SUMMARY
A high-throughput RNA interference (RNAi) screen targeting 542 genes of the human kinome was used to discover regulators of RNAi. Here we report that the proto-oncogene Akt-3/PKBγ (Akt3) phosphorylates Argonaute 2 (Ago2) at Ser387 which down-regulates cleavage and up-regulates translational repression of endogenous microRNA (miRNA)-targeted mRNAs. We further demonstrate that Akt3 co-immunoprecipitates with Ago2 and that phosphorylation of Ago2 at Ser387 facilitates its interaction with GW182 and localization to cytoplasmic P-bodies, where miRNA-targeted mRNAs are thought to be stored and degraded. Therefore, Akt3-mediated phosphorylation of Ago2 is a molecular switch between target mRNA cleavage and translational repression activities of Ago2.
Keywords: Akt, Ago2, RNAi, microRNA, translational repression, target cleavage, P-body, phosphorylation, high-throughput screen
INTRODUCTION
RNAi is both a natural pathway and an invaluable biomedical tool used to silence gene expression (Tiemann and Rossi, 2009). RNAi is triggered by small double-stranded RNAs (dsRNAs) that recruit a multiprotein Ago-containing RNA-induced silencing complex (RISC) to mRNAs with binding sites for these small RNAs. When small interfering RNAs (siRNAs) or miRNAs bind to target mRNAs with perfect sequence complementarity in mammals, Ago2 endonucleolytically cleaves these mRNAs. When siRNAs or miRNAs bind to target mRNAs with imperfect sequence complementarity, target mRNAs undergo translational repression and mRNA destabilization (Wightman et al., 1993). The majority of mammalian miRNAs are imperfectly complementary to their mRNA targets and thus are thought to mediate translational repression and mRNA destabilization [reviewed in (Filipowicz et al., 2008)].
Recent data suggest that a larger number of miRNA-targeted mRNAs may be repressed by Ago2-mediated endonucleolytic cleavage than previously thought. In addition to the two most studied examples, HoxB8 (Yekta et al., 2004) and Rtl1 (Davis et al., 2005), new reports describe widespread miRNA-directed mRNA cleavage in a variety of human and mouse tissues and potential new mechanisms driving miRNA-mediated cleavage. In one study, deep sequencing of the RNA degradome in HeLa cells and complex brain tissue revealed 23 miRNA-directed cleavage targets (Bracken et al., 2011). In another, a new class of miRNA target sites comprising 11–12 contiguous Watson-Crick pairs in the center of the miRNA biding site was found that may enable miRNA-mediated mRNA cleavage (Shin et al., 2010). Collectively these data imply that the degree of sequence complementarity between a small RNA silencing trigger and its target may not be the sole mechanistic determinant of Ago-mediated action.
To functionally define mechanistic determinants regulating human RNAi repression, we engineered cells lines that specifically report on shRNA-mediated mRNA repression rather than cleavage. We then performed an RNAi screen targeting the human protein kinome, seeking secondary RNAi pathway modulators. Suppression of the proto-oncogene Akt3/PKBγ disrupted shRNA-mediated repression to the same degree as suppression of known RNAi effector genes. Characterization of this activity revealed that Akt3 positively regulates translational repression of the miR-21 targets PTEN (Meng et al., 2007) and PDCD4 (Asangani et al., 2008) but negatively regulates target mRNA cleavage of the known cleavage targets ATPAF1 (Karginov et al., 2010), PLEKHM1 (Karginov et al., 2010), PFKFB1 (Liu et al., 2010), and HoxB8 (Yekta et al., 2004). These effects depended upon Akt3-mediated Ago2 phosphorylation. Akt3 knockdown reduced phosphoAgo2 (pAgo2) levels and disrupted Ago2 interaction with GW182 and localization to P-bodies. Interestingly, Akt3-mediated phosphorylation of Ago2 did not affect RISC loading, consistent with previous observations that passenger strand cleavage and target mRNA cleavage are mechanistically distinct (Wang et al., 2009; Yoda et al., 2010). Similar though smaller effects were observed for Akt1 and Akt2, indicating that the entire family of Akt proto-oncogenes coordinates mRNA cleavage and repression activities of Ago2. Our data is most consistent with Ago2 phosphorylation functioning as a molecular switch between these two Ago2 effector functions.
RESULTS
A High-Throughput RNAi Screen Identifies Akt3 as a Regulator of Translational Repression
To screen for protein kinase regulators of small RNA function, we engineered HeLa cells to stably co-express the firefly luciferase gene (FLuc) and a perfect match shRNA targeting the FLuc ORF (called D1, Figure 1A; Figure S1A). Analysis of single cell-sorted clones confirmed suppression of FLuc protein. Paradoxically, quantification by qPCR revealed minimal changes in FLuc mRNA levels (Figure S1B), indicating that the FLuc shRNA acts via mRNA repression and not via mRNA cleavage. Because Ago2 is the only Ago reportedly capable of loading perfectly complementary siRNA duplexes (Wang et al., 2009), Ago2 was the only Ago protein capable of repressing FLuc, which we confirmed (Figure S1C).
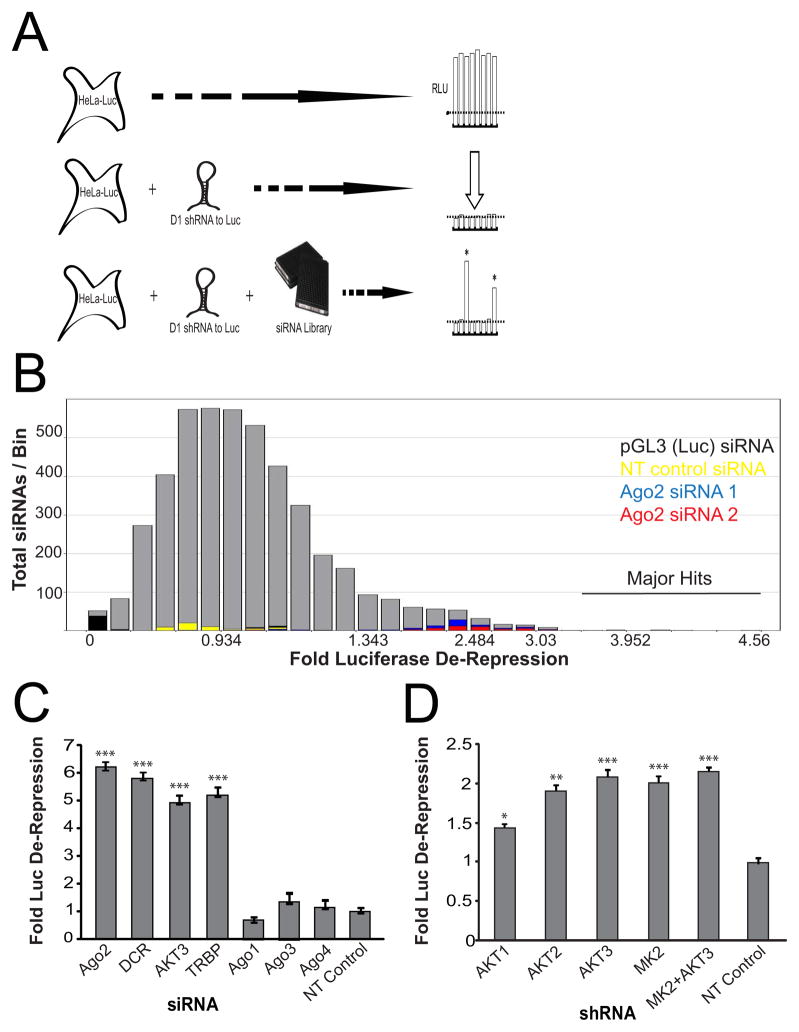
Figure 1. Primary Screening and Reconfirmation of Novel Regulators of siRNA-Mediated Gene Silencing.
(A) Schematic diagram of the primary screen to identify novel regulators of RNAi. siRNA-mediated knockdown of only those genes acting as positive regulators of RNAi elicited luciferase de-repression. (B) A histogram showing the results of the primary RNAi genomics screen. (C) Reconfirmation of the main hit, Akt3, relative to known miRNA effector proteins. (D) Confirmation that all three Akt isoforms positively regulate RNAi. HeLa-Luc-D1 cells were transduced with lentiviruses expressing shRNAs to Akt1, Akt2, Akt3, MAPKAPK2 (MK2) or a non-targeting control (NT) and assayed for FLuc de-repression. Error bars indicate SD. *P ≤ .05, **P ≤ .01, ***P ≤ .001. See also Figure S1.
To identify regulators of shRNA-mediated repression in this system, we screened HeLa cells transduced with the D1 shRNA against 2,168 unique siRNAs targeting 542 human protein kinase genes. Technically, knockdown of the positive control yielded an average screen-wide FLuc de-repression of 2.5-fold (Figure 1B). A normal activity distribution of siRNAs was observed, suggesting that hit activities are unlikely to derive from FLuc siRNA competitive displacement from RISC. From the screen we identified Akt3/PKBγ as the strongest positive regulator of FLuc repression. Multiple siRNAs against Akt3 yielded 5-fold de-repression of FLuc, similar to the activities of known RNAi pathway components Ago2, Dicer, and TRBP (Figure 1C, Figures S1D-F). Although Akt1 and Akt2 demonstrate partial redundancy with Akt3 in several signaling pathways [reviewed in (Dummler and Hemmings, 2007)], these kinases were marginal hits in the primary screen; as was MAPKAPK2, which we had previously shown could phosphorylate Ago2 (Zeng et al., 2008). Upon secondary reconfirmation we observed de-repression of FLuc by shRNAs targeting Akt1 or Akt2 but more significant de-repression by those targeting Akt3 or MK2 (Figure 1D). Notably, knockdown of MK2 and Akt3 together did not lead to synergistic de-repression (Figure 1D), suggesting functional redundancy or complementary mode of action in regulating repression. While a small number of other protein kinases scored as primary screening hits (Table S1), weak de-repression, single-hitter siRNAs, or failed secondary re-confirmation led us to focus exclusively on Akt-based regulation of mRNA repression.
Akt3 Phosphorylates Ago2 at Ser387
To determine whether Akt3 directly interacts with Ago2, we performed co-immunoprecipitation (IP) assays. His-tagged human Akt1, Akt2 and Akt3 were immunoprecipitated from HeLa cells and the immunoprecipitates probed for Ago2. High levels of Ago2 associated with Akt3, moderate levels with Akt1, and detectable levels with Akt2 (Figure 2A, IP: top panel). In a similar experiment, Akt3 co-precipitated with Ago2 but also with Akt1 and Akt2, suggesting that Akt1 and Akt2 may indirectly bind to Ago2 via Akt3 (data not shown). Together, these data demonstrate physical interaction between Akt3 and Ago2.
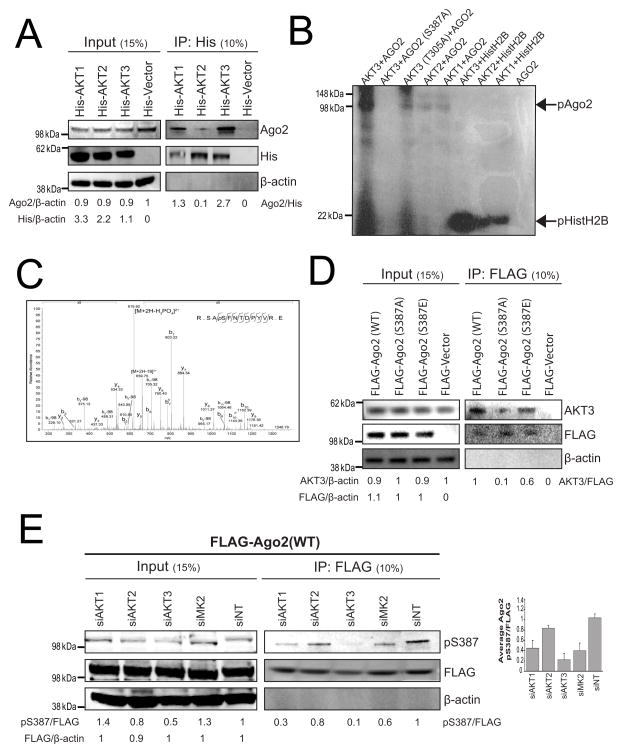
Figure 2. Akt3 Interacts With and Phosphorylates Ago2 at S387.
(A) Akt and Ago2 physically interact in cells. His-Akt-expressing lysates and IP extracts were immunoblotted for Ago2, His or β-actin. Relative amounts of Ago2 protein are indicated using His-Vector as a reference for input and His-Akt1 as a reference for IP. (B) Kinase reaction showing Akt3 phosphorylating Ago2 at S387. Purified His-tagged Akt1, Akt2, Akt3 or a kinase-impaired Akt3 mutant (T305A) were incubated with purified WT or S387A Ago2 in the presence of 32P-γ-ATP, and resolved on a denaturing gel. Histone H2B was used as a positive control for Akt phosphorylation. (C) The MS/MS spectrum of the Ago2 SApSFNTDPYVR phosphopeptide from the kinase reaction in (B). Prominent y- and b- ions are shown and [M+2H-H3PO4]2+ = 620. (D) Akt3 binds Ago2 at the S387 site. HeLa cells were transiently transfected with FLAG-tagged Ago2 constructs and lysates and IP extracts were immunoblotted for Akt3, FLAG or β-actin. Relative amounts of Akt3 protein are indicated using FLAG vector as a reference for input and FLAG-Ago2 (WT) as a reference for IP. (E) Knockdown of Akt3 reduces phosphorylation of Ago2 S387. Akt knockdown HeLa cell lysates and IP fractions were immunoblotted for Ago2-pS387, FLAG or β-actin. Relative amounts of pS387 protein are indicated using siNT as a reference for input and IP. The experiment was repeated four times and the average pS387 levels along with standard deviations as error bars are shown in the inset graph. See also Figure S2.
To directly test whether Akt3 phosphorylates Ago2, we purified a panel of recombinant wild-type and mutant Akt and Ago2 proteins (Figure S2) and assessed Akt phosphoryltransferase activity toward Ago2 in vitro. Akt3 exhibited the highest Ago2 phosphoryltransferase activity, though Akt1 and Akt2 showed detectable activity as well (Figure 2B). Previous work suggested that a T305A mutation in the Akt3 catalytic domain inactivates its kinase activity (Masure et al., 1999). In contrast, we observed reduced but detectable phosphoryltransferase activity of this mutant, suggesting that T305 is important but not essential for Akt3 phosphoryltransferase activity to Ago2. To determine which Ago2 residue(s) are phosphorylated by Akt3, we subjected purified Ago2 from the Akt3 phosphoryltransferase reaction (Figure 2B) to fractionated peptide mass spectrometry (LC/MS), showing that S387 is the lone phosphorylation site for Akt3 (Figure 2C). Mutating Ago2 S387 to a phospho-null alanine (S387A) blocked Akt3-mediated phosphorylation in vitro (Figure 2B), confirming that Ago2 S387 is the lone phosphorylation site for Akt3. To assess cellular binding between Akt3 and various Ago2 S387 mutant constructs, we introduced FLAG-tagged WT, S387A, or phospho-dominant S387E Ago2 into HeLa cells, starved and stimulated the cells with serum plus insulin to encourage Akt activity, immunoprecipitated the Ago2 proteins, and assayed the IP fractions for Akt3. Strong Akt3 binding was detected in the WT and S387E Ago2 precipitates but only weak binding in the phospho-null Ago2 S387A (Figure 2D).
To assess Akt phosphorylation of Ago in HeLa cells, we knocked down the three Akt isoforms or MK2, introduced a FLAG-tagged Ago2 construct, immunoprecipitated Ago2, and probed the IP reactions with a pS387-Ago2-specific antibody (Figure 2E). In this cellular context, reduction of Akt3 most robustly reduced pS387-Ago2 levels, though the relatively weak involvement of MK2 could be attributed to less efficient MK2 knockdown (Figure S1F). Taken together, these results demonstrate that the proto-oncogene Akt3 binds to and phosphorylates Ago2 on residue S387.
Knockdown of Akt3 Relieves miRNA-mediated Repression
To initially assess the impact of Ago2 S387 phosphorylation on mRNA repression we first employed an FLuc reporter engineered with six imperfect binding sites in its 3′ UTR for an siRNA to CXCR4 (called FL6X, Figure 3A, top). HeLa cells were co-transfected with this reporter, the CXCR4 siRNA, WT or mutant Ago2 ORF variants (Figure S3A), and an siRNA targeting the 3′UTR of endogenous Ago2. Ectopic re-expression of WT Ago2 modestly restored repression of FLuc as expected while the Ago2-S387A mutant, which cannot be phosphorylated by Akt, robustly de-repressed the FLuc repression reporter (Figure 3A). Ectopic expression of the phospho-dominant Ago2-S387E mutant, in contrast, significantly increased the FLuc repression, suggesting that S387 phosphorylation promotes Ago2-mediated repression. To ensure that S387 mutation did not affect Ago2 small RNA loading, we performed IPs using wild-type and mutant Ago2 proteins followed by northern blotting for CXCR4 siRNAs and endogenous miRNAs. We did not observe any differences in loading of CXCR4 siRNAs or endogenous microRNAs (let-7a, miR-21, miR-17, miR-29a) onto Ago2-S387 mutant proteins compared to the WT Ago2 (Figure S3B).
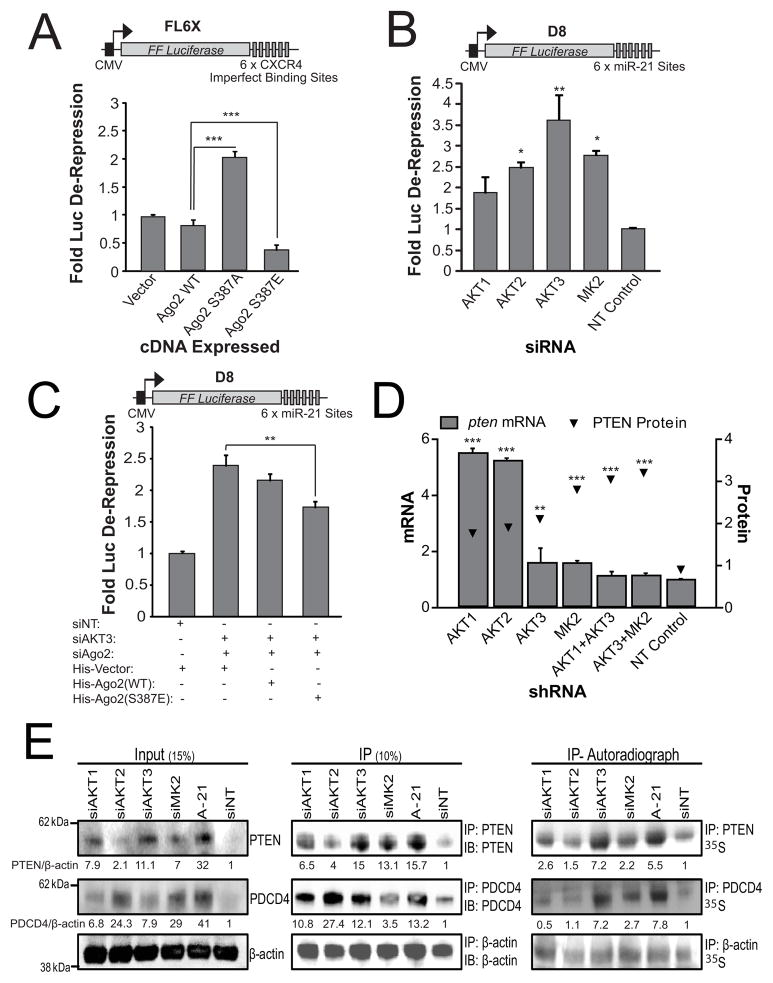
Figure 3. Knockdown of Akt Reduces Ago2-mediated mRNA Repression.
(A) Ago2 S387 phosphorylation positively affects CXCR4 siRNA-mediated repression. HeLa cells harboring endogenous Ago2 knockdown and expressing ectopic Ago2 mutants were assayed for CXCR4 siRNA targeting to the FL6X mRNA repression reporter. (B) Knockdown of Akt3 significantly disrupts miR-21-mediated repression in HeLa-D8 cells. (C) Phospho-dominant Ago2-S387E mutant partially rescues the Akt knockdown phenotype. HeLa-D8 cells were co-transfected with siRNAs targeting Akt3 and Ago2 and Ago2 constructs. (D) Knockdown of Akt3 increases PTEN protein but not pten mRNA levels in HeLa cells. Bars indicate pten mRNA levels (normalized to GAPDH) and triangles represent pten protein levels relative to the NT control sample. (E) Knockdown of Akt3 up-regulates translation of miR-21 targets PTEN and PDCD4 in HEK293T cells measured by 35S methionine incorporation. Relative amounts of PTEN and PDCD4 proteins are indicated using siNT as a reference for input and IP. Error bars indicate SD. *P ≤ .05, **P ≤ .01, ***P ≤ .001. See also Figure S3.
We next assessed the effects of Akt3-mediated Ago2 phosphorylation on mRNA repression in HeLa cells engineered to stably express FLuc containing six imperfect miR-21 binding sites in its 3′UTR (called D8, Figure 3B, top). miR-21 is expressed between 10,000 and 60,000 copies per HeLa cell (Lim et al., 2003) and robustly represses FLuc expression in D8 cells. This effect can be blocked by addition of a miR-21 antagonist, which elicits time-dependent up-regulation of FLuc protein without concomitant increase in FLuc mRNA (Figure S3C). In the D8 cell line, knockdown of Akt1, Akt2, Akt3 (Figure S3E), and MK2 de-repressed FLuc by approximately, 1.9, 2.5, 3.5, and 2.7 fold, respectively (Figure 3B). Combined Akt3/Ago2 knockdown de-repressed FLuc 2.4-fold (Figure 3C, column 2). In contrast, ectopic expression of WT or S387E Ago2 ORFs in D8 cells depleted of endogenous Akt3 and Ago2 restored FLuc repression (Figure 3C). Importantly, rescue by the Ago2-S387E mutant was greater than rescue by WT Ago2, indicating S387 phosphorylation of Ago2 increases repression. Surprisingly, the Ago2-S387A mutant also restored FLuc repression comparable to WT Ago2 (Figure S3D), implying that S387 phosphorylation promotes but is not required for miR-21-mediated repression (see Discussion). To control for possible impact of Akt depletion on Ago2 miRNA loading we transfected an Ago2-FLAG construct into D8 cells harboring Akt or MK2 knockdowns, immunoprecipitated Ago2, and analyzed Ago2-associated miRNAs by northern blotting. We observed no differences in CXCR4 siRNA or endogenous let-7a or miR-21 loading onto Ago2 (Figure S3F), suggesting that the observed responses genuinely reflect Akt modulation of Ago2-mediated repression.
To assess the Akt’s role in regulating repression of endogenous miRNA targets we focused on the miR-21 targets PTEN (Meng et al., 2007) and PDCD4 (Asangani et al., 2008). In HeLa cells, Akt3 or MK2 knockdown doubled PTEN protein levels within 24 hours without affecting pten mRNA levels, demonstrating a requirement for these kinases in maintaining translation repression (Figure 3D). Knockdown of Akt1 or Akt2, in contrast, de-repressed both pten mRNA and PTEN protein levels, indicative of a more canonical signaling pathway (Figure 3D). To rule out miR-21-dependent Akt regulatory effects, we analyzed cellular levels of other miRNA-targeted proteins upon Akt knockdown, observing coordinate de-repression of the let-7 targets RAS (Johnson et al., 2005) and IMP-1 (Boyerinas et al., 2008) (Figure S3G). We further tested Renilla Luciferase (RL) reporter vectors containing let-7 target sequences in their 3′UTRs side-by-side with the FL6X reporter (containing six imperfect CXCR4 siRNA binding sites within its 3′UTR, Figure 3A, top). Curiously, alleviation of repression of KRAS using these reporters was less sensitive than data observed from western blots. However, significant derepression of the RL-KRAS reporter as well as the FL6X reporter were observed in cells harboring Akt3 knockdown (Figure S3H). Surprisingly, we observed only mild derepression of the RL-HMGA2 construct in Akt knockdown cells (Figure S3H).
To test Akt’s impact on the translation rates of miRNA-targeted mRNAs we transfected HEK293Tcells, which express high levels of Akt3 (Su et al., 2004; Wu et al., 2009) (Figure S3I), with siRNAs against Akt1, Akt2, Akt3, or MK2, or a miR-21 antagomir, pulsed cells with 35S-methionine and 35S-cysteine for 45 minutes to label newly-translated proteins, and then immunopreciptated and autoradiographed the miR-21 targets PTEN and PDCD4. By western, PTEN repression was most strongly relieved by knockdown of Akt1 or Akt3 and PDCD4 repression by knockdown of Akt2 or Akt3 (Figure 3E, input), in all cases equivalent to treatment with the miR-21 antagomir. Measured by 35S autoradiography, levels of newly-translated, immunoprecipitated PTEN and PDCD4 were most strongly de-repressed by knockdown of Akt3, with loss of Akt1 and Akt2 eliciting negligible effects on translation (Figure 3E, IP-Autoradiograph). To ensure that Akt knockdowns did not exert effects on general cellular translation rates, IP of β-actin was used as a control (Figure 3E). Together, these data suggest that Akt3 and MK2 directly regulate miRNA-mediated target repression whereas Akt1 and Akt2 may regulate miRNA-mediated repression in a more canonical manner.
Akt3 Promotes P-body Localization of Ago2
P-bodies are multi-megadalton RNA-protein complexes that function in mRNA surveillance and may associate with miRNAs, miRNA-targeted mRNAs and Ago2 [reviewed in (Eulalio et al., 2007a; Parker and Sheth, 2007)]. MK2 phosphorylation of Ago2 at S387 has been shown to facilitate Ago2 localization to these complexes (Zeng et al., 2008) and Ago2 is known to bind the P-body marker GW182 (Jakymiw et al., 2005). We therefore sought to assess Akt’s impact on the physical association of Ago2 and GW182. In control HeLa cells, immunoprecipitation of FLAG-tagged Ago2 yielded high levels GW182 (Figure 4A, IP: top panel). In HeLa cells knocked down for Akt1, Akt2 or Akt3 however, this interaction was disrupted by more than 60% (Figure 4A, IP: top panel). To assess the importance of the Ago2 S387 residue to this association, we introduced the FLAG-tagged Ago2 S387 mutants into HeLa cells, immunoprecipitated, and again assessed GW182 levels. We observed that introduction of the phospho-dominant mutant S387E Ago2 did not significantly affect the Ago2-GW182 interaction, though introduction of the phospho-null S387A Ago2 reduced the Ago2-GW182 interaction by 70% (Figure 4B, IP: top panel). Ago2 S387 phosphorylation status was confirmed by western blotting of IP fractions using the pS387-Ago2-specific antibody (Figure 4B).
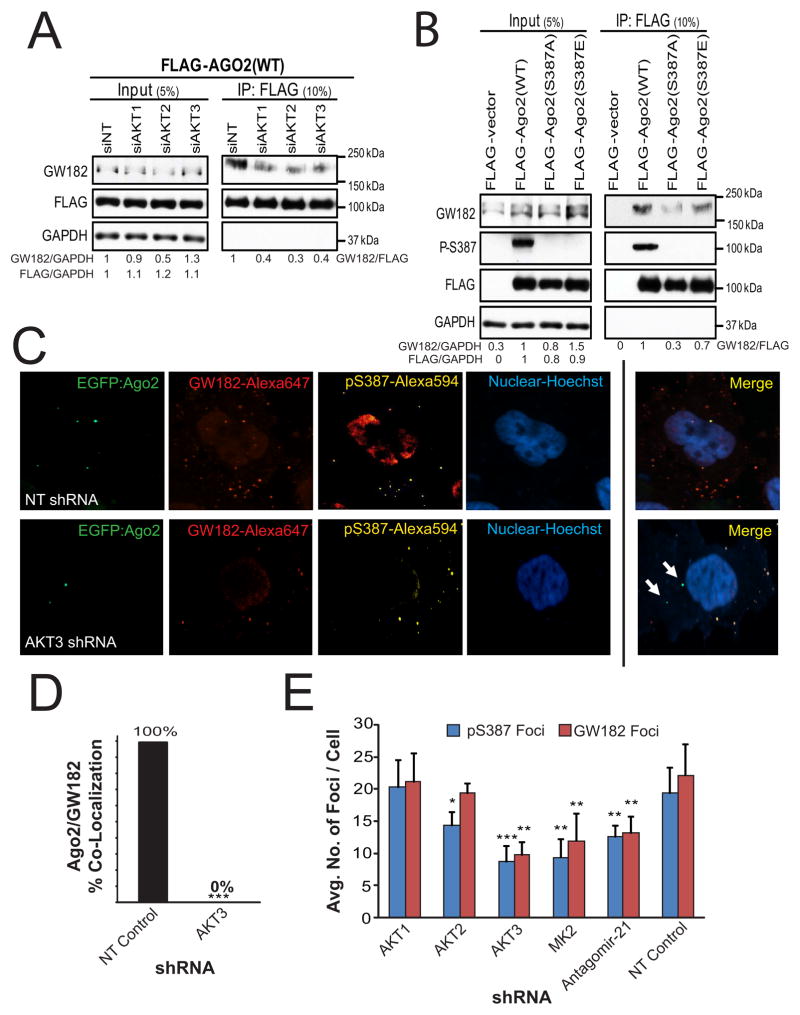
Figure 4. Akt-mediated Phosphorylation of Ago2 S387 Promotes Interactions of Ago2 with GW182 and P-bodies.
(A) Akt positively regulates Ago2-GW182 interactions. FLAG-Ago2 and GW812 were co-immunoprecipitated in Akt knockdown HeLa cells. Relative amounts of GW182 protein are indicated using siNT as a reference for input and IP. (B) S387 phosphorylation status guides Ago2-GW182 interactions. Lysates and IP fractions from HeLa cells expressing FLAG-Ago2 constructs were analyzed for GW182 interaction and pS387 levels. Relative amounts of GW182 protein are indicated using FLAG-Ago2 (WT) as a reference. (C) HeLa-EGFP-Ago2 cells show disrupted Ago2/GW182 co-localization upon knockdown of Akt3. Representative HeLa-EGFP-Ago2 cells stained with immunofluorescent antibodies to GW182, Ago2-pS387 and Hoechst; EGFP represents endogenous Ago2. White arrows indicate Ago2 not co-localized with GW182 bodies. (D) Co-localization quantifications from (C). (E) Knockdown of Akt3 decreases total P-body formation in cells. Quantitation of phospho-Ago2 and GW182 foci in cells from (C); foci were enumerated on a confocal microscope with n = 10 cells. Error bars indicate SD. *P ≤ .05, **P ≤ .01, ***P ≤ .001. See also Figure S4.
To visualize the Ago2-GW182 interaction in cells and further explore the consequences of Ago2 S387 phosphorylation on this interaction, we performed immunofluorescence co-localization studies using a HeLa cell line stably expressing an endogenous EGFP-Ago2 fusion protein. In this line EGFP could be used to assess Ago2 localization and antibodies to assess endogenous GW182 and pS387-Ago2 localization. GW182 and either Ago2 or pS387-Ago2 proteins were found to co-localize in control cells (Figure 4C, top panels) whereas knockdown of Akt3 abolished Ago2 co-localization with GW182 (Figure 4C, bottom panels and Figure 4D). Intriguingly, Akt3 knockdown also reduced the average number of pS387-Ago2 and GW182 foci per cell (n>10 cells) by 50% (Figure 4E). Akt1 or Akt2 knockdown marginally reduced Ago2 co-localization with GW182 (Figure S4A), and knockdown of neither Akt1 nor Akt2 reduced the number of GW182 foci. These data show that Akt3 (and MK2) may promote both the formation of P-bodies and Ago2 recruitment to them. To confirm the role of S387 phosphorylation on the Ago2-GW182 interaction, we ectopically expressed EGFP-WT, -S387A, or -S387E Ago2 fusion cDNAs in HeLa cells and analyzed GW182 co-localization by immunofluorescence. We observed strong associations in cells between GW182 and WT Ago2 and between GW182 and the S387E Ago2 mutant (Figure S4B, top and bottom panels). In contrast the interaction between GW182 and the S387A Ago2 mutant was reduced 45% (Figure S4B, middle panels and Figure S4C). Thus, immunofluorescence co-localization analyses corroborated our IP results, collectively implying a requirement for Akt3 kinase activity, specifically on S387, for Ago2-GW182 interaction and Ago2 P-body localization.
Akt3-Mediated Phosphorylation of Ago2 Down-Regulates Target mRNA Cleavage
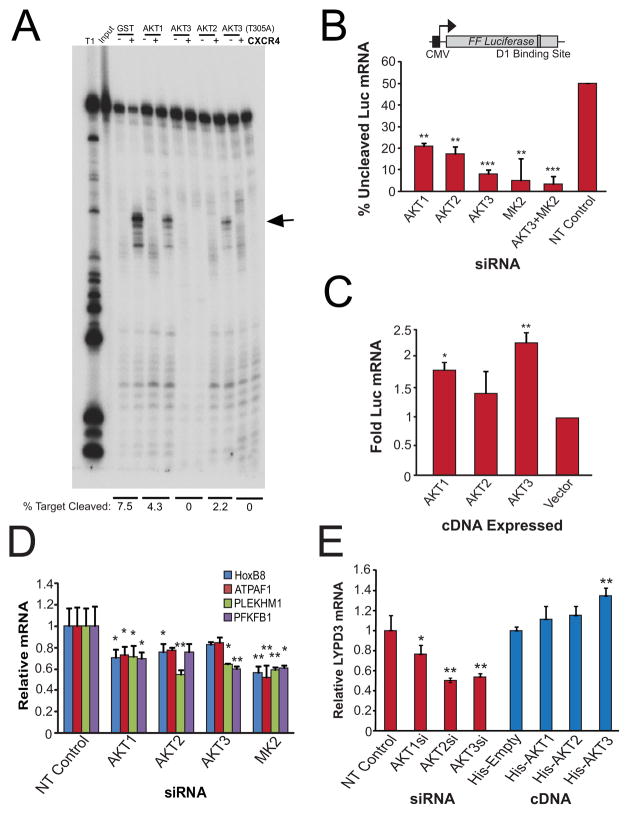
To directly assess the effects of Akt-mediated phosphorylation of Ago2 on the capacity of Ago2 to catalyze target mRNA cleavage, we performed coupled in vitro kinase and target mRNA cleavage assays. Recombinant human Ago2 and Akt proteins were incubated with ATP, CXCR4 siRNA, and a radiolabeled mRNA with one perfectly-complementary CXCR4 binding site and the resulting RNA reaction products were resolved by SDS-PAGE. Unphosphorylated Ago2 cleaved approximately 7.5% of the target mRNA (Figure 5A). Phosphorylation of Ago2 by Akt1 or Akt2 reduced target mRNA cleavage by 43% and 71%, respectively. Phosphorylation of Ago2 by Akt3 abolished target mRNA cleavage. Consistent with our previous observations that mRNA cleavage and passenger strand cleavage activities are distinct biochemical reactions (Wang et al., 2009), Akt3-mediated phosphorylation of Ago2 had no effect on passenger strand cleavage (Figure S5A). Interestingly, the Akt3-T305A mutant, which reduced but did not eliminate Ago2 S387 phosphorylation (Figure 2B), also completely abolished the target mRNA cleavage activity of Ago2 (Figure 5A). These data likely indicate that the residual kinase activity of Akt3-T305A is sufficient to phosphorylate Ago2 and thus inhibit its endonuclease activity and/or that the Akt3-T305A mutant binds to Ago2 and represses its endonuclease activity.
Figure 5. Akt3 Negatively Regulates Ago2-Mediated Cleavage of miRNA-targeted mRNAs.
(A) Akt phosphorylation of Ago2 reduces target mRNA cleavage activity of Ago2 in vitro. Purified Ago2 was incubated with purified Akt proteins in the presence of ATP. The target mRNA cleavage activity of the phosphorylated Ago2 was assayed by incubating the Ago2-Akt mixture with the antisense strand of CXCR4 siRNA and 5′-labeled CXCR4 substrate. Specific cleavage products are indicated with an arrow based on the ladder generated from partial digestion of input substrate by RNase T1 (T1). (B) Knockdown of Akt or MK2 results in increased siRNA-mediated target mRNA cleavage. HeLa-Luc cells with Akt knockdown were transfected with 1nM of an siRNA against the Luc ORF (D1) (NT; represents 50% Luc mRNA cleavage). RNA was analyzed for uncleaved Luc mRNA. (C) Overexpression of Akt results in decreased siRNA-mediated target mRNA cleavage. HeLa-Luc cells over-expressing Akt cDNAs were assayed in as (B). (D) Akt knockdown increases cleavage of endogenous miRNA-targeted mRNAs in HeLa-D8 cells. (E) Cytoplasmic levels of the miR target LYPD3 are subject to tight coordination with cellular Akt levels in A375 melanoma cells. Error bars indicate SD. *P ≤ .05, **P ≤ .01, ***P ≤ .001. See also Figure S5.
In order to distinguish between these two possibilities we performed coupled in vitro kinase and target mRNA cleavage assays in the presence of the Akt inhibitor KP372-1 (Mandal et al., 2005)(Figure S5B). In this assay Akt3 phosphorylation of wild type Ago2 reduced target cleavage by 25% (compare lanes 2 and 6). Inhibition of Akt activity by KP372-1 alleviated that reduction by 6% (compare lanes 2 and 4). These effects were not observed when we used the Ago2 S387A mutant (Figure S5B), implicating S387 as the key catalytic amino acid of the reaction. It should be noted that the presence of KP372-1 in either the WT or S387A Ago2 reactions reduced cleavage by up to 19%, making its effects on Akt3 inhibition more significant. These results confirm that kinase activity of Akt3 is required to inhibit mRNA cleavage activity of Ago2.
We next tested the effects of Akt-mediated Ago2 phosphorylation on the activity of mRNA cleavage reporters in cells. Transfection of 1nM of a synthetic perfect match siRNA (D1) targeting the FLuc ORF in HeLa cells reduced FLuc mRNA levels by 50% measured by qPCR (Figure 5B, NT Control), consistent with Ago2-mediated FLuc mRNA cleavage. Knockdown of Akt1 increased this FLuc mRNA cleavage to 70%, Akt2 to 75%, and Akt3 to more than 90% (Figure 5B, Figure S1F). Knockdown of MK2 alone or in combination with Akt3 increased FLuc mRNA cleavage to more than 95%. We then used an FLuc reporter containing one perfect CXCR4 siRNA binding site in its 3′UTR (called FL1P, Figure S5C, top). Knockdown of Akt2 or Akt3 increased CXCR4 siRNA-mediated FLuc mRNA cleavage by 20% (Figure S5C).
Similar to our in vitro kinase/cleavage assays, Akt1 or Akt3 over-expression decreased CXCR4 siRNA-mediated FLuc mRNA cleavage by up to twofold (Figure 5C, Figure S5D). These results suggest that Akt3- and, to a lesser extent, Akt1- and Akt2-mediated phosphorylation of Ago2 reduce siRNA-mediated target mRNA cleavage.
Akt Regulates Cleavage of Endogenous miRNA Targets
Finally we sought to assess the effects of Akt on endogenous miRNA-mediated target mRNA cleavage. We analyzed mRNA levels of HoxB8, repressed by miR-196-mediated cleavage (Yekta et al., 2004), ATPAF1, repressed by miR-151-5p-mediated cleavage (Karginov et al., 2010), PLEKHM1, repressed by miR-106b-mediated cleavage (Karginov et al., 2010), and PFKFB1, repressed by let-7-mediated cleavage (Liu et al., 2010). Knockdown of Akt1, Akt2, Akt3, and MK2 in HeLa cells increased cleavage of these targets as measured by qPCR, though the degree of enhancement varied in a target- and kinase-dependent fashion (Figure 5D, Figure S5E). Similar knockdowns in A375 melanoma cells enhanced LYPD3 mRNA cleavage [repressed by by miR-151-5p cleavage (Karginov et al., 2010)], whereas ectopic expression of each Akt isoform reduced LYPD3 mRNA cleavage (Figure 5E, Figure S5D, Figure S5G). To prove that the reduction of target mRNAs was specific to repression by particular miRNAs, we transfected antagomirs to miR-151-5p and miR-106b into cells harboring knockdown of the Akt isoforms or MK2 and assessed ATPAF1, LYPD3 and PLEKHM1 mRNA levels (Figure S5E, Figure S5F). Inhibition of miR-151-5p or miR-106b abolished the positive effects of Akt knockdown on endogenous mRNA cleavage targets ATPAF1, LYPD3 and PLEKHM (Figure S5F), ruling out indirect effects of Akt knockdown on general mRNA stability or transcription. Thus, all Akt isoforms are capable of down-regulating cleavage of endogenous miRNA-targeted mRNAs.
DISCUSSION
Few processes as ancient, central, and complex as small RNA silencing avoid some form of secondary regulation. Coordinate regulation of ancient processes and the pathways underlying them may have proven highly adaptive over time (Jeong et al., 2000; Lee et al., 2007). Our screen of the human protein kinome was originally designed to identify regulators of siRNA-mediated mRNA cleavage. Curiously, the use of a perfect match shRNA targeting FLuc resulted in FLuc mRNA repression and not cleavage. Previous reports show that perfect sequence complementarity between an siRNA and target mRNA is not always sufficient to direct target mRNA cleavage [reviewed in (Valencia-Sanchez et al., 2006)]. Other reports have computationally and experimentally demonstrated that miRNAs can target ORFs and 5′UTRs of target mRNAs (Lal et al., 2009; Thomas et al., 2010). Our data agree with these findings, showing that a perfectly-complementary siRNA targeting the ORF of a target mRNA can mediate robust repression. Therefore, our screen identified kinase regulators of repression by an siRNA targeting an ORF.
Only knockdown of Akt3 de-repressed reporter mRNAs comparable to knockdown of key RNAi effector genes Ago2, Dicer, and TRBP in the primary screen. However, other kinases including Akt1 and Akt2 de-repressed reporter mRNAs in follow-up experiments. We cannot discount the possibility that inefficient knockdown or cell-specific expression may have generated false negatives in the primary screen, masking equally or more important kinases involved in regulating mRNA repression. For example, MK2, which phosphorylates the same residue of Ago2 as Akt3 (Zeng et al., 2008) and is expressed in HeLa cells, did not score in the primary screen, though it did score in most secondary and orthogonal experiments in HeLa and other cell types.
Recently published data implicates serine phosphorylation of Ago2 in the regulation of RISC loading (Rudel et al., 2011; Zeng et al., 2008). Additionally, tyrosine phosphorylation of Ago2 at residue 529 has been implicated in negative regulation of RISC loading (Rudel et al., 2011). In contrast we observed that, in vitro, Ago2 phosphorylation by Akt does not affect RISC loading as measured by passenger strand cleavage (Figure S5A). In cells, neither Akt nor Ago2 S387 phosphorylation affects RISC loading because Ago2 association with antisense strands of endogenous miRNAs and siRNAs is unchanged as assessed by Ago2 immunoprecipitations (Figures S3B/F). Because Ago2 phosphorylation decreased mRNA cleavage activity but increased translational repression of several miRNA targets, our data is most consistent with Ago2 phosphorylation functioning as a molecular switch between effector complex functions rather than regulating RISC loading and passenger strand cleavage.
Akt3 phosphorylation of Ago2 was specific to residue S387 and did not occur on any of the C-terminal serine residues (data not shown). S387 phosphorylation was predominantly catalyzed by Akt3 since knockdown of Akt3 resulted in a 90% loss of pS387-Ago2 in cells (Figure 2E). The phospho-null S387A Ago2 mutant demonstrated variable rescue of the loss of FLuc repression in Ago2 3′UTR knockdown cells (Compare Figures 3A and S3D), possibly indicating that S387 phosphorylation promotes but is not required for Ago2-mediated repression. For instance, we were still able to detect GW182 association with the Ago2-S387A mutant (Figure 4B) indicating partial functionality. On the other hand, the phospho-dominant (S387E) Ago2 mutant significantly restored the reduced FLuc repression in Ago2 3′UTR knockdown cells (Figures 3A and 3B). It is unclear how phosphorylation of Ago2 at S387 up-regulates translational repression and down-regulates endonucleolytic cleavage. S387 resides between the PAZ and MID domains of Ago2. It is possible that S387 phosphorylation induces a conformational change affecting the orientation of the PIWI domain, thereby reducing mRNA cleavage and increasing mRNA repressive activities of Ago2.
Akt3-mediated phosphorylation inhibited Ago2-mediated cleavage of target mRNAs in vitro and in cells. There are far fewer endogenous miRNA targets repressed by endonucleolytic cleavage than by translational repression and/or mRNA destabilization. It is interesting to note, however, that cleavage-dependent mRNA targets are enriched for developmentally-regulated and oncogenesis-promoting genes such as HoxB8, ATPAF1, PLEKHM1, PFKFB1 and LYPD3. Akt is an oncogene whose exact oncogenic mechanism remains unknown [reviewed in (Restuccia and Hemmings, 2010)]. It is possible that as a tumor converts to anaerobic glycolysis, increased Akt up-regulates PFKFB1 and the ATPase subunit ATPAF1, enhancing respiration, and HoxB8, activating key developmental genes. In this study, Akt3 knockdown de-repressed translation of endogenous miR-21-targeted mRNAs. For example, knockdown of Akt3 significantly increased translation rates but not mRNA levels of the miR-21 targets PTEN and PDCD4 (Figure 3F), suggesting that Akt3 normally represses translation of these miRNA targets. Akt2 was recently shown to contribute to PTEN silencing through the upregulation of miR-21 transcripts in a variety of carcinoma cells under hypoxic conditions (Polytarchou et al., 2011), indicating that Akt isoforms may also increase oncogenic miRNA abundance as well as function. It is therefore tempting to speculate that oncogenic Akt upregulates miRNA-mediated translational repression of tumor suppressors such as PTEN and PDCD4 while down-regulating miRNA-mediated cleavage of oncogenic mRNAs such as HoxB8 (Figure 6).
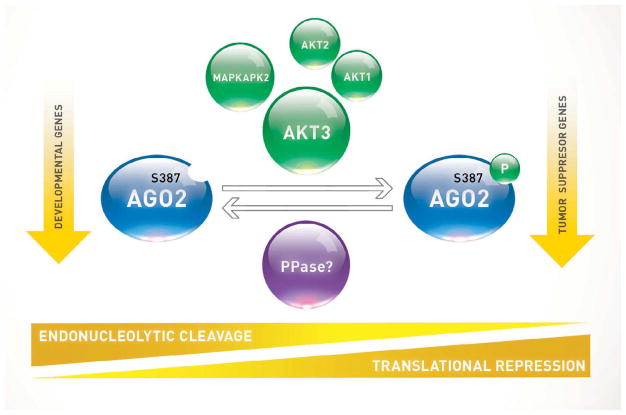
Figure 6. Akt3-Mediated Phosphorylation of Ago2 is a Molecular Switch Between Target mRNA Cleavage and Translational Repression.
Ago2 is capable of cleaving miRNA-targeted mRNAs or directing them for translational repression, which is dependent on its S387 phosphorylation status. This is dictated by Akt3, MK2 or to a lesser extent Akt1 or Akt2. Ago2 unphosphorylated at S387 maintains high endonucleolytic cleavage activity which results in suppressed expression of developmental genes. Ago2 phosphorylated at S387 switches to high translational repression which results in suppressed expression of tumor suppressor genes.
One outcome of miRNA targeting can be translocation of Agos, miRNAs, and target mRNAs to P-bodies (Jagannath and Wood, 2009; Liu et al., 2005a; Liu et al., 2005b). Our previous work implicated Ago2 phosphorylation at S387 in P-body localization (Zeng et al., 2008). Consistent with this report, our new data demonstrate that Akt3 is necessary for Ago2 phosphorylation, mRNA repression, and association with GW182, an important component of cytoplasmic P-bodies (Eulalio et al., 2009; Jagannath and Wood, 2009; Liu et al., 2005b). Although P-body formation may not be a prerequisite for mRNA degradation (Hu et al., 2009; Stoecklin et al., 2006), P-bodies may recruit ribonucleoprotein complexes that reinforce repression by shielding target mRNAs from the translation machinery (Eulalio et al., 2007b). For PTEN and PDCD4 target mRNAs, our data demonstrate that Akt3 and MK2 regulate translational repression without significantly affecting mRNA destabilization. For other miRNA-targeted mRNAs, our data do not discriminate whether phosphorylation of Ago2 leads first to translational repression of mRNA targets and then translocation to P-bodies and target mRNA destabilization, or whether phosphorylation of Ago2 leads directly to target mRNA translocation to P-bodies and target mRNA destabilization without translational repression.
Our immunofluorescence data demonstrate that most Ago2 protein co-localizes with GW182 protein. However, Ago2 S387 phosphorylation increases its GW182 association and P-body localization, while loss of S387 phosphorylation decreases its GW182 association by 55–70% (Compare Figures 4B and S4C). We note that the pS387 antibody used in our immunofluorescence assay did not recognize the EGFP-Ago2 fusion protein. It is conceivable that the N-term EGFP tag altered Ago2 conformation reducing recognition by the pS387 antibody. Our observations also imply the existence of a regulatory loop between pAgo2 and GW182 where the presence of one directly affects the level of the other, consistent with previously published data regarding P-body formation (Eulalio et al., 2007b). The causes and consequences of their interdependent expression are not known at this time but merit more thorough investigation.
Collectively, these data describe a mechanism enabling secondary regulation of the key RISC component Ago2 via phosphorylation by the proto-oncogene Akt. Ago2 S387 phosphorylation toggles Ago2 activity between small RNA-directed mRNA target cleavage and translational repression, directs its subcellular localization, and governs its interactions with the P-body protein GW182. Given the central role of Akt in coordinating extracellular stimuli and cellular growth signaling, it will be intriguing to identify the intracellular or extracellular stimuli that elicit specific Ago2-mediated gene silencing and the deeper mechanistic implications of toggling between mRNA cleavage and translational repressive activities.
EXPERIMENTAL PROCEDURES
All experimental procedures were performed according to manufacturers’ instructions unless otherwise noted.
Cell Culture, Transfections and Transductions
HeLa-D1 (stable line expressing firefly luciferase transgene and shRNA targeting firefly ORF), HeLa-D8 (stable line expressing firefly luciferase transgene harboring 6 perfect miR-21 binding sites in the 3′UTR and Renilla luciferase transgene with no miRNA binding sites), HeLa-EGFP-Ago2 [stable line expressing Ago2 fused with an N-terminal EGFP knocked into the endogenous Ago2 locus (Leung et al., 2006)], A375 (malignant melanoma), and HEK293T (human embryonic kidney cells expressing the large T-antigen of simian virus 40) were all maintained according to standard growth conditions in DMEM (Dulbecco’s modified Eagle’s medium, Gibco). Transfections were performed using Lipofectamine2000 (Invitrogen) for DNA plasmids. For siRNA transfections, 25nM of siRNA was used and cells were assayed at 48 hours post-transfection (unless otherwise indicated). For DNA transfections, 1 μg of DNA/mL of medium was used. Cells were allowed to express recombinant proteins for 48 hours before harvesting and protein extraction. HeLa-Luc cells were transduced with shRNA-expressing VSV-G pseudotyped lentiviruses in the presence of 6 μg/mL of polybrene overnight.
High Throughput Genomics Screening and Data Analysis
A library of 2176 Dicer-substrate siRNAs targeting 542 human protein kinase genes (IDT) was screened to identify siRNAs that inhibit RNAi in HeLa-Luc cells expressing an shRNA to Luc. Screening methods are described in (Chanda et al., 2003).
Luciferase Assays
Cells were plated in 96-well white bottom plates, grown to confluency and read using the Dual-Glo Luciferase Assay System (Promega) on a CLIPR Imager (Molecular Devices). Data was analyzed using CLIPR SoftMax Pro software (MDS Analytical Technologies). For calculating statistical significance and P values the Student’s Paired T-Test was used.
Translational Repression Reporter Assays
For mRNA cleavage versus translational repression reporter experiments, HEK293T cells were first transfected with siRNAs against endogenous genes, then transfected with FL6X (6 imperfect CXCR4 siRNA binding sites within the firefly luciferase 3′UTR) and RL0X (no CXCR4 siRNA binding sites). Then CXCR4 siRNAs (25nM) were transfected and the plates were incubated overnight before being read using the Dual-Glo Luciferase Assay System (Promega). Levels of luciferase derepression were calculated as FLuc/RLuc and normalized to non-targeting (NT) siRNA control samples.
Real-Time Quantitative PCR
Total RNA from cell lines was purified using Trizol reagent (Invitrogen). Purified RNA (1–10 μg) was reverse transcribed using either the cDNA High Capacity Archive kit (Applied Biosystems) or random hexamer primers with SuperScript III (Invitrogen) after DNase I treatment. Real-time PCR was performed using either Taqman primer/probe sets or Platinum SYBR Green qPCR SuperMix-UDG with ROX (Invitrogen) on an ABI 7900 amplification system (Applied Biosystems). Taqman primer/probe set information can be found in supplementary material. Gene Expression cycling thresholds (Cts) were analyzed using SDS 7000 1.1 RQ software (Applied Biosystems) and normalized to endogenous Gapdh.
Immunoprecipitations and Western Blots
IPs were performed with either anti-His-Tag mAb sepharose bead conjugates (4079s, Cell Signaling Technology) or anti-FLAG agarose (A2220, Sigma-Aldrich) pre-blocked with BSA. PTEN IPs were performed using anti-PTEN-N-term (sc-133197, Santa Cruz Biotechnology Inc.), PDCD4 IPs were performed using anti-PDCD4 (ab45124, Abcam), βactin IPs were performed using anti-βactin (ab8226, Abcam) and the Pierce Classic I.P. Kit (Pierce). Cross-linking, when employed, was performed using 2 mM DTSSP (Thermo Scientific).
Western blot analyses were performed according to standard methods using denaturing gels (Invitrogen) and the iBlot Transfer System (Invitrogen). Antibody information can be found in supplementary material. Westerns were resolved with SuperSignal® West Dura Extended Duration Substrate (Thermo Scientific) on a VersaDoc Imaging System (BioRad). Exposures were analyzed using the Quantity-One Imaging Software (Bio-Rad).
Expression and Purification of Recombinant Proteins and Kinase Assays
cDNAs encoding the S873A or S387E mutant Ago2s or the T305A mutant Akt3 were generated by Quickchange PCR (Stratagene). Wild-type Akt1, Akt2 and Akt3 human cDNAs were cloned into the pDEST-26 Gateway-compatible vector (His-tag, Invitrogen). His-tagged proteins were purified using the His-Bind® Purification Kit (Novagen). Wild-type and mutant Ago2 proteins were expressed and purified as described previously (MacRae et al., 2008).
Kinase assays were performed according to standard methods using 500 ng of Akt protein with 5 μg of Ago2 protein in 1x kinase buffer (Cell Signaling Technologies) in the presence of 10 μCi γ-32P-ATP (Perkin Elmer) and 50 μM cold ATP. 10 μg of purified recombinant Histone H2B (New England Biolabs) was used as positive control substrate for Akt phosphorylation.
In vitro Target mRNA Cleavage Assays
Bacterially-expressed Ago2 was incubated with Akt for one hour at 37°C and the cleavage activity of the phosphorylated Ago2 was assayed by incubating the Ago-Akt mixture with siCXCR4 and 5′-labeled CXCR4 substrate RNA. The cleavage products were resolved by PAGE and exposed to a Storm Phosphoimager (GE Healthcare). The specific cleavage products were indicated with an arrow based on the ladder generated from partial digestion of input substrate by RNase T1 (T1). Cleavage efficiency (%) was quantified by the equation: (intensity of cleavage products/intensity of total input) × 100.
Immunofluorescence Assays
For the analysis of Ago2 or phospho-Ago2 cytoplasmic association with the P-body component GW182, either stable or transient HeLa-EGFP-Ago2 cells were used, stained and imaged according to standard methods. Akt knockdown cells and Ago2 mutant expressing cells were expanded in culture for 36 hrs before being starved overnight, then stimulated with FBS and insulin (40ng/mL) for 20 min. Antibody and laser information can be found in supplementary material. All images were captured on a Perkin Elmer UltraVIEW confocal system mounted on an Olympus IX81 microscope using a 40x water objective. Images were processed using Volocity 3D Image Analysis Software (Perkin Elmer).
Translation Rate Assays
For the analysis of translational repression of miR-21 targets, HEK293T Akt knockdown cells were expanded in culture for 36 hrs before being serum starved overnight. Cells were stimulated with FBS and insulin (40 ng/mL) for 30 min to encourage protein translation before the addition of 35S-methionine and 35S-cysteine (0.5 mCi total, Easy Tag Express Protein Labeling Mix, Perkin Elmer) for 45 min. Approximately 500 μg of cell lysates was immunoprecipitated using the Pierce Classic I.P. Kit (Pierce) using 2–5 μg of antibodies. IP protein gels were exposed to a phosphoimaging screen for 3–5 days before analysis on a Storm Phosphoimager (GE Healthcare). Autoradiograph band intensities were quantified using the ImageQuant 5.2 software package (GE Healthcare) and scored relative to the NT control.
Supplementary Material
HIGHLIGHTS.
Akt3 interacts with and phosphorylates Ago2 at Ser387.
Phosphorylation of Ago2 increases translational repression of pten and pdcd4.
Phosphorylation of Ago2 decreases mRNA cleavage of the hoxb8 oncogene.
Akt3-mediated phosphorylation of Ago2 is an RNAi molecular switch.
Acknowledgments
We wish to thank Richard G. Brusch for assistance with confocal microscopy, Angelica Romero for synthesis of viruses, Genevieve Welch for running the human kinome siRNA screen, Anthony Marelli for assistance with cloning Ago2 cDNAs, Myleen N. Medina for synthesis of AKT shRNA plasmids, Daniel E. Mason for performing LC/MS and Lauren C. Fox for generation of Figure 6. This work was supported by a Distinguished Young Scholars Award from the W.M. Keck Foundation to C.D.N.
The abbreviations used are
- RNAi
RNA interference
- Ago2
Argonaute-2
- P-bodies
processing bodies
- RISC
RNA-induced silencing complex
- siRNA
small interfering RNA
- shRNA
short hairpin RNA
- miRNA
microRNA
- MK2
MAPKAPK2
- LC/MS
liquid chromatography–mass spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–2591. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- Bracken CP, Szubert JM, Mercer TR, Dinger ME, Thomson DW, Mattick JS, Michael MZ, Goodall GJ. Global analysis of the mammalian RNA degradome reveals widespread miRNA-dependent and miRNA-independent endonucleolytic cleavage. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda SK, White S, Orth AP, Reisdorph R, Miraglia L, Thomas RS, DeJesus P, Mason DE, Huang Q, Vega R, et al. Genome-scale functional profiling of the mammalian AP-1 signaling pathway. Proc Natl Acad Sci U S A. 2003;100:12153–12158. doi: 10.1073/pnas.1934839100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E, Caiment F, Tordoir X, Cavaille J, Ferguson-Smith A, Cockett N, Georges M, Charlier C. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr Biol. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- Dummler B, Hemmings BA. Physiological roles of PKB/Akt isoforms in development and disease. Biochem Soc Trans. 2007;35:231–235. doi: 10.1042/BST0350231. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007a;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007b;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Tritschler F, Izaurralde E. The GW182 protein family in animal cells: New insights into domains required for miRNA-mediated gene silencing. Rna-a Publication of the Rna Society. 2009;15:1433–1442. doi: 10.1261/rna.1703809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannath A, Wood MJA. Localization of Double-stranded Small Interfering RNA to Cytoplasmic Processing Bodies Is Ago2 Dependent and Results in Up-Regulation of GW182 and Argonaute-2. Molecular Biology of the Cell. 2009;20:521–529. doi: 10.1091/mbc.E08-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK. Disruption of GW bodies impairs mammalian RNA interference. Nature cell biology. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Karginov FV, Cheloufi S, Chong MM, Stark A, Smith AD, Hannon GJ. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CT, Risom T, Strauss WM. Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol. 2007;26:209–218. doi: 10.1089/dna.2006.0545. [DOI] [PubMed] [Google Scholar]
- Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Fan J, Zeng W, Zhou Y, Ingvarsson S, Chen H. Quantitative analysis of miRNA expression in several developmental stages of human livers. Hepatol Res. 2010;40:813–822. doi: 10.1111/j.1872-034X.2010.00683.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nature cell biology. 2005a;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JD, Rivas FV, Wohlschlegel J, Yates JR, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nature cell biology. 2005b;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci U S A. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Kim S, Younes MN, Jasser SA, El-Naggar AK, Mills GB, Myers JN. The Akt inhibitor KP372-1 suppresses Akt activity and cell proliferation and induces apoptosis in thyroid cancer cells. British journal of cancer. 2005;92:1899–1905. doi: 10.1038/sj.bjc.6602595. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Masure S, Haefner B, Wesselink JJ, Hoefnagel E, Mortier E, Verhasselt P, Tuytelaars A, Gordon R, Richardson A. Molecular cloning, expression and characterization of the human serine/threonine kinase Akt-3. Eur J Biochem. 1999;265:353–360. doi: 10.1046/j.1432-1327.1999.00774.x. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Polytarchou C, Iliopoulos D, Hatziapostolou M, Kottakis F, Maroulakou I, Struhl K, Tsichlis PN. Akt2 regulates all Akt isoforms and promotes resistance to hypoxia through induction of miR-21 upon oxygen deprivation. Cancer Res. 2011 doi: 10.1158/0008-5472.CAN-11-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restuccia DF, Hemmings BA. From man to mouse and back again: advances in defining tumor AKTivities in vivo. Dis Model Mech. 2010;3:705–720. doi: 10.1242/dmm.004671. [DOI] [PubMed] [Google Scholar]
- Rudel S, Wang Y, Lenobel R, Korner R, Hsiao HH, Urlaub H, Patel D, Meister G. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res. 2011;39:2330–2343. doi: 10.1093/nar/gkq1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol. 2010;17:1169–1174. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- Tiemann K, Rossi JJ. RNAi-based therapeutics-current status, challenges and prospects. EMBO Mol Med. 2009;1:142–151. doi: 10.1002/emmm.200900023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Wang B, Li S, Qi HH, Chowdhury D, Shi Y, Novina CD. Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat Struct Mol Biol. 2009;16:1259–1266. doi: 10.1038/nsmb.1712. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Sankala H, Zhang X, Graves PR. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J. 2008;413:429–436. doi: 10.1042/BJ20080599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.