 1.
Primers used in this study.
| Gene (Genbank accession) |
Experiment | Primer (5′-3′) | Product size (bp) |
Comments |
|---|---|---|---|---|
|
gnmt
(NM_212816.1) |
qPCR | F: TCATTGACCACCGCAACTATG R: GTAGTCCAGTGTGATCATGTG |
149 | Spans exon 4 and 5. |
|
dnmt1
(NM_131189.1) |
qPCR | F: GGGCTACCAGTGCACCTTTG R: GATGATAGCTCTGCGTCGAGTC |
76 | Spans exon 26 and 27. |
|
vasa
(NM_131057) |
qPCR | F: CAGACAAGTTGGATCAAGAAGGAA R: GGCGGCGGCACATAAAC |
64 | Spans exon 15 and 16. |
|
rassf1 (total)
(NM_001004550.1) |
qPCR | F: TGTGCTGGACCCAATGAGAA R: TTATCACGAGCCAGAGTGTATCG |
176 | Spans exon 5 and 6. |
|
rassf1-001
(NM_003335970.1) |
qPCR | F: AGCGACAACGCGAATGAAG R: GGCAGAAAGATGAATTCCTCAAG |
60 | Within exon 1 |
|
tert
(NM_001083866) |
qPCR | F: GAAAGCCGGTTCTGCTGGA R: AGAGTGAACGCCAAGACCTC |
142 | Span exon 1 and 2. |
|
c-jun
(NM_199987.1) |
qPCR | F: CGCTTTCTCTCAGCATGACAGT R: GATTGAGCGTCATGTTGTGTTTC |
74 | Does not span exons. |
|
c-myca
(NM_131412.1) |
qPCR | F: GAAACAATTCTGGAACGGCATT R: AGGTTGAGTCTGTCCTTGCTGAT |
168 | Spans exon 1 and 2. |
|
18S rRNA
(FJ915075.1) |
qPCR | F: TGGTTAATTCCGATAACGAACGA R: CGCCACTTGTCCCTCTAAGAA |
95 | Internal control. |
|
vasa
a (NM_131057) |
BS-seq | F: ATTGGGAATTTTAGTAGTATATTGATAGT R:TTTAATTTAAAACCTCATTAATCTAAATCA |
207 | Contains 5 CpGs in the promoter |
|
rassf1-CGI 1
(NM_001004550.1) |
BS-seq | F: TTGTTTTTTTAAGAATGAGTTTAAT R: AACAAAAAAATAAATTCCTCAAA |
314 | Contains 26 CpGs crosses the TSS1. |
|
rassf1-CGI 2
(NM_001004550.1) |
BS-seq | F: AGTGATTTATGTTTGTGGATATGTAA R: TATACAAACCCCAAATAAACTCTCC |
438 | Contains 17 CpGs crosses the TSS2. |
|
tert
a (NM_001083866) |
BS-seq | F: ATAGTAGGATAGGGTTTTGGTTTTG R: CCTTCAATTCTTCAAAAATTAACTC |
233 | Contains 13 CpGs in the promoter. |
|
c-jun
(NM_199987.1) |
BS-seq | F: TGAGGGTATTGGTTGAGTTGTA R: AAAAATCCTCTCCAATTTCCTCT |
425 | Contains 36 CpGs in the gene. |
|
c-myca
(NM_131412.1) |
BS-seq | F: TAAGTGTTAAAATGTTGGTGAGTG R:TACTATCAAACATCAATTCCTTCC |
444 | Contains 26 CpGs in the gene. |
2.5 Quantitative reverse transcription real time PCR (qPCR)
RNA was isolated with RNAzol (Molecular Research Center, Cincinnati, OH) and purified with RNeasy Mini Kit (Qiagen, Valencia, CA) by following the manufacturer’s protocols. Total RNA (250 ng) was reverse transcribed to double stranded cDNA libraries by using TaqMan® Reverse Transcription Reagents (Applied Biosystems). qPCR primers were designed with Primer Express® Software v2.0 (Applied Biosystems) (Table 1). Relative abundance of target genes to 18S rRNA transcripts was determined by qPCR with SYBR®Green in a GeneAmp 7500 Sequence Detection System (Applied Biosystems). Statistical differences between treatments were determined on the linearized 2−ΔCT values. Amplification efficiencies of the target genes and 18S rRNA primer pairs were tested to ensure that they were not statistically different. In addition to melt curve analyses, all the target gene qPCR products were confirmed by sequencing.
2.6 GNMT and DNMT enzyme activity assay
Cytosolic and nuclear proteins were extracted with NE-PER® Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Waltham, MA) and quantified with Bio-rad Protein Assay using BSA as a standard. The GNMT enzymatic activity in cytosolic extract was determined using the method of Cook and Wagner (1984) with some modifications (Fang et al. 2010), and performed in duplicate. Briefly, the GNMT assay mixture (100 μl) contained 0.1 M Tris HCl (pH 7.4), 5 mM DTT, 10 mM glycine, 1 mM S-adenosyl-L-(methyl-3H) methionine (0.02 μCi/reaction) + cold S-adenosylmethionine (SAM), and 180-250 μg sample protein and was incubated at 25°C for 60 min. Unreacted SAM was removed by the addition of activated charcoal and an aliquot of the subsequent supernatant was subjected to liquid scintillation counting.
Total DNMT enzyme activities (de novo and maintenance) in nuclear extracts were measured with the EpiQuik DNA Methyltransferase Activity/Inhibition Assay Kit (Epigentek Group) by following the manufacturer’s protocol. The OD value was measured with a HTS 7000 Bioassay Reader. Three biological samples were used per treatment and each sample was measured in triplicate.
2.7 Statistical analysis
Results were analyzed with GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA) and presented as mean ± SEM. Statistical differences between treatment groups were determined using student t-test (p < 0.05) or one-way ANOVA followed by Neuman–Keulls post hoc test (p < 0.05). For qPCR, statistical differences between treatments were determined on the linearized 2−ΔCT values. Fold change of gene expression was calculated with the 2−ΔΔCT method.
3. Results
3.1 BaP decreased survival and hatching efficiency in zebrafish embryos
In the 2.4 μg/L BaP treated group, the survival rate was significantly decreased by 38.5% compared to controls (Figure 1A). In the 0.24 and 24 μg/L BaP treated groups, the average survival rates were non-significantly reduced by 25.0%. Death typically occurred within the first 24 hours of exposure. After that period, the survival rate was stable until 72 hpf.
Figure 1.
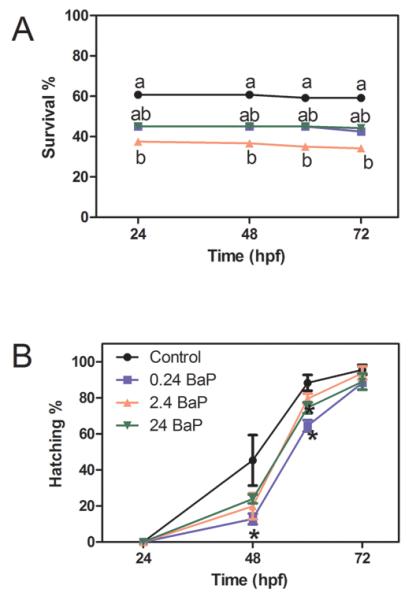
BaP effects on the embryo survival rate (A) and hatching efficiency (B). Embryos were exposed to waterborne BaP (0-24 μg/L) from 2.5 to 72 hpf (n=4 pools, 30 embryos/pool; *, P<0.05; different letters indicate significantly different).
At 48 hpf, fewer embryos hatched in the 0.24 μg/L BaP treated groups compared to control (Figure 1B). At 60 hpf, fewer embryos hatched in the groups treated with 0.24 or 24 μg/L BaP. However, there were no differences in the hatching percentages among all the treatment groups at 72 hpf. Therefore, although BaP delayed zebrafish hatching, the overall hatching percentage was not affected. Deformities were observed in BaP treated larvae, but the incidence was not statistically different from control (data not shown).
3.2 BaP decreased global DNA methylation
Constitutively, 5.2% of cytosines were methylated in zebrafish at 96 hpf. BaP exposure at 2.4 μg/L for 96 hours reduced 5-methylcytosine content by 25.0% compared to control methylation (Figure 2), which was significantly different from control by using student t-test (P=0.04). BaP at 24 μg/L significantly reduced 5-methylcytosine content by 44.8% compared to the control group and by 36.6% compared to the 0.24 μg/L BaP treated group (P=0.02, one-way ANOVA and Neumann Keuls multiple comparison tests).
Figure 2.
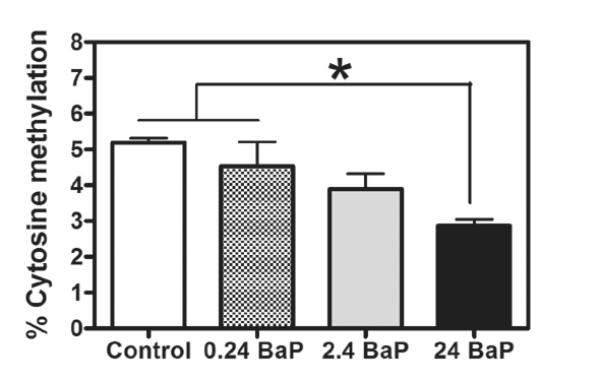
BaP effects on global DNA methylation in zebrafish larvae. Methylation was measured with the Methylamp™ Global DNA Methylation Quantification Kit (Epigentek Group). Embryos were exposed to waterborne BaP (0-24 μg/L) from 2.5 to 96 hpf (* p<0.05, n=3 pools, 20 embryos/pool).
3.3 BaP decreased vasa promoter methylation and increased vasa gene expression
Waterborne 24 μg/L BaP exposure from 2.5 to 96 hpf significantly decreased the overall methylation percentage by 17.2% in the five CpG sites measured in vasa promoter (Figure 3A and Supplemental Figure 1A). The reduction of methylation was not specific to any single CpG site (Supplemental Figure 1B). Consistent with promoter DNA demethylation, BaP exposure at 24 μg/L from 2.5 to 96 hpf significantly increased the vasa mRNA expression by 33.0% (Figure 3B).
Figure 3.
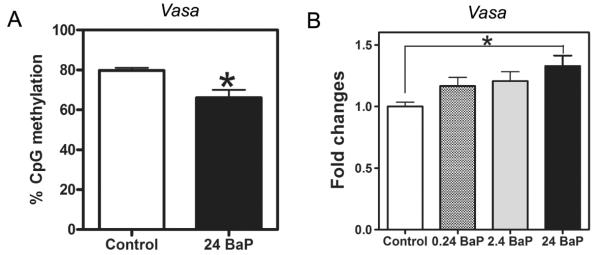
BaP effects on vasa promoter methylation percentage (A) and gene expression (B). Fold change of vasa expression was normalized to 18S rRNA expression and relative to controls (n=6 pools, 20-30 embryos/pool; * P<0.05).
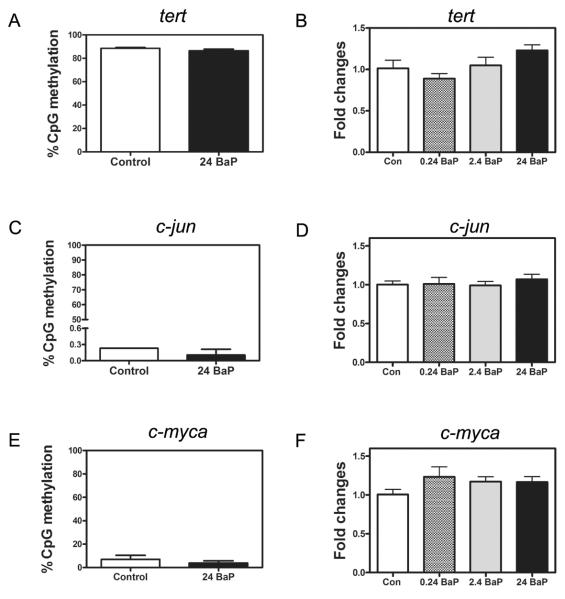
3.4 BaP effects on methylation and gene expression of rassf1, tert, c-jun and c-myca
BaP treatment (24 μg/L) did not significantly change CpG methylation patterns or percentage in rassf1 CGI-1 (Figure 4A and Supplemental Figure 2A), rassf1 CGI-2 (Figure 4B and Supplemental Figure 2B), tert (Figure 5A and Supplemental Figure 3), c-jun (Figure 5C and Supplemental Figure 4) or c-myca (Figure 5E and Supplemental Figure 5). Gene expression of total rassf1 (Figure 4C), tert (Figure 5B), c-jun (Figure 5D) and c-myca (Figure 5F) was also not changed in the zebrafish that were exposed to BaP. However, the transcript rassf1-001 was significantly increased by 49% by BaP at 24 μg/L (Figure 4D).
Figure 4.
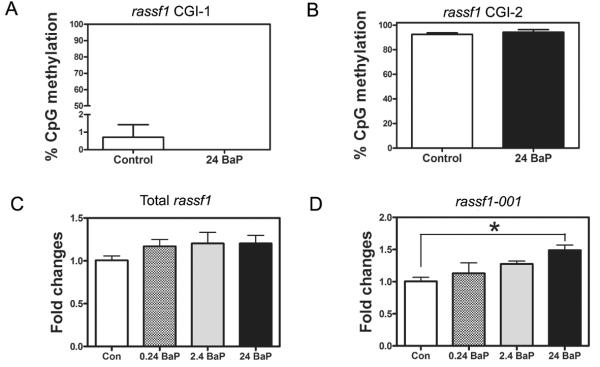
BaP effects on rassf1 DNA methylation percentage and gene expression. (A) and (B) show the methylation percentage of CpG island 1 (CGI 1) and CGI 2, which are located in the first and second promoter, respectively. (C) and (D) show the gene expression of total rassf1 and rassf1-001, respectively. Fold change of gene expression was normalized to 18S rRNA expression and relative to control gene expression (n=3 pools, 20 larvae/pool).
Figure 5.
BaP effects on the methylation percentage of tert (A), c-jun (C), and c-myca (E) and on the gene expression of tert (B), c-jun (D), and c-myca (F). Gene expression was normalized to 18S rRNA expression and relative to control gene expression (n=3 pools, 20 larvae/pool).
3.5 BaP effects on gene expression and enzyme activity of DNMT1 and GNMT
Neither expression of dnmt1 (Figure 6A) nor gnmt (Figure 6B) was affected in zebrafish larvae that were exposed to BaP from 2.5 to 48, 60 or 96 hpf. The total DNMT enzyme activity includes the activities from all DNMT iso-zymes in zebrafish, namely DNMT 1, 3, 4, 5, 6, 7 and 8. BaP exposure did not change the total DNMT enzyme activity in the nuclear extracts of larvae at 96 hpf (Figure 7A). BaP at 0.24 μg/L increased GNMT activity by ~2-fold compared to the activity in control zebrafish at 96 hpf (Figure 7B). This induction was significantly different from control by using student t-test (P=0.035). BaP exposure at 2.4 and 24 μg/L non-significantly increased GNMT enzyme activity by 51% and 75%, respectively.
Figure 6.
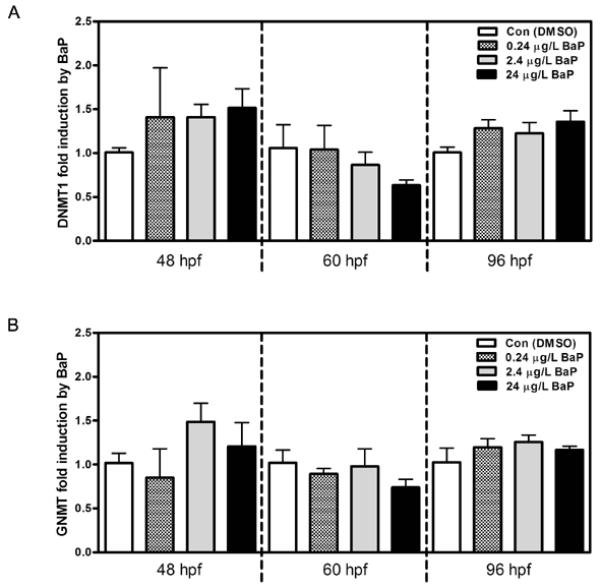
BaP effects on dnmt1 (A) and gnmt (B) mRNA expression in zebrafish embryos at 48, 60 and 96 hpf. Each sample was measured by qPCR in duplicate. Bars ± SEM indicate fold change of gene expression normalized to 18S rRNA expression and relative to individual time-point controls (n=3 pools, 20-30 embryos/pool).
Figure 7.
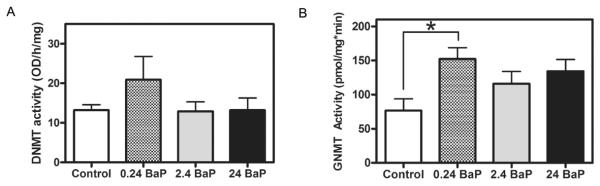
BaP effects on total DNMT (A) and GNMT (B) activity in zebrafish larvae at 96 hpf. Enzyme activity assay was tested in 3 times for each biological sample. Each bar ± SEM represents n = 3 pools, 20 embryos/pool (* P<0.05).
4. Discussion
BaP is teratogenic to zebrafish embryos with LC50 of 5.1 μM (1285 μg/L) and EC50 (the dosage that causes teratogenic changes in 50% of the animals) of 0.52 μM (131 μg/L) (Weigt et al. 2011). The pure water solubility of BaP ranges from 2.3 (Agency for Toxic Substances and Disease Registry (ATSDR 1995)) to 4 μg/L (Mackay and Shiu 1977). However, BaP is highly lipophilic and likely to accumulate to higher concentrations in the lipid-rich ova. In this study, we used environmentally relevant concentrations of BaP at 0.24, 2.4 and 24 μg/L for all the embryonic exposures. Decreased survival rate and delayed embryo hatching indicated that BaP caused adverse developmental consequences as has been previously reported in BaP-exposed zebrafish (Weigt et al. 2011; Incardona et al. 2011). Similarly, in mice, in utero exposure to BaP leads to long-term adverse effects including cardiovascular dysfunction (Jules et al. 2012), ovotoxicity (Sobinoff et al. 2012), testicular malformation (Mohamed et al. 2010), and cancer (Turusov et al. 1990).
Increasing evidence shows that prenatal exposure to different chemicals leads to abnormal DNA methylation patterns (Heindel 2006; Singh and Li 2012). In this study, BaP caused global loss of DNA methylation in the exposed zebrafish larvae (Figure 2). This demethylation effect supports reports of DNA hypomethylation triggered by air pollution or cigarette smoke both of which can contain BaP as one of their major toxic constituents (Baccarelli and Bollati 2009; Ma et al. 2011). DNA hypomethylation may increase the vulnerability to many diseases by altering gene expression, elevating mutation rates, increasing genome instability, or triggering apoptosis (Kisseljova and Kisseljov 2005). Importantly, altered DNA methylation patterns can be stably inherited during DNA replication and can mediate persistent toxicological consequences in subsequent generations (Guerrero-Bosagna and Skinner 2012; Skinner et al. 2012; Szyf 2011; Szyf 2012).
To further study the BaP demethylation effects, five specific genes, i.e. vasa, rassf1, tert, c-jun and c-myca, were selected for bisulfite sequencing analysis. These genes were selected based on three criteria: 1) they are essential for embryogenesis (Coussens et al. 2006; Hilberg et al. 1993; Soucek and Evan 2010), 2) their expression is disrupted by exposure to high doses of BaP (Fields et al. 2004; Qin and Meng 2006), and/or 3) altered methylation or expression of these genes is involved in carcinogenesis (Jochum et al. 2001; Nesbit et al. 1999; Nowak 2000).
Vasa is a germ cell specific gene and its expression can be disrupted by exposure to polycyclic aromatic hydrocarbons (Kee et al. 2010). Effects on embryonic vasa methylation and expression may compromise developmental success and affect fertility in the offspring (Kee et al. 2010). We found that BaP exposure significantly reduced vasa promoter methylation and increased vasa mRNA expression in 96 hpf zebrafish larvae (Figure 3A and 3B). This indicates that demethylation in the vasa promoter is permissive to gene activation, suggesting an inverse relationship of vasa promoter methylation and gene expression. Because vasa is required for primordial germ cell differentiation and migration, the alteration in epigenetic regulation of vasa gene expression may affect the number, distribution, and migration of primordial germ cells, leading to reproductive toxicities (Gruidl et al. 1996; Kuznicki et al. 2000; Li et al. 2009; Weidinger et al. 2003).
Our previous study found that BaP exposure to Fundulus heteroclitus (killifish) larvae leads to CYP1 gene activation and increased incidence of liver lesions and tumors in adulthood (Wang et al. 2010). Altered DNA methylation is implicated in carcinogenesis in zebrafish (Mirbahai et al. 2011a; Mirbahai et al. 2011b). It is critical to investigate whether early life BaP exposure can cause aberrant DNA methylation patterns in cancer-related genes, persistently affect gene expression, and lead to carcinogenesis in later life.
The tumor suppressor gene rassf1 was selected because it is one of the most frequently silenced genes by promoter hypermethylation in many cancer types (Richter et al. 2009; van der Weyden and Adams 2007). Expression of rassf1a is predominantly controlled by DNA methylation, but it also can be inactivated by gene deletions or point mutations (Pan et al. 2005). In mammals, rassf1 has multiple transcript variants that utilize two different CGI promoters (Hesson et al. 2007). Similarly, zebrafish rassf1 has five transcripts that are possibly regulated by three CpG rich promoters. Based on the cDNA alignment with the genomic DNA, it could be predicted that control of rassf1-001 is by the first promoter, rassf1-201 and rassf1-203 is by the second promoter, and rassf1-002 and rassf1-202 is by the third promoter. Because the transcripts that are regulated by the first and second promoters have the highest similarity with human rassf1a and rassf1c, we selected them for DNA methylation analysis. Notably, the first promoter is frequently reported to be hypermethylated in many types of cancers and is responsible for silencing rassf1a (Dammann et al. 2000). On the contrary, the methylation of the second promoter is not affected in cancer, and thus, rassf1c is usually not deactivated (Li et al. 2004; Vos et al. 2000). In this study, BaP did not alter DNA methylation in either of these two promoters. Although the total rassf1 transcription was not changed, rassf1-001, which is the most similar transcript to human rassf1a, was significantly increased by BaP at 24 μg/L. This indicates that other mechanisms besides DNA methylation could be mediating the BaP effects on the expression of rassf1-001.
tert is a catalytic subunit of the enzyme telomerase, which is essential for telomerase activity (Liu et al. 2000). As a proto-oncogene, its upregulation is involved in tumorigenesis (Nowak 2000; Sirera et al. 2011). Epigenetic mechanisms, including DNA methylation and histone modulation, are important for regulation of the expression of tert promoter (Kyo et al. 2008). In this study, however, neither the DNA methylation nor gene expression of tert was affected by the BaP exposure.
Like tert, in zebrafish larvae, in vivo BaP exposure did not change c-jun expression or DNA methylation. c-jun is involved in many different mechanisms of oncogenesis (Jochum et al. 2001; Vogt 2001). c-jun expression is upregulated in chemically-induced tumors through hypomethylation in the c-jun promoter (Tao et al. 2000). Several studies have shown that high concentrations of BaP were able to induce c-jun expression in vitro (primary human macrophages, 2 μM (504.6 μg/L) BaP) (Sparfel et al. 2010) and in vivo (Wistar rats, 3 mg/animal) (Qin and Meng 2006). BPDE activated the JNK pathway and subsequently activated the downstream activity of c-jun (Dreij et al. 2010). Another study found that low dose BPDE (0.05 μM) exposure inhibited c-jun expression in normal human amnion epithelial cells (Lu et al. 2010).
c-myc is a universal transcription regulator that participates in the activation or repression of approximately 10-15% of all genes (Dang et al. 2006; Patel et al. 2004). c-myc is overexpressed in most tumor cells (Adhikary et al. 2005; Leder et al. 1986; Nesbit et al. 1999) and is associated with altered DNA methylation status. For example, in colorectal cancers and hepatocellular carcinomas, upregulation of c-myc is correlated with DNA hypomethylation in the third exon (Sharrard et al. 1992; Shen et al. 1998). In chemical-induced liver tumors, elevated expression of c-myc mRNA and protein was associated with hypomethylation in the c-myc promoter (Tao et al. 2000). Expression of c-myc can be disrupted by BaP or BPDE exposure. For example, in human bronchial epithelial cells, BaP exposure at 0.03-3 μM (7.6-756.9 μg/L) dose-dependently increased c-myc expression (Fields et al. 2004). In human placental choriocarcinoma JEG-3 cells, BaP exposure at 10 μM (2523.1 μg/L) decreased c-myc mRNA expression by 61% (Zhang and Shiverick 1997). Additionally, BPDE exposure from 0.01 to 3 μM decreased c-myc expression in normal human cell lines (Akerman et al. 2004; Lu et al. 2009). Notably, all of these effects were at concentrations significantly higher than used in this study. Lower BaP concentrations can be the reason that the c-myca expression and gene body methylation were not changed (Figure 5E and 5F).
In order to identify the mechanism of BaP induced global hypomethylation, we measured the gene expression and enzyme activity of two important methyltransferases, namely DNMT and GNMT. In zebrafish, eight isozymes of DNMT are known. All the isozymes except DNMT2 are involved in DNA methylation. In this study, we focused on gene expression of the best known isozyme in zebrafish, which is dnmt1. In embryos/larvae exposed to BaP, alteration of dnmt1 mRNA expression was not seen (Figure 6A). Similarly, total DNMT activity (including the seven DNMT isozymes) was not affected (Figure 7A). This result is contradictory to the previous findings that BPDE, the metabolite of BaP, was capable of inhibiting DNMT activity in vitro (Pfeifer et al. 1984; Wilson and Jones 1983; Wilson and Jones 1984; Wojciechowski and Meehan 1984). The explanation for the discrepancy can be that high doses of BPDE (1251.5 or 1650 μg/L) were used in the cell experiments. Not only were our BaP doses lower (0.24-24 μg/L), the majority of BaP would be metabolically deactivated, and the expected BPDE concentration in embryos would be very low. Another possible reason is that we were using whole embryo extracts for the DNMT assay. Although BaP did not affect the overall DNMT activity, the binding of BaP or BPDE to DNA may change the accessibility and activity of DNMT locally, leading to DNA demethylation (Wojciechowski and Meehan 1984).
Similar to dnmt, zebrafish gnmt mRNA expression was not affected by the same BaP waterborne exposure up to 96 hpf (Figure 6B). However, BaP at 2.4 μg/L significantly increased GNMT enzyme activity (Figure 7B). GNMT is the enzyme responsible for the transfer of a methyl group from SAM to glycine forming S-adenosylhomocysteine and sarcosine; thus it regulates the SAM/SAH ratio, which is a metabolic indicator of cellular methylation capacity. Evidence has shown the relationship between altered GNMT expression, SAM concentrations, DNA methylation, and gene expression (Lu and Mato 2012; Luka et al. 2006; Martinez-Chantar et al. 2008). When GNMT activity was increased by BaP, it could be expected that the SAM supply would be decreased in the embryos, and this could be an explanation for the DNA hypomethylation (Figure 2).
Notably, the GNMT results from zebrafish were different from the results of our previous work in Fundulus where GNMT mRNA was increased and enzyme activity was decreased by BaP (Fang et al. 2010). In fact, we found that the constitutive GNMT enzyme activity was about 7.7-fold higher in zebrafish compared to the activity in Fundulus during early development (data not shown). The lower GNMT activity and subsequently higher SAM concentrations in Fundulus may protect this fish against the demethylating effects induced by BaP, which may contribute to Fundulus’ ability to tolerate PAH exposure (Wills et al. 2009).
In summary, BaP is a demethylating agent for global and gene specific DNA methylation status in zebrafish larvae at environmentally relevant concentrations. The reduction in DNA methylation is possibly mediated by increased GNMT activity. BaP did not cause gene specific loss of methylation in rassf1, tert, c-jun and c-myca, but it significantly decreased DNA methylation in the vasa promoter and subsequently increased vasa mRNA expression at environmentally relevant concentrations. Future studies are needed to assess the long-term phenotypic changes caused by BaP-induced DNA hypomethylation in early life.
Supplementary Material
Highlights.
BaP decreased global DNA methylation in zebrafish larvae
BaP decreased vasa promoter methylation and increased vasa gene expression
BaP did not change methylation or gene expression of rassf1, tert, c-jun, or c-myca
GNMT, but not DNMT, enzyme activity was moderately increased by BaP
BaP is an epigenetic modifier of DNA methylation status in zebrafish larvae
Acknowledgement
We would like to thank Fanny Liu from USDA-ARS Genomics and Bioinformatics Research Unit for her work on plasmid isolation and sequencing. We also want to thank all the fish feeders.
This project was supported in parts by Grant Numbers R01ES012710, R03ES018962 and R21ES019940 from the National Institute of Environmental Sciences. It was also partly supported by South Central Technology Transfer Award from Society of Toxicology, and Graduate Student Council Research Grant from the University of Mississippi, 6402-21310-003-00 and project of the Agricultural Research Service. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Environmental Health Science, the National Institutes of Health, or the Agricultural Research Service.
Abbreviations
- BaP
Benzo[a]pyrene
- BPDE
Anti-7β,8α-dihydroxy-9α,10α-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene
- CpG
Cytosine phosphate guanine
- CGI
CpG island
- dnmt
DNA methyltransferase
- DMSO
Dimethyl sulfoxide
- hpf
Hours post fertilization
- gnmt
Glycine N-methyltransferase
- rassf1
Ras association domain family member 1
- SAM
S-adenosylmethionine
- tert
Telomerase reverse transcriptase
- TSS
Transcriptional start site
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, Kogel U, Scheffner M, Helin K, Eilers M. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Akerman GS, Rosenzweig BA, Domon OE, McGarrity LJ, Blankenship LR, Tsai CA, Culp SJ, MacGregor JT, Sistare FD, Chen JJ, Morris SM. Gene expression profiles and genetic damage in benzo(a)pyrene diol epoxide-exposed TK6 cells. Mutat. Res. 2004;549:43–64. doi: 10.1016/j.mrfmmm.2003.11.013. [DOI] [PubMed] [Google Scholar]
- ATSDR Toxicological profile for polycyclic aromatic hydrocarbons (PAHs) 1995;217 [PubMed] [Google Scholar]
- Augoustides-Savvopoulou P, Luka Z, Karyda S, Stabler SP, Allen RH, Patsiaoura K, Wagner C, Mudd SH. Glycine N -methyltransferase deficiency: a new patient with a novel mutation. J. Inherit. Metab Dis. 2003;26:745–759. doi: 10.1023/B:BOLI.0000009978.17777.33. [DOI] [PubMed] [Google Scholar]
- Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr. Opin. Pediatr. 2009;21:243–251. doi: 10.1097/mop.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A. Environmental epigenetics. Heredity. 2010;105:105–112. doi: 10.1038/hdy.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am. J. Respir. Crit Care Med. 2009;180:462–467. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook RJ, Wagner C. Glycine N-methyltransferase is a folate binding protein of rat liver cytosol. Proc. Natl. Acad. Sci. U. S. A. 1984;81:3631–3634. doi: 10.1073/pnas.81.12.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens M, Yamazaki Y, Moisyadi S, Suganuma R, Yanagimachi R, Allsopp R. Regulation and effects of modulation of telomerase reverse transcriptase expression in primordial germ cells during development. Biol. Reprod. 2006;75:785–791. doi: 10.1095/biolreprod.106.052167. [DOI] [PubMed] [Google Scholar]
- Damiani LA, Yingling CM, Leng S, Romo PE, Nakamura J, Belinsky SA. Carcinogen-induced gene promoter hypermethylation is mediated by DNMT1 and causal for transformation of immortalized bronchial epithelial cells. Cancer Res. 2008;68:9005–9014. doi: 10.1158/0008-5472.CAN-08-1276. [DOI] [PubMed] [Google Scholar]
- Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat. Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin. Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- Dreij K, Rhrissorrakrai K, Gunsalus KC, Geacintov NE, Scicchitano DA. Benzo[a]pyrene diol epoxide stimulates an inflammatory response in normal human lung fibroblasts through a p53 and JNK mediated pathway. Carcinogenesis. 2010;31:1149–1157. doi: 10.1093/carcin/bgq073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Dong W, Thornton C, Willett KL. Benzo[a]pyrene effects on glycine N-methyltransferase mRNA expression and enzyme activity in Fundulus heteroclitus embryos. Aquat. Toxicol. 2010;98:130–138. doi: 10.1016/j.aquatox.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields WR, Desiderio JG, Leonard RM, Burger EE, Brown BG, Doolittle DJ. Differential c-myc expression profiles in normal human bronchial epithelial cells following treatment with benzo[a]pyrene, benzo[a]pyrene-4,5 epoxide, and benzo[a]pyrene-7,8-9,10 diol epoxide. Mol. Carcinog. 2004;40:79–89. doi: 10.1002/mc.20023. [DOI] [PubMed] [Google Scholar]
- Gruidl ME, Smith PA, Kuznicki KA, McCrone JS, Kirchner J, Roussell DL, Strome S, Bennett KL. Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 1996;93:13837–13842. doi: 10.1073/pnas.93.24.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Bosagna C, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of phenotype and disease. Mol. Cell Endocrinol. 2012;354:3–8. doi: 10.1016/j.mce.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Zhao Z. Comparative analysis of CpG islands in four fish genomes. Comp Funct. Genomics. 2008:565631. doi: 10.1155/2008/565631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindel JJ. Role of exposure to environmental chemicals in the developmental basis of reproductive disease and dysfunction. Semin. Reprod. Med. 2006;24:168–177. doi: 10.1055/s-2006-944423. [DOI] [PubMed] [Google Scholar]
- Herbstman JB, Tang D, Zhu D, Qu L, Sjodin A, Li Z, Camann D, Perera FP. Prenatal exposure to polycyclic aromatic hydrocarbons, benzo[a]pyrene-DNA adducts, and genomic DNA methylation in cord blood. Environ. Health Perspect. 2012;120:733–738. doi: 10.1289/ehp.1104056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesson LB, Cooper WN, Latif F. The role of RASSF1A methylation in cancer. Dis. Markers. 2007;23:73–87. doi: 10.1155/2007/291538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilberg F, Aguzzi A, Howells N, Wagner EF. c-jun is essential for normal mouse development and hepatogenesis. Nature. 1993;365:179–181. doi: 10.1038/365179a0. [DOI] [PubMed] [Google Scholar]
- Hu W, Feng Z, Tang MS. Preferential carcinogen-DNA adduct formation at codons 12 and 14 in the human K-ras gene and their possible mechanisms. Biochemistry. 2003;42:10012–10023. doi: 10.1021/bi034631s. [DOI] [PubMed] [Google Scholar]
- Incardona JP, Linbo TL, Scholz NL. Cardiac toxicity of 5-ring polycyclic aromatic hydrocarbons is differentially dependent on the aryl hydrocarbon receptor 2 isoform during zebrafish development. Toxicol. Appl. Pharmacol. 2011;257:242–249. doi: 10.1016/j.taap.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Jochum W, Passegue E, Wagner EF. AP-1 in mouse development and tumorigenesis. Oncogene. 2001;20:2401–2412. doi: 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, Huang Z, Hoyo C, Midttun O, Cupul-Uicab LA, Ueland PM, Wu MC, Nystad W, Bell DA, Peddada SD, London SJ. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ. Health Perspect. 2012;120:1425–1431. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jules GE, Pratap S, Ramesh A, Hood DB. In utero exposure to benzo(a)pyrene predisposes offspring to cardiovascular dysfunction in later-life. Toxicology. 2012;295:56–67. doi: 10.1016/j.tox.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee K, Flores M, Cedars MI, Reijo Pera RA. Human primordial germ cell formation is diminished by exposure to environmental toxicants acting through the AHR signaling pathway. Toxicol. Sci. 2010;117:218–224. doi: 10.1093/toxsci/kfq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseljova NP, Kisseljov FL. DNA demethylation and carcinogenesis. Biochemistry (Mosc.) 2005;70:743–752. doi: 10.1007/s10541-005-0179-z. [DOI] [PubMed] [Google Scholar]
- Kuznicki KA, Smith PA, Leung-Chiu WM, Estevez AO, Scott HC, Bennett KL. Combinatorial RNA interference indicates GLH-4 can compensate for GLH-1; these two P granule components are critical for fertility in C. elegans. Development. 2000;127:2907–2916. doi: 10.1242/dev.127.13.2907. [DOI] [PubMed] [Google Scholar]
- Kyo S, Takakura M, Fujiwara T, Inoue M. Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci. 2008;99:1528–1538. doi: 10.1111/j.1349-7006.2008.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder A, Pattengale PK, Kuo A, Stewart TA, Leder P. Consequences of widespread deregulation of the c-myc gene in transgenic mice: multiple neoplasms and normal development. Cell. 1986;45:485–495. doi: 10.1016/0092-8674(86)90280-1. [DOI] [PubMed] [Google Scholar]
- Li J, Wang F, Protopopov A, Malyukova A, Kashuba V, Minna JD, Lerman MI, Klein G, Zabarovsky E. Inactivation of RASSF1C during in vivo tumor growth identifies it as a tumor suppressor gene. Oncogene. 2004;23:5941–5949. doi: 10.1038/sj.onc.1207789. [DOI] [PubMed] [Google Scholar]
- Li M, Hong N, Xu H, Yi M, Li C, Gui J, Hong Y. Medaka vasa is required for migration but not survival of primordial germ cells. Mech. Dev. 2009;126:366–381. doi: 10.1016/j.mod.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Lindeman LC, Winata CL, Aanes H, Mathavan S, Alestrom P, Collas P. Chromatin states of developmentally-regulated genes revealed by DNA and histone methylation patterns in zebrafish embryos. Int. J. Dev. Biol. 2010;54:803–813. doi: 10.1387/ijdb.103081ll. [DOI] [PubMed] [Google Scholar]
- Liu Y, Snow BE, Hande MP, Yeung D, Erdmann NJ, Wakeham A, Itie A, Siderovski DP, Lansdorp PM, Robinson MO, Harrington L. The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo. Curr. Biol. 2000;10:1459–1462. doi: 10.1016/s0960-9822(00)00805-8. [DOI] [PubMed] [Google Scholar]
- Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiol Rev. 2012;92:1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Shao J, Li H, Yu Y. Early whole-genome transcriptional response induced by benzo[a]pyrene diol epoxide in a normal human cell line. Genomics. 2009;93:332–342. doi: 10.1016/j.ygeno.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Lu X, Shao J, Li H, Yu Y. Temporal gene expression changes induced by a low concentration of benzo[a]pyrene diol epoxide in a normal human cell line. Mutat. Res. 2010;684:74–80. doi: 10.1016/j.mrfmmm.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Luka Z, Capdevila A, Mato JM, Wagner C. A glycine N-methyltransferase knockout mouse model for humans with deficiency of this enzyme. Transgenic Res. 2006;15:393–397. doi: 10.1007/s11248-006-0008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YT, Collins SI, Young LS, Murray PG, Woodman CB. Smoking initiation is followed by the early acquisition of epigenetic change in cervical epithelium: a longitudinal study. Br. J. Cancer. 2011;104:1500–1504. doi: 10.1038/bjc.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay D, Shiu W. Aqueous solubility of polynuclear aromatic hydrocarbons. J. Chem. Eng. 1977;(Data 22):399–402. [Google Scholar]
- Martinez-Chantar ML, Vazquez-Chantada M, Ariz U, Martinez N, Varela M, Luka Z, Capdevila A, Rodriguez J, Aransay AM, Matthiesen R, Yang H, Calvisi DF, Esteller M, Fraga M, Lu SC, Wagner C, Mato JM. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology. 2008;47:1191–1199. doi: 10.1002/hep.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirbahai L, Williams TD, Zhan H, Gong Z, Chipman JK. Comprehensive profiling of zebrafish hepatic proximal promoter CpG island methylation and its modification during chemical carcinogenesis. BMC. Genomics. 2011a;12:3. doi: 10.1186/1471-2164-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirbahai L, Yin G, Bignell JP, Li N, Williams TD, Chipman JK. DNA methylation in liver tumorigenesis in fish from the environment. Epigenetics. 2011b;6:1319–1333. doi: 10.4161/epi.6.11.17890. [DOI] [PubMed] [Google Scholar]
- Mohamed e., Song WH, Oh SA, Park YJ, You YA, Lee S, Choi JY, Kim YJ, Jo I, Pang MG. The transgenerational impact of benzo(a)pyrene on murine male fertility. Hum. Reprod. 2010;25:2427–2433. doi: 10.1093/humrep/deq205. [DOI] [PubMed] [Google Scholar]
- Nesbit CE, Tersak JM, Prochownik EV. Myc oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- Nowak JA. Telomerase, cervical cancer, and human papillomavirus. Clin. Lab Med. 2000;20:369–382. [PubMed] [Google Scholar]
- Pan ZG, Kashuba VI, Liu XQ, Shao JY, Zhang RH, Jiang JH, Guo C, Zabarovsky E, Ernberg I, Zeng YX. High frequency somatic mutations in RASSF1A in nasopharyngeal carcinoma. Cancer Biol. Ther. 2005;4:1116–1122. doi: 10.4161/cbt.4.10.2023. [DOI] [PubMed] [Google Scholar]
- Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat. Rev. Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Grunberger D, Drahovsky D. Impaired enzymatic methylation of BPDE-modified DNA. Carcinogenesis. 1984;5:931–935. doi: 10.1093/carcin/5.7.931. [DOI] [PubMed] [Google Scholar]
- Qin G, Meng Z. The expressions of protooncogenes and CYP1A in lungs of rats exposed to sulfur dioxide and benzo(a)pyrene. Regul. Toxicol. Pharmacol. 2006;45:36–43. doi: 10.1016/j.yrtph.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Raizis AM, Schmitt F, Jost JP. A bisulfite method of 5-methylcytosine mapping that minimizes template degradation. Anal. Biochem. 1995;226:161–166. doi: 10.1006/abio.1995.1204. [DOI] [PubMed] [Google Scholar]
- Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim. Biophys. Acta. 2009;1796:114–128. doi: 10.1016/j.bbcan.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Sadikovic B, Haines TR, Butcher DT, Rodenhiser DI. Chemically induced DNA hypomethylation in breast carcinoma cells detected by the amplification of intermethylated sites. Breast Cancer Res. 2004;6:R329–R337. doi: 10.1186/bcr799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadikovic B, Rodenhiser DI. Benzopyrene exposure disrupts DNA methylation and growth dynamics in breast cancer cells. Toxicol. Appl. Pharmacol. 2006;216:458–468. doi: 10.1016/j.taap.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Sharrard RM, Royds JA, Rogers S, Shorthouse AJ. Patterns of methylation of the c-myc gene in human colorectal cancer progression. Br. J. Cancer. 1992;65:667–672. doi: 10.1038/bjc.1992.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Fang J, Qiu D, Zhang T, Yang J, Chen S, Xiao S. Correlation between DNA methylation and pathological changes in human hepatocellular carcinoma. Hepatogastroenterology. 1998;45:1753–1759. [PubMed] [Google Scholar]
- Singh S, Li SS. Epigenetic effects of environmental chemicals bisphenol a and phthalates. Int. J. Mol. Sci. 2012;13:10143–10153. doi: 10.3390/ijms130810143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirera R, Bremnes RM, Cabrera A, Jantus-Lewintre E, Sanmartin E, Blasco A, Del PN, Rosell R, Guijarro R, Galbis J, Sanchez JJ, Camps C. Circulating DNA is a useful prognostic factor in patients with advanced non-small cell lung cancer. J. Thorac. Oncol. 2011;6:286–290. doi: 10.1097/JTO.0b013e31820189a5. [DOI] [PubMed] [Google Scholar]
- Skinner MK, Mohan M, Haque MM, Zhang B, Savenkova MI. Epigenetic transgenerational inheritance of somatic transcriptomes and epigenetic control regions. Genome Biol. 2012;13:R91. doi: 10.1186/gb-2012-13-10-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobinoff AP, Pye V, Nixon B, Roman SD, McLaughlin EA. Jumping the gun: smoking constituent BaP causes premature primordial follicle activation and impairs oocyte fusibility through oxidative stress. Toxicol. Appl. Pharmacol. 2012;260:70–80. doi: 10.1016/j.taap.2012.01.028. [DOI] [PubMed] [Google Scholar]
- Soucek L, Evan GI. The ups and downs of Myc biology. Curr. Opin. Genet. Dev. 2010;20:91–95. doi: 10.1016/j.gde.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparfel L, Pinel-Marie ML, Boize M, Koscielny S, Desmots S, Pery A, Fardel O. Transcriptional signature of human macrophages exposed to the environmental contaminant benzo(a)pyrene. Toxicol. Sci. 2010;114:247–259. doi: 10.1093/toxsci/kfq007. [DOI] [PubMed] [Google Scholar]
- Suter M, Ma J, Harris A, Patterson L, Brown KA, Shope C, Showalter L, Abramovici A, agaard-Tillery KM. Maternal tobacco use modestly alters correlated epigenome-wide placental DNA methylation and gene expression. Epigenetics. 2011;6:1284–1294. doi: 10.4161/epi.6.11.17819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M. The early-life social environment and DNA methylation. Clin. Genet. 2012;81:341–349. doi: 10.1111/j.1399-0004.2012.01843.x. [DOI] [PubMed] [Google Scholar]
- Szyf M. The implications of DNA methylation for toxicology: toward toxicomethylomics, the toxicology of DNA methylation. Toxicol. Sci. 2011;120:235–255. doi: 10.1093/toxsci/kfr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L, Yang S, Xie M, Kramer PM, Pereira MA. Hypomethylation and overexpression of c-jun and c-myc protooncogenes and increased DNA methyltransferase activity in dichloroacetic and trichloroacetic acid-promoted mouse liver tumors. Cancer Lett. 2000;158:185–193. doi: 10.1016/s0304-3835(00)00518-8. [DOI] [PubMed] [Google Scholar]
- Tommasi S, Kim SI, Zhong X, Wu X, Pfeifer GP, Besaratinia A. Investigating the epigenetic effects of a prototype smoke-derived carcinogen in human cells. PLoS. One. 2010;5:e10594. doi: 10.1371/journal.pone.0010594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turusov VS, Nikonova TV, Parfenov Y. Increased multiplicity of lung adenomas in five generations of mice treated with benz(a)pyrene when pregnant. Cancer Lett. 1990;55:227–231. doi: 10.1016/0304-3835(90)90123-f. [DOI] [PubMed] [Google Scholar]
- van der Weyden L, Adams DJ. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim. Biophys. Acta. 2007;1776:58–85. doi: 10.1016/j.bbcan.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt PK. Jun, the oncoprotein. Oncogene. 2001;20:2365–2377. doi: 10.1038/sj.onc.1204443. [DOI] [PubMed] [Google Scholar]
- Vos MD, Ellis CA, Bell A, Birrer MJ, Clark GJ. Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J. Biol. Chem. 2000;275:35669–35672. doi: 10.1074/jbc.C000463200. [DOI] [PubMed] [Google Scholar]
- Wang L, Camus AC, Dong W, Thornton C, Willett KL. Expression of CYP1C1 and CYP1A in Fundulus heteroclitus during PAH-induced carcinogenesis. Aquat. Toxicol. 2010;99:439–447. doi: 10.1016/j.aquatox.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger G, Stebler J, Slanchev K, Dumstrei K, Wise C, Lovell-Badge R, Thisse C, Thisse B, Raz E. Dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr. Biol. 2003;13:1429–1434. doi: 10.1016/s0960-9822(03)00537-2. [DOI] [PubMed] [Google Scholar]
- Weigt S, Huebler N, Strecker R, Braunbeck T, Broschard TH. Zebrafish (Danio rerio) embryos as a model for testing proteratogens. Toxicology. 2011;281:25–36. doi: 10.1016/j.tox.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Wills LP, Zhu S, Willett KL, Di Giulio RT. Effect of CYP1A inhibition on the biotransformation of benzo[a]pyrene in two populations of Fundulus heteroclitus with different exposure histories. Aquat. Toxicol. 2009;92:195–201. doi: 10.1016/j.aquatox.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson VL, Jones PA. Inhibition of DNA methylation by chemical carcinogens in vitro. Cell. 1983;32:239–246. doi: 10.1016/0092-8674(83)90514-7. [DOI] [PubMed] [Google Scholar]
- Wilson VL, Jones PA. Chemical carcinogen-mediated decreases in DNA 5-methylcytosine content of BALB/3T3 cells. Carcinogenesis. 1984;5:1027–1031. doi: 10.1093/carcin/5.8.1027. [DOI] [PubMed] [Google Scholar]
- Wojciechowski MF, Meehan T. Inhibition of DNA methyltransferases in vitro by benzo[a]pyrene diol epoxide-modified substrates. J. Biol. Chem. 1984;259:9711–9716. [PubMed] [Google Scholar]
- Zhang L, Shiverick KT. Benzo(a)pyrene, but not 2,3,7,8-tetrachlorodibenzo-p-dioxin, alters cell proliferation and c-myc and growth factor expression in human placental choriocarcinoma JEG-3 cells. Biochem. Biophys. Res. Commun. 1997;231:117–120. doi: 10.1006/bbrc.1997.6053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.