SUMMARY
CDP-diacylglycerol (CDP-DAG) is central of the phospholipid biosynthesis pathways in cells. A prevailing view is that only one CDP-DAG synthase named Cds1 is present in both the endoplasmic reticulum (ER) and mitochondrial inner membrane (IM), and mediates generation of CDP-DAG from phosphatidic acid (PA) and CTP. However, we demonstrate here by using yeast Saccharomyces cerevisiae as a model organism that Cds1 resides in the ER but not in mitochondria, and that Tam41, a highly conserved mitochondrial maintenance protein, directly catalyzes the formation of CDP-DAG from PA in the mitochondrial IM. We also find that inositol depletion by overexpressing an arrestin-related protein Art5 partially restores the defects of cell growth and CL synthesis in the absence of Tam41. The present findings unveil the missing step of the cardiolipin synthesis pathway in mitochondria as well as the flexibile regulation of phospholipid biosynthesis to respond to compromised CDP-DAG synthesis in mitochondria.
INTRODUCTION
Phospholipids, major components of cellular membranes are mainly generated via sequential modifications of PA by multiple phospholipid-synthetic enzymes located in various cellular compartments such as the ER, Golgi, and mitochondria (Henry et al., 2012; Osman et al., 2011; van Meer et al., 2008). PA is converted to an important intermediate CDP-DAG by CDP-DAG synthase by using a nucleotide CTP (Shen et al., 1996). Then phospholipid-synthetic pathways are branched into several different pathways, one of which leads to synthesis of cardiolipin (CL), a mitochondria-specific phospholipid crucial for optimal mitochondrial functions (Joshi et al., 2009; Claypool et al., 2009). The phospholipid synthetic pathway is well conserved between yeast and mammals. For the synthesis of CL in yeast Saccharomyces cerevisiae, CDP-DAG is first converted to phophatidylglycerol phosphate (PGP) by Pgs1 PGP synthase (Chang et al., 1998a), and then PGP is dephosphorylated to form phophatidylglycerol (PG) by Gep4 PGP phosphatase (Osman et al., 2010). PG and CDP-DAG are coupled to form CL by Crd1 cardiolipin synthase (Tuller et al., 1998; Chang et al., 1998b). For maturation of CL, Cld1 cardiolipin-specific phospholipase and Taz1 acyltransferase remodel CL acyl-chains (Beranek et al., 2009; Gu et al., 2004). All these CL synthetic enzymes are located in the mitochondrial IM or matrix while ER-resident Cho1 PS synthase and Pis1 PI synthase convert CDP-DAG to PS (phosphatidylserine) and PI (phosphatidylinositol) by using serine and inositol, respectively (Henry et al., 2012; Osman et al., 2011). In mammalian cells, PS is synthesized from pre-existing PC or PE and serine through the base-exchange reaction (Kuge and Nishijima, 1997; Vance and Tasseva, 2013). In S. cerevisiae, only one CDP-DAG synthase, Cds1, was identified for formation of CDP-DAG (Shen et al., 1996). Cds1 is conserved from bacteria to human and its enzymatic activities are reportedly present in both the ER and mitochondrial IM (Kuchler et al., 1986; Kelly et al., 1987). However the precise cellular localization of Cds1 has not yet been determined so far.
In this study, we carefully analyzed the cellular localization of Cds1 and found that Cds1 is exclusively present in the ER, indicating existence of another CDP-DAG synthase in mitochondria. We further show that Tam41, a mitochondrial IM protein facing the matrix (Tamura et al., 2006), functions as a novel CDP-DAG synthase. Our findings solve a long-standing mystery of how cells produce a key intermediate phospholipid, CDP-DAG, in distinct organelles and provide insight into the CL synthetic pathway in mitochondria.
RESULTS
Cds1 functions in the ER
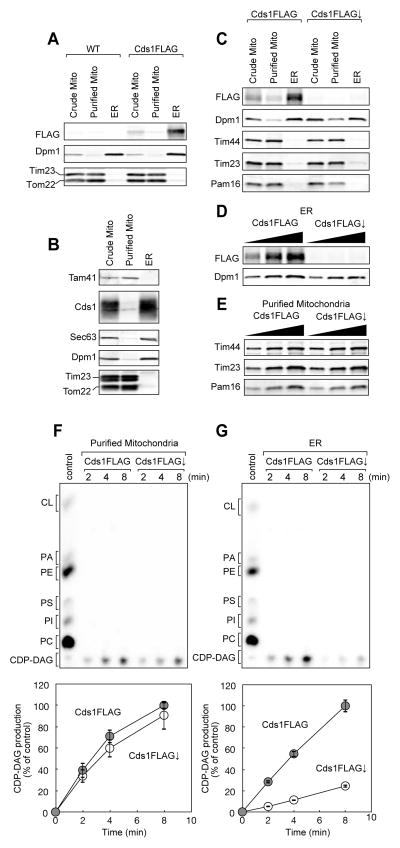
To examine the cellular location of Cds1, we purified the ER and mitochondrial fractions from yeast cells expressing FLAG-tagged Cds1 (Cds1FLAG) and performed immunoblotting of Cds1 with the anti-FLAG antibody (Figure 1A). Cds1FLAG was, like an ER protein marker Dpm1, mainly found in the ER, but not in highly purified mitochondria while mitochondrial proteins Tim23 and Tom22 were enriched in the purified mitochondrial fraction (Figure 1A). We also confirmed by using anti-Cds1 antibodies that authentic Cds1 is enriched in the ER (Figure 1B). These results indicate that Cds1 is an ER-resident protein, arguing against the prevailing view of the dual localization of Cds1 in the ER and mitochondria. Rather, they suggest the existence of a distinct CDP-DAG synthase in mitochondria. To test this possibility, we utilized the yeast strain expressing Cds1FLAG under the control of the TetO7 promoter (Mnaimneh et al., 2004), and examined CDP-DAG synthase activities of mitochondria purified from the cells with or without Cds1FLAG depletion by addition of doxycycline. We confirmed by using the anti-FLAG antibody that the expression level of Cds1FLAG was significantly decreased in the ER fraction upon Cds1FLAG depletion while levels of mitochondrial proteins such as Tim44, Tim23, and Pam16 were not affected (Figures 1C-1E). Then we monitored generation of CDP-DAG by incubating PA with purified mitochondria or ER fraction, which were solubilized with Triton X100 in the presence of [α-32P]CTP (Figures 1F and 1G). Mitochondria with and without Cds1FLAG depletion showed similar CDP-DAG synthase activities while production of CDP-DAG was dramatically decreased in the Cds1FLAG-depleted ER fractions. This strongly suggests that mitochondria possess a CDP-DAG synthase that is distinct from Cds1.
Figure 1. Cds1 is an ER-resident protein.
Crude and purified mitochondria and ER fractions were prepared from (A, B) wild-type and Cds1FLAG or (C) Cds1FLAG-expressing (Cds1FLAG) and Cds1FLAG-depleted (Cds1FLAG↓) cells and analyzed by immunoblotting with the indicated antibodies. (D, E) ER and purified mitochondria fractions isolated from Cds1FLAG and Cds1FLAG↓ cells were analyzed by immunoblotting with the indicated antibodies. (F, G) Phosphatidic acid (PA) and purified mitochondria or ER fractions solubilized with Triton X100 were incubated for the indicate time in the presence of 32P-CTP. Phospholipids were then extracted and analyzed by TLC. Amounts of CDP-DAG generated after 8 min incubation in wild-type cells is set to 100%. Values are mean ± SEM (n=3).
Tam41 is a CDP-DAG synthase in mitochondria
What is the identity of the mitochondrial protein responsible for the observed CDP-DAG synthase activity? The putative mitochondrial CDP-DAG synthase is likely present in the IM or matrix because it is reportedly protease insensitive even after rupturing the mitochondrial outer membrane (OM; Kuchler et al., 1986). Besides, loss of a mitochondrial CDP-DAG synthase should lead to significant reductions in the CL level as well as accumulations of PA, the precursor of CDP-DAG. On the basis of these considerations, we reasoned that Tam41 could be a potential candidate. Tam41 is a peripheral IM protein facing the matrix, and was originally identified as a maintenance protein for the IM translocator, the TIM23 complex (Tamura et al., 2006; Gallas et al., 2006), which mediates translocation of presequence-containing precursor proteins across or into the IM. tam41Δ cells exhibit defects in cell growth at elevated temperature and in mitochondrial protein import, likely due to the destabilized TIM23 complex (Tamura et al., 2006; Gallas et al., 2006). This destabilization of the TIM23 complex is associated with loss of CL, which also destabilizes several IM protein complexes, and with concomitant accumulation of PA (Kutik et al., 2008).
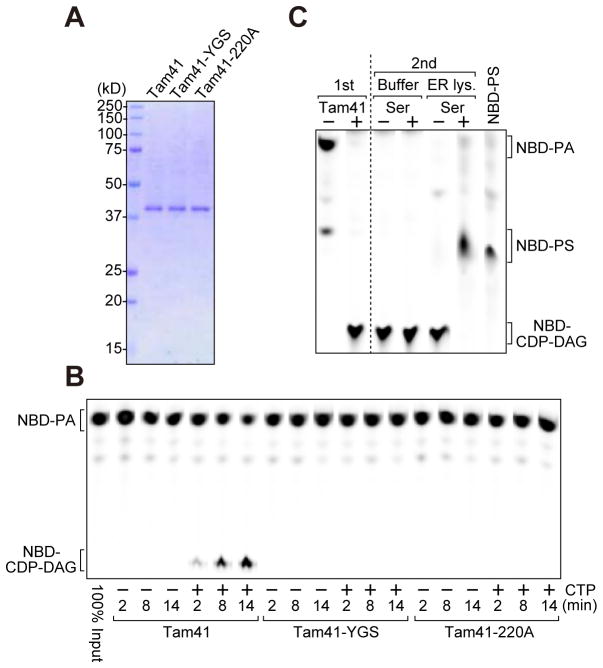
We thus asked if Tam41 takes the role of the mitochondrial CDP-DAG synthase. For this purpose, we overexpressed C-terminally hexahistidine-tagged Tam41 under the GAL1 promoter in yeast cells, and purified it with the Ni-NTA agarose resin followed by ion-exhange chromatography using SP-shepharose (Figure 2A, Tam41; Figure S1A). Then we measured its CDP-DAG synthase activity by using a fluorescence-labeled substrate, nitrobenzoxadiazole (NBD)-PA (Figure 2B). When we incubated NBD-PA and purified Tam41, NBD-PA was efficiently consumed and an additional lipid product with smaller migration similar to CDP-DAG on the TLC plate accumulated in a CTP-dependent manner (Figure 2B, Tam41). As a negative control, we used Tam41 mutants (Figure 2A, Tam41-220A and Tam41-YGS) with the single and triple mutations, D220A and Y130A/G131A/S132A, respectively, and confirmed that these mutations totally abolish the enzymatic activities (Figure 2B, Tam41-220A, Tam41-YGS). This result is consistent with our previous observation that Tam41-220A and Tam41-YGS cannot complement the growth defects of the tam41Δ cells (Harada et al., 2010; In this reference, the Art5 mutant Y130A/G131A/S132A was mistakenly indicated as Y131A/G132A/S133A). To further confirm the identity of the lipid product in Figure 2B as CDP-DAG, we tested if it could be converted to a subsequent phospholipid such as PS in the lipid synthetic pathway. We first incubated 10 μM NBD-PA with purified Tam41 in the presence of the excess amount of CTP (1 mM) for 30 min until NBD-PA was completely consumed, so that the lipid product was exclusively present (Figure 2C, 1st, Tam41+). Then, the lipid procuct was incubated with the Triton X100-solubilized ER fraction containing Cho1 PS synthase in the presence of serine (Figure 2C, 2nd, ER lysate, Ser+), which resulted in generation of a phospholipid co-migrating with NBD-PS. This indicates that the accumulating lipid product with a slower migration was indeed CDP-DAG (Figure 2C). Taken together, we conclude that Tam41 directly catalyzes the conversion of PA to CDP-DAG in the presence of CTP. Yeast cells have thus two distinct CDP-DAG synthases, Cds1 and Tam41, residing in the ER and mitochondrial IM, respectively, for phospholipid biosynthesis.
Figure 2. Tam41 is a mitochondrial CDP-DAG synthase.
(A) Purified Tam41-His6 and its mutants were analyzed by SDS-PAGE followed by CBB-staining. (B) NBD-PA and purified Tam41 and Tam41 mutants (Tam41-YGS and Tam41-220A) were incubated for the indicated periods of time at 30°C in the presence or absence of CTP. Phospholipids were then extracted and analyzed by TLC. Note that Tam41 appears to belong to the superfamily of nucleotidetransferase (NTase) fold proteins containing conserved active site residues; hG[GS], [DE]h[DE]h and h[DE]h (h indicates a hydrophobic amino acid) (Kuchta et al., 2009). Tam41-YGS mutant has mutations in the hG[GS] motif, which may has a role in holding substrates within the active site, while it is not clear if Asp220 functions in the active site since intact [DE]h[DE]h and h[DE]h motifs are missing in Tam41. (C) CDP-DAG generated in vitro was further incubated with solubilized ER fractions (ER lys.) in the presence or absence of serine. See also Figure S1.
Identificaiton of Tam41 as a mitochondrial CDP-DAG synthase means that four CL synthetic enzymes, Tam41, Pgs1, Gep4 and Crd1, are in operation in yeast mitochondria. Interestingly, although yeast mutants lacking one of these enzymes are not able to produce CL in common, they are phenotypically different. tam41Δ and pgs1Δ cells showed severe growth defects on fermentable (Figure S1B, YPD) or non-fermentable (Figure S1C, YPGE) media at high temperature as compared with gep4Δ and crd1Δ cells, and the growth defects of gep4Δ cells were stronger than those of crd1Δ cells. As reported previously (Kutik et al., 2008; Gerbert et al., 2009; Osman et al., 2010), blue-native PAGE analyses showed that those CL synthesis mutants have a slightly smaller TOM40 complex in the OM and smaller TIM23 and TIM22 complexes in the IM (Figure S1D). The respiratory-chain supercomplex consisting of complexes III and IV and the TOM40 complex were partly destabilized in tam41Δ and pgs1Δ mitochondria, and a part of complex V fell apart in pgs1Δ mitochondria (Figure S1D). In tam41Δ and pgs1Δ mitochondria, but not in gep4Δ or crd1Δ mitochondria, the steady-state levels of cytochrome c1 (Cyt c1) of complex III and cytochrome oxidase subunit IV (Cox4) of complex IV were decreased (Figures S1D and S1E). These phenotypic differences are probably caused by accumulation of different intermediate phospholipids such as PA, CDP-DAG, PGP or PG in tam41Δ, pgs1Δ, gep4Δ or crd1Δ cells, respectively, in mitochondria.
Enzymological properties of Tam41
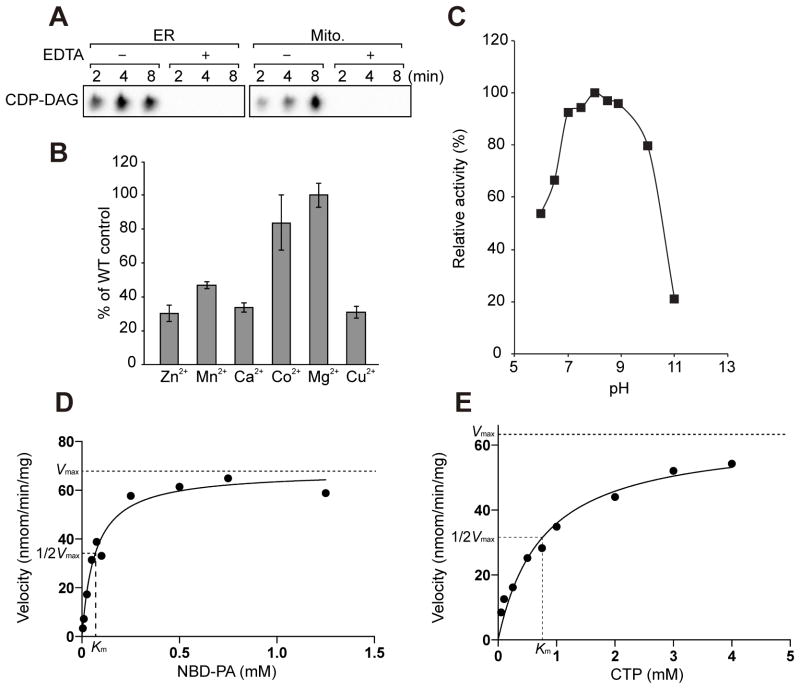
Next, we characterized the enzymological properties of purified Tam41 CDP-DAG synthase. First, we tested the requirement of divalent metal ions. The Cds1 CDP-DAG synthase activity requires Mg2+ and is inhibited by EDTA (Kelly and Carman, 1987; Nigou and Besra, 2002; Fig. 3A). Similarly, highly purified mitochondria containing Tam41 CDP-DAG synthase were not able to produce CDP-DAG in the presence of EDTA (Figure. 3A). Dependence of the Tam41 CDP-DAG activity on different divalent metal ions was, however, not exactly the same as that of the Mycobacterium smegmatis Cds1 CDP-DAG synthase activity (Nigou and Besra, 2002) (Figure 3B). For example, Tam41, but not M. smegmatis Cds1, exhibits the CDP-DAG synthase activity in the presence of Co2+ (~80% of the case for Mg2+) and Cu2+ (~30% of the case for Mg2+). Next, we followed the pH dependence of the Tam41 CDP-DAG synthase activity in the range of pH 6–11, and found that the enzymatic activity is maximal at around pH 7–9 (Figure 3C).
Figure 3. Enzymological analyses of Tam41.
(A) CDP-DAG synthase activities of highly purified ER and mitochondria fractions were measured in the presence of 10 mM EDTA. (B) Purified Tam41 and NBD-PA were incubated in the presence of 3 mM divalent metal ions for 5 min. The CDP-DAG synthase activity detected in the presence of Mg2+ was set to 100%. Values are mean ± SEM (n=3). (C) pH profile of Tam41 CDP-DAG synthase. Tam41 CDP-DAG activity was determined at varying pH. The buffer used are 50 mM PIPES-NaOH pH 6.0 and 6.5; 50 mM Tris-HCl pH 7.0, 7.5, 8.0, 8.5 and 8.9; and 50 mM CAPS-NaOH pH 10 and 11. The CDP-DAG activity obtained at pH 8.0 was set to 100%. (D, E) Kinetic analyses of Tam41 CDP-DAG synthase on the concentrations of NBD-PA and CTP. NBD-PA and purified Tam41 were incubated for 10 min at 30°C. 2 mM CTP or 250 μM NBD-PA was used for (D) and (E) experiments, respectively. Curve fitting was performed with Prism 6.0 (GraphPad software).
By measuring the Tam41 CDP-DAG synthase activities at different concentrations of NBD-PA and CTP, we confirmed that the CDP-DAG synthase activities follow normal saturation kinetics and analyzed the data to obtain enzymatic kinetic parameters, Km for NBD-PA and CTP and Vmax (Figures 3D and 3E). The obtained Km value for NBD-PA was 0.067±0.01 mM, which is smaller than the Km values for PA for Cds1 from yeast (0.50 mM), E. coli (0.28 mM), Plasmodium falciparum (0.90 mM) and M. smegmatis (0.61 mM) (Kelly and Carman, 1987; Martin et al., 2000; Nigou and Besra, 2002; Sparrow and Raetz, 1985). The small Km value for NBD-PA for Tam41 may well reflect the higher affinity of Tam41 for PA than Cds1 CDP-DAG synthases, although it may be perhaps due to the nature of the unnatural phospholipid, NBD-PA, which contains a relatively short acyl chain in addition to the normal length of the acyl chain, as compared with PA. The Km value for CTP was 0.76±0.14 mM, which is similar to those for Cds1 from yeast (1.0 mM) and E. coli (0.58 mM) (Kelly and Carman, 1987; Sparrow and Raetz, 1985). The Vmax value was 68±3.0 nmol/min/mg, which is smaller than that for yeast Cds1 (4,700 nmol/min/mg) (Kelly and Carman, 1987).
Decreased inositol level restores the growth defects of tam41Δ cells
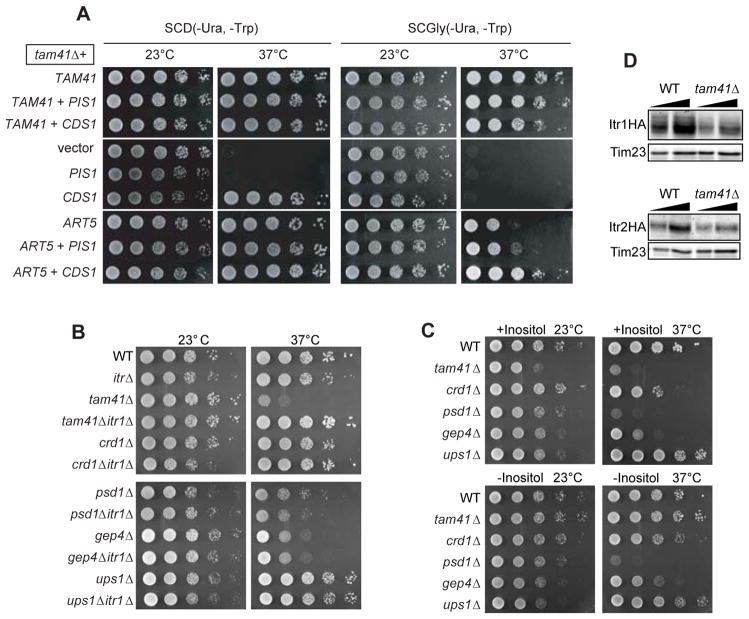
Recently, we identified Art5, a member of the ART (arrestin-related trafficking adaptor) protein family (Lin et al., 2008) as a multicopy suppressor for the temperature-sensitive tam41-ts as well as tam41Δ mutants (Harada et al., 2010). Overexpression of Art5 restored the temperature-sensitive growth defects of tam41Δ cells completely on fermentable media (Figure 4A, SCD), but only partly on non-fermentable media, which renders the cell growth dependent on mitochondrial respiration (Figure 4A, SCGly) (Harada et al., 2010).
Figure 4. Decreased inositol uptake restores the growth defects of tam41Δ cells.
(A) Serial dilutions of tam41Δ cells with a multi-copy plasmid harboring the TAM41, PIS1, CDS1 or ART5 gene or empty vector were spotted onto glucose- or glycerol-containing media and cultivated for 2 or 4 days, respectively. (B, C) Serial dilutions of the indicated yeast strains were spotted on SCD plates at 23 or 37°C (B) or on Synthetic minimal plates with or without inositol at 23 or 37°C (C). (D) Whole cell extracts prepared from wild-type and tam41Δ cells expressing C-terminally HA-tagged Itr1 or Itr2 were analyzed by immunoblotting with the antibodies against HA-tag and Tim23.
Art5 mediates regulated degradation of the inositol transporter Itr1 in response to exogenous inositol (Lin et al., 2008; Nikko et al., 2009). This raises a possibility that overexpression of Art5 perturbs phospholipid biosynthetic pathways through an altered cellular level of inositol. Indeed, PI levels decreased when Art5 was overexpressed (Figure 5A). We thus tested the effects of deletion of the ITR1 gene on the growth of tam41Δ and several mutant strains with decreased CL levels, crd1Δ (Chang et al. 1998b), gep4Δ (Osman et al., 2010), and ups1Δ (Tamura et al., 2009; Osman et al., 2009) and with a decreased PE level, psd1Δ (Clancey et al., 1993) (Figure 4B). Ups1 was recently found to mediate intramitochondrial transport of PA (Connerth et al., 2012). The growth defects of tam41Δ at elevated temperature were suppressed by the simultaneous deletion of the ITR1 gene (Figure 4B). In contrast, the weak growth defects of crd1Δ, gep4Δ, psd1Δ, or ups1Δ cells were not suppressed by the ITR1 deletion (Figure 4B). We then depleted inositol from the culture medium instead of deletion of the ITR1 gene. Depletion of inositol restored the growth defects of tam41Δ cells, but not of crd1Δ, psd1Δ, gep4Δ, or ups1Δ cells (Figure 4C). Steady-state levels of inositol transporters Itr1 and Itr2 decrease in tam41Δ cells likely in order to adopt the circumstances of compromised CL synthesis (Figure 4D). Since the Art5 overexpression, depletion of inositol, and deletion of the ITR1 inositol transporter gene suppresses the growth defects of tam41Δ cells specifically and in similar manners, overexpression of Art5 likely compensates the lack of Tam41 through decrease in the cellulear level of inositol.
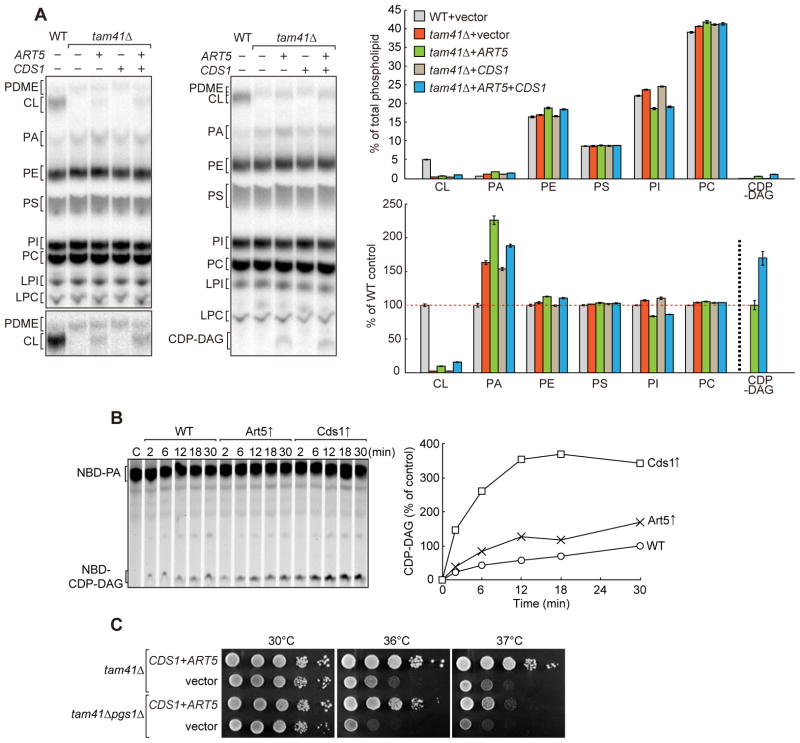
Figure 5. Phospholipid profiles of tam41Δ cells upon overexpression of Art5 or Cds1.
(A) WT and tam41Δ cells with a multi-copy plasmid harboring the indicated gene or empty vector were cultivated in YPD containing 32P at 30°C. Total phospholipids were extracted from whole cells and separated on TLC for 2h (left) or 4h (right). A part of TLC plate showing PDME and CL is shown with high contrast (left below). Amounts of each lipid relative to total phospholipids (above) and to those in WT cells (except for CDP-DAG) (below) were determined. In the latter representation, the amount of CDP-DAG relative to that in the “tam41Δ+ART5” strain (CDP-DAG) was shown. Values are mean ± SEM (n=3). (B) CDP-DAG synthetic activities in ER fractions. Triton X100-solubilized ER fractions prepared from wild-type cells with or without overexpression of Art5 or Cds1 were incubated with 100 μM NBD-PA in the presence of 2 mM CTP for the indicated times at 30°C. Amounts of CDP-DAG generated after 30 min incubation in wild-type ER fraction is set to 100%. (C) Overexpression of Cds1 and Art5 rescues growth defects of tam41Δpgs1Δ cells. Serial dilutions of tam41Δ and tam41Δpgs1Δ cells with multi-copy plasmids expressing both Cds1 and Art5 or with empty vectors were spotted onto YPD and cultivated for 3 days. See also Figure S2
Overexpression of Art5 alters phospholipid profiles in tam41Δ cells
Interestingly, overexpression of Cds1, the ER CDP-DAG synthase, rescued temperature-sensitive growth defects of tam41Δ cells lacking the mitochondrial CDP-DAG synthase on fermentable media (Figure 4A, SCD), although it did not on non-fermentable media (Figure 4A, SCGly). Besides, overexpression of both Cds1 and Art5 synergetically rendered tam41Δ cells capable of growing on non-fermentable as well as fermentable media completely at 37°C (Figure 4A). This suggests the possibility of the uncharacterized delivery route of CDP-DAG from the ER to mitochondria, which could become prominent upon Cds1 overexpression and then result in restoration of the CL level in mitochondria. However, the effects of Cds1 or Art5 overexpression is complicated since the CL level was not restored in tam41Δ cells by Cds1 overexpression alone (Figure 5A). Instead, overexpression of Art5 led to detectable accumulations of CL and CDP-DAG in tam41Δ cells (Figure 5A) despite that overexpression of Cds1 alone increased production of CDP-DAG in the ER more significantly than Art5 overexpression in vitro (Figure 5B). These results suggest that overproduction of CDP-DAG alone in the ER is not sufficient to accumulate detectable amounts of CDP-DAG and CL, or in other words that the transport of CDP-DAG from the ER to mitochondria is therefore, if any, inefficient.
Importantly, overexpression of both Cds1 and Art5 rescued growth defects of pgs1Δtam41Δ cells, which are presumably unable to produce CL (Figure 5C). This suggests that restoration of the CL level may not be the sole mechanism for the retrieval of the defective growth of tam41Δ cells by Art5 overexpression or inositol depletion. In relation to this, we note that, when Art5 was overexpressed, the levels of phospholipids other than CL were also perturbed; CDP-DAG, PA and PE levels increased while the PI level decreased. Since CL is an acidic phospholipid that favors local non-bilayer structures, acidic phospholipids CDP-DAG and PA and non-bilayer-forming lipid PE could functionally substitute the roles of CL in part (Gohil et al., 2005; Joshi et al., 2012; Rand and Sengupta, 1972; van den Bring-van der Laan et al., 2004). Therefore not only CL restoration but overall changes in the phospholipid profile including the levels of CDP-DAG, PA, and PE may give positive effects on cell growth in the absence of Tam41.
DISCUSSION
In this study, we found that two CDP-DAG synthases, Cds1 and Tam41, function in distinct organelles, the ER and mitochondria, respectively, in yeast (Figure 6). Eukaryotic cells require two CDP-DAG synthases in the ER and mitochondria for efficient phospholipid synthesis due to inefficient CDP-DAG transport between the ER and mitochondria.
Figure 6. Phospholipid biosynthesis leading to CL, PI, and PS in mitochondria and the ER.
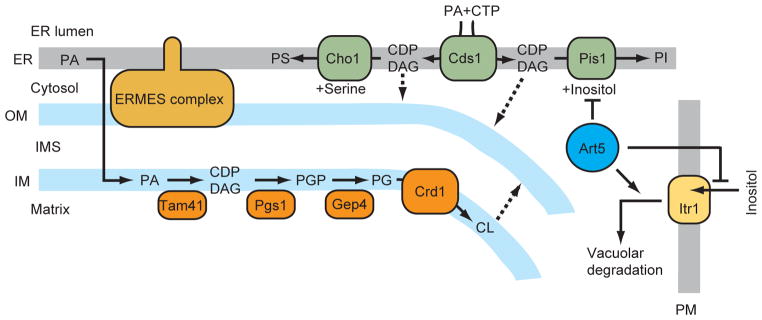
Broken lines indicate putative transport routes. OM, mitochondrial outer membrane; IM, mitochondrial inner membrane; PM, plasma membrane.
Both Tam41 and Cds1 are highly conserved proteins, yet their origins seem to be different. Our sequence analysis and a previous study (Kuchta et al., 2009) suggest that the N-terminal portion of Tam41 may possess the NTase (Nucleotide Transferase) fold, which is consistent with the CDP-DAG synthase function of Tam41, although the NTase activity of Tam41 and its family members had not been predicted. In contrast, Cds1 exhibits the CDP-DAG synthase i.e. NTase activity, yet it does not contain the NTase fold. While orthologs of Tam41 are present exclusively in eukaryotes (Figure 7A), potential homologs of Tam41 are well conserved in proteobacteria including a-proteobacteria, which are close to the presumable endosymbiotic origin of mitochondria (Figure 7C) (Gray et al., 1999). On the other hand, orthologs of Cds1 can be widely found in prokaryotes as well as eukaryotes (Figure 7B). Therefore Cds1 was likely derived from the ancient eukaryotes that took up mitochondrial ancestors, and that eukaryotic cells have conserved two distinct CDP-DAG synthases, Tam41 and Cds1, after establishment of mitochondria. Possible evolutionary constraints behind this could be that, while PA is transported from the ER membrane to the mitochondrial IM efficiently (Connerth et al., 2012), transport of CDP-DAG between the ER membrane and mitochondrial IM may be inefficient, if any, as discussed later. Cds1 and Tam41 have therefore developed their own commitment to lipid biosynthetic pathways operating in two evolutionary distinct organelles, the ER and mitochondria, during evolution of eukaryotic cells.
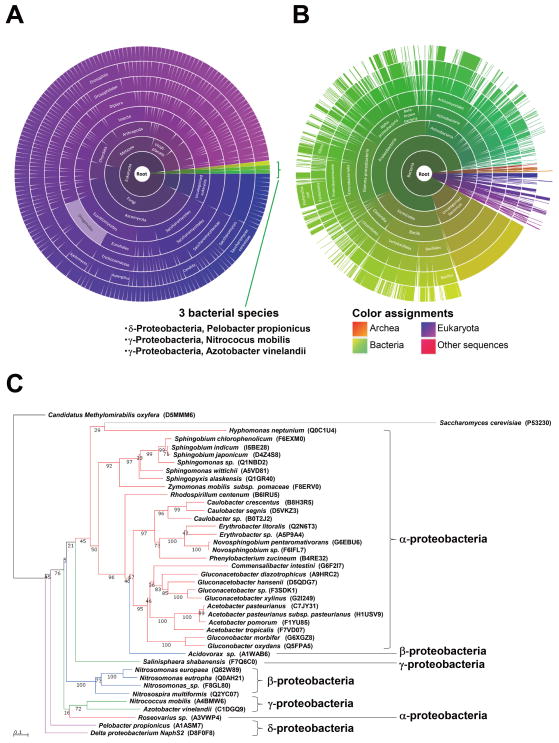
Figure 7. Taxonomic distribution of Tam41 and Cds1 homologs.
Two sunburst charts adopted from Pfam show the distribution of PF09139 including Tam41 (A) and PF01148 including Cds1 (B) across species. Segments shown in purple represent the proteins in eukaryotes and green ones represent bacterial proteins. Detailed explanations of color assignments are given in a rectangle. Names of three bacterial species that have homologous proteins of PF09139 are shown. (C) Evolutionary relationships of yeast Tam41 and its homologs in bacteria. The tree was colored to depict subdivisions of proteobacteria. Confidence values from 1000 re-samplings are shown.
The present study indicates that Art5 overexpression rescues the growth defects of tam41Δ cells through decrease in inositol uptake. Previous studies have shown that the Ino2-Ino4 heterodimer binds to UASINO (inositol sensitive upstream activating element) and activate transcription of a number of phospholipid synthetic enzymes including Cds1, Cho1, Psd1 and Cho2/Opi3 for the CDP-DAG, PS, PE and PC synthases, respectively, upon depletion of inositol (Henry et al., 2012; Greenberg and Lopes, 1996). Besides, it was also reported that the Pgs1 enzymatic activiy can be up-regulated through reduced phospholyration of Pgs1 in response to inositol depletion (He and Greenberg, 2004). Therefore, inositol depletion could likely compensate for the loss of Tam41 by transcriptional control of the proteins involved in phospholipid biosynthesis and/or transport.
To examine whether CDP-DAG has an ability to be transported into mitochondria from the other organelles like the ER by crossing the aqueous environment, we conducted in vitro CDP-DAG transport assay using mitochondria isolated from wild-type and mutant cells lacking one of the four CL synthetic enzymes, Tam41, Pgs1, Gep4, and Crd1, and liposomes containing NBD-CDP-DAG as reported previously (Tamura et al., 2012). Interestingly, NBD-CDP-DAG was taken up by mitochondria and was converted to other NBD-containing substances (likely lipids and other acyl-containing species) in wild-type, tam41Δ, gep4Δ, and crd1Δ mitochondria (Figure S2A). In contrast, NBD-CDP-DAG associated with mitochondria was hardly consumed in the absence of Pgs1, which converts CDP-DAG to PGP. The Pgs1-dependent conversion of CDP-DAG indicates that externally added CDP-DAG is imported into mitochondria, crossing the OM and IM to reach the matrix side where Pgs1 is located. When NBD-PA instead of NBD-CDP-DAG was added in the in vitro lipid transport assay, NBD-PA was also converted to other NBD-containing substances including CDP-DAG, which was not consumed efficiently in pgs1Δ mitochondria (Figure S2B). Note that the Gep4-catalyzing reaction may not work properly in this assay system since possible NBD-PGP species accumulates at similar levels between gep4Δ and crd1Δ mutant strains. Although these results may suggest the potential ability of CDP-DAG to be transported from the ER to mitochondria across the aqueous environment, we should be aware that conjugation of the NBD moiety to the short acyl chain of CDP-DAG may change the property of CDP-DAG, thereby affecting the efficiency in uptake of NBD-CDP-DAG by mitochondria. Indeed, our results on cellular phospholipid profiles suggest that such CDP-DAG transport pathway should be, if any, very inefficient in terms of CL synthesis (Fig. 5A).
Possible inefficient transport of CDP-DAG from the ER to mitochondria may further require facilitated transport systems in vivo. Perhaps Art5 overexpression or inositol depletion, but not the Cds1 overexpression alone, may accelerate the facilitated transport of CDP-DAG from the ER to mitochondria. Since tam41Δ cells show synthetic growth defects with deletion of an ERMES (ER-Mitochondria Encounter Structure) subunit, Mdm12 (Figure S2C), CDP-DAG transport between the ER and mitochondria may involve the ERMES complex, which is proposed to facilitate phospholipid exchange between the ER and mitochondria by tethering the two organelles (Kornmann et al., 2009). Clearly, more precise analyses of the mitochondrial lipid contents should be required to characterize the possible inefficient CDP-DAG transport from the ER to mitochondria.
In conclusion, the present study has revealed that a CDP-DAG synthase Tam41 operates in the CL biosynthesis pathway in mitochondria, and thus it fills the last missing link for the CL synthetic pathway in mitochondria. Although depletion of Tam41 blocks the CL synthetic pathway, cells have a mechanism to circumvent the lack of Tam41 by likely modulating the cellular level of inositol, which may affect the CDP-DAG transport from the ER to mitochondria and/or phospholipid profiles in mitochondria. It will be of great interest to explore in future studies how the change in the cellular inositol level leads to optimization of the CL and other phospholipid levels in mitochondria to maintain mitochondrial functional homeostasis.
EXPERIMENTAL PROCEDURES
Yeast strains and plasmids
Yeast strains used in this work are listed in Table S1. Gene disruption was performed by genomic gene replacement with the CgHIS3 or CgTRP1 genes amplified from pCgHIS3 or pCgTRP1 (Kitada et al., 1995). The gene for Cds1FLAG was constructed by homologous recombination of FY833 or YSC1180-7428809 (Open Biosystems) with the FLAG-KanMX6 or FLAG-HIS3 gene cassettes amplified from pFA6a-FLAG-kanMX6 or pFA6a-FLAG-HIS3 (Tamura et al., 2010).
pYES2-CT-Tam41, an URA3-2μ plasmid that expresses C-terminally hexahistidine-tagged Tam41 under the GAL1 promoter was constructed as follows. The TAM41 gene was amplified from pRS314-Tam41 (Tamura et al., 2006) using primers YES2TAM41F (AGG GAA TAT TAA GCT ATG TTA CGA GTT TCT GAA AATG GT) and YES2TAM41R (GCC CTC TAG ACT CGA AGC TTC TCC TCA TCG ATT TTA GTT T). The PCR product was ligated into HindIII/XhoI-digested pYES2/CT (Invitrogen) by using In-Fusion HD Cloning Kit (Clontech).
Antibodies
For detection of FLAG epitope, anti-FLAG M2 monoclonal antibody (Sigma) or anti-DYKDDDDK tag, monoclonal antibody (1E6) (Wako Pure Chemical Industries, Ltd.) was used. HA epitope was detected using anti-HA-tag mAb (TANA2) (Medical & Biological Laboratories Co., Ltd.). Anti-Dpm1 yeast monoclonal antibody (5C5A7) was purchased from Invitrogen. Purified Cds1 antibodies were a gift from Nikolaus Pfanner (University of Freiburg). Preparation of anti-Tam41 antibodies was described previously (Tamura et al., 2006).
Growth conditions
Cells were grown in YPD (1% yeast extract, 2% polypeptone and 2% glucose), YPLac (1% yeast extract, 2% polypeptone and 2% lactate), YPGE (1% yeast extract, 2% polypeptone, 3% glycerol and 2% ethanol), YPGalSuc (1% yeast extract, 2% polypeptone, 2% galactose and 2% sucrose), SD (0.67% yeast nitrogen base without amino acids and 2% glucose), SCD (0.67% yeast nitrogen base without amino acids, 0.5% casamino acids and 2% glucose), or SCGly (0.67% yeast nitrogen base without amino acids, 0.5% casamino acids and 2% glycerol) media. SD, SCD, and SCGly media were supplemented appropriately with 20 μg/ml each of adenine sulfate, L-tryptophan, L-histidine, methionine, and uracil and 30 μg/ml each of L-leucine and L-lysine. Synthetic minimal plates with or without inositol were prepared as described (Sherman, 1991).
Purification of Tam41 from yeast cells
Yeast cells with pYES2-CT-Tam41 were cultivated in YPGalSuc and crude mitochondrial fractions were prepared. The mitochondria were solubilized in DDM buffer (1% n-dodecyl β-D-maltoside, 20 mM Tris-HCl pH 7.5, 300 mM KCl, 2 mM MgCl2, 10% glycerol, 10 mM imidazole) and subjected to Ni-NTA affinity purification. After washing the Ni-NTA resin with Wash buffer (1% Triton-X100, 20 mM Tris-HCl pH 7.5, 2 mM MgCl2, 2 mM β-mercaptoethanol, 30 mM imidazole) with 300 mM and 50 mM KCl, respectively, proteins were eluted with Elution buffer (0.2% Triton-X100, 20 mM Tris-HCl pH 7.5, 80 mM KCl, 2 mM MgCl2, 10% glycerol, 250 mM imidazole). Eluted proteins were dialyzed against Dialysis buffer (0.2 % Triton X-100, 20 mM Tris-HCl pH 7.5, 10% glycerol, 2 mM β-mercaptoethanol) and subjected to ion-exchange chromatography with SP-sepharose to obtain Tam41. Tam41 was eluted with 100 mM NaCl (0.2 % Triton X-100, 20 mM Tris-HCl pH 7.5, 10% glycerol, 2 mM β-mercaptoethanol, 100 mM NaCl).
Phospholipid analysis
To prepare total phospholipids from whole cells, overnight cultures of yeast cells were = 0.01 in 1 ml of YPD containing 5 μCi/ml 32Pi and cultivated for 20 h at diluted to OD600 30°C. Phospholipids were extracted from cells and subjected to TLC analyses using chloroform/ethanol/water/triethylamine (30:35:7:35) as a solvent (Vaden et al., 2005). Labeled phospholipids were analyzed by radioimaging with a Typhoon 9200 (GE Healthcare) or PharosFX Plus Molecular Imager (Bio-Rad).
Cell fractionation
Yeast cells were grown in YPGE and treated with Zymolyase 20T (Seikagaku Corp.). The resulting spheroplasts were homogenized and centrifuged at 1,500 × g, and the supernatant was centrifuged at 12,000 × g to obtain crude mitochondria. The post-mitochondrial supernatant was centrifuged at 30,000 × g to get rid of residual mitochondria and further centrifuged at 40,000 × g to obtain the microsome (ER) fraction. The crude mitochondria fraction was subjected to a step sucrose gradient or an Optiprep density-gradient (Axis-Shield) to obtain highly purified mitochondria as described previously (Meisinger et al., 2000) or according to manufacturer’s instructions.
Assays for CDP-DAG synthase activity
Unless otherwise stated, to measure the CDP-DAG synthase activity, 10 μM NBD-PA (Avanti Polar Lipids, catalog number 810174) was incubated with either 2 mg/ml purified mitochondria, 1 mg/ml ER or 250 nM purified Tam41 in 50 μl of assay buffer (20 mM Tris-HCl pH 7.5; 80 mM KCl; 2 mM MgCl2; 0.2% Triton X-100; 10% glycerol) with either 740 kBq (6.7 pmol)/ml 32P-CTP or 1 mM CTP for the indicated time periods at 30°C. After the incubation, 750 μl of 2:1 chloroform/methanol was added to the samples and vortexed for 15 min to extract lipids. A hundred μl of water was then added to the sample and vortexed for 5 min. The organic phase was separated by centrifugation at 400 × g for 5 min, dried in a speedvac and then resuspended in 80 μl of chloroform. Twenty μl of the samples were loaded and analyze by TLC using chloroform/ethanol/water/triethylamine (30:35:7:35) or chloroform/methanol/acetic acid/water (50:28:4:8) as a solvent (Nigou and Besra, 2002; Vaden et al., 2005). Phospholipids were analyzed by radioimaging or fluorescence imaging with a Typhoon 9200 (GE Healthcare) or PharosFX Plus Molecular Imager (Bio-Rad).
Phylogenetic analysis
Forty bacterial homologous sequences of Tam41 were detected by Jackhmmer (Finn et al., 2011) with the UniProt database, by using three bacterial homologs (Figure 4A) in Pfam 26.0 (Finn et al., 2010) as query sequences. A multiple alignment of those sequences and yeast Tam41 was calculated by MAFFT version 6 (Katoh and Toh, 2008) with the L-INS-i strategy and homologs with the BLAST E-values < 10−10 from SwissProt. A phylogenetic tree of them was constructed using the neighbor joining method under the JTT model with ungapped sites of the multiple alignment (http://mafft.cbrc.jp/alignment/software/).
Statistical Analysis
Experimental data are presented as means ± SEM.
Supplementary Material
Acknowledgments
We thank Dr. Nikolaus Pfanner for antibodies against Cds1. We are grateful to the members of the Endo lab for valuable discussions. KI is a research fellow of Japan Society for the Promotion of Science (JSPS). We acknowledge support of this work by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) and the grant for CREST from Japan Science and Technology Agency (JST) to TE, the grant of Platform for Drug Discovery, Informatics, and Structural Life Science from the MEXT to KT, and the grant of NIH (GM089853) to HS.
Footnotes
Supplemental information includes two figures and one table and can be found with this article online at xxx.
References
- Beranek A, Rechberger G, Knauer H, Wolinski H, Kohlwein SD, Leber R. Identification of a cardiolipin-specific phospholipase encoded by the gene CLD1 (YGR110W) in yeast. J Biol Chem. 2009;284:11572–11578. doi: 10.1074/jbc.M805511200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Heacock PN, Clancey CJ, Dowhan W. The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. J Biol Chem. 1998a;273:9829–9836. doi: 10.1074/jbc.273.16.9829. [DOI] [PubMed] [Google Scholar]
- Chang SC, Heacock PN, Mileykovskaya E, Voelker DR, Dowhan W. Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. J Biol Chem. 1998b;273:14933–14941. doi: 10.1074/jbc.273.24.14933. [DOI] [PubMed] [Google Scholar]
- Clancey CJ, Chang SC, Dowhan W. Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant. J Biol Chem. 1993;268:24580–24590. [PubMed] [Google Scholar]
- Claypool SM. Cardiolipin, a critical determinant of mitochondrial carrier protein assembly and function. Biochim Biophys Acta. 2009;1788:2059–2068. doi: 10.1016/j.bbamem.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerth M, Tatsuta T, Haag M, Klecker T, Westermann B, Langer T. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science. 2012;337:815–818. doi: 10.1126/science.1225625. [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallas MR, Dienhart MK, Stuart RA, Long RM. Characterization of Mmp37p, a Saccharomyces cerevisiae mitochondrial matrix protein with a role in mitochondrial protein import. Mol Biol Cell. 2006;17:4051–4062. doi: 10.1091/mbc.E06-04-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert N, Joshi AS, Kutik S, Becker T, McKenzie M, Guan XL, Mooga VP, Stroud DA, Kulkarni G, Wenk MR, et al. Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: implications for Barth syndrome. Curr Biol. 2009;19:2133–2139. doi: 10.1016/j.cub.2009.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohil VM, Thompson MN, Greenberg ML. Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine and cardiolipin biosynthetic pathways in Saccharomyces cerevisiae. J Biol Chem. 2005;280:35410–35416. doi: 10.1074/jbc.M505478200. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Greenberg ML, Lopes JM. Genetic regulation of phospholipid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:1–20. doi: 10.1128/mr.60.1.1-20.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Valianpour F, Chen S, Vaz FM, Hakkaart GA, Wanders RJ, Greenberg ML. Aberrant cardiolipin metabolism in the yeast taz1 mutant: a model for Barth syndrome. Mol Microbiol. 2004;51:149–158. doi: 10.1046/j.1365-2958.2003.03802.x. [DOI] [PubMed] [Google Scholar]
- Harada Y, Tamura Y, Endo T. Identification of yeast Art5 as a multicopy suppressor for the mitochondrial translocator maintenance protein Tam41. Biochem Biophys Res Commun. 2010;392:228–233. doi: 10.1016/j.bbrc.2010.01.024. [DOI] [PubMed] [Google Scholar]
- He Q, Greenberg ML. Post-translational regulation of phosphatidylglycerolphosphate synthase in response to inositol. Mol Microbiol. 2004;53:1243–1249. doi: 10.1111/j.1365-2958.2004.04202.x. [DOI] [PubMed] [Google Scholar]
- Henry SA, Kohlwein SD, Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190:317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AS, Thompson MN, Fei N, Huttemann M, Greenberg ML. Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. J Biol Chem. 2012;287:17589–17597. doi: 10.1074/jbc.M111.330167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AS, Zhou J, Gohil VM, Chen S, Greenberg ML. Cellular functions of cardiolipin in yeast. Biochim Biophys Acta. 2009;1793:212–218. doi: 10.1016/j.bbamcr.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Kelley MJ, Carman GM. Purification and characterization of CDP-diacylglycerol synthase from Saccharomyces cerevisiae. J Biol Chem. 1987;262:14563–14570. [PubMed] [Google Scholar]
- Kitada K, Yamaguchi E, Arisawa M. Cloning of the Candida glabrata TRP1 and HIS3 genes, and construction of their disruptant strains by sequential integrative transformation. Gene. 1995;165:203–206. doi: 10.1016/0378-1119(95)00552-h. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler K, Daum G, Paltauf F. Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. J Bacteriol. 1986;165:901–910. doi: 10.1128/jb.165.3.901-910.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchta K, Knizewski L, Wyrwicz LS, Rychlewski L, Ginalski K. Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 2009;37:7701–7714. doi: 10.1093/nar/gkp854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge O, Nishijima M. Phosphatidylserine synthase I and II of mammalian cells. Biochim Biophys Acta. 1997;1348:151–156. doi: 10.1016/s0005-2760(97)00137-9. [DOI] [PubMed] [Google Scholar]
- Kutik S, Rissler M, Guan XL, Guiard B, Shui G, Gebert N, Heacock PN, Rehling P, Dowhan W, Wenk MR, et al. The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. J Cell Biol. 2008;183:1213–1221. doi: 10.1083/jcb.200806048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Martin D, Gannoun-Zaki L, Bonnefoy S, Eldin P, Wengelnik K, Vial H. Characterization of Plasmodium falciparum CDP-diacylglycerolsynthase, a proteolytically cleaved enzyme. Mol Biochem Parasitol. 2000;110:93–105. doi: 10.1016/s0166-6851(00)00260-7. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Sommer T, Pfanner N. Purification of Saccharomcyes cerevisiae mitochondria devoid of microsomal and cytosolic contaminations. Anal Biochem. 2000;287:339–342. doi: 10.1006/abio.2000.4868. [DOI] [PubMed] [Google Scholar]
- Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Nigou J, Besra GS. Cytidinediphosphate-diacylglycerol synthesis in Mycobacterium smegmatis. Biochem J. 2002;367:157–162. doi: 10.1042/BJ20020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E, Pelham HR. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic. 2009;10:1856–1867. doi: 10.1111/j.1600-0854.2009.00990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Haag M, Potting C, Rodenfels J, Dip PV, Wieland FT, Brugger B, Westermann B, Langer T. The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J Cell Biol. 2009;184:583–596. doi: 10.1083/jcb.200810189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Haag M, Wieland FT, Brugger B, Langer T. A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. EMBO J. 2010;29:1976–1987. doi: 10.1038/emboj.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Voelker DR, Langer T. Making heads or tails of phospholipids in mitochondria. J Cell Biol. 2011;192:7–16. doi: 10.1083/jcb.201006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand RP, Sengupta S. Cardiolipin forms hexagonal structures with divalent cations. Biochim Biophys Acta. 1972;255:484–492. doi: 10.1016/0005-2736(72)90152-6. [DOI] [PubMed] [Google Scholar]
- Shen H, Heacock PN, Clancey CJ, Dowhan W. The CDS1 gene encoding CDP-diacylglycerol synthase in Saccharomyces cerevisiae is essential for cell growth. J Biol Chem. 1996;271:789–795. doi: 10.1074/jbc.271.2.789. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sparrow CP, Raetz CR. Purification and properties of the membrane-bound CDP-diglyceride synthetase from Escherichia coli. J Biol Chem. 1985;260:12084–12091. [PubMed] [Google Scholar]
- Tamura Y, Endo T, Iijima M, Sesaki H. Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J Cell Biol. 2009;185:1029–1045. doi: 10.1083/jcb.200812018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Harada Y, Yamano K, Watanabe K, Ishikawa D, Ohshima C, Nishikawa S, Yamamoto H, Endo T. Identification of Tam41 maintaining integrity of the TIM23 protein translocator complex in mitochondria. J Cell Biol. 2006;174:631–637. doi: 10.1083/jcb.200603087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Iijima M, Sesaki H. Mdm35p imports Ups proteins into the mitochondrial intermembrane space by functional complex formation. EMBO J. 2010;29:2875–2887. doi: 10.1038/emboj.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Onguka O, Itoh K, Endo T, Iijima M, Claypool SM, Sesaki H. Phosphatidylethanolamine Biosynthesis in Mitochondria: Phosphatidylserine (PS) trafficking is independent of a PS decarboxylase and intermembrane space proteins Ups1p and Ups2p. J Biol Chem. 2012;287:43961–43971. doi: 10.1074/jbc.M112.390997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller G, Hrastnik C, Achleitner G, Schiefthaler U, Klein F, Daum G. YDL142c encodes cardiolipin synthase (Cls1p) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS Lett. 1998;421:15–18. doi: 10.1016/s0014-5793(97)01525-1. [DOI] [PubMed] [Google Scholar]
- Vaden DL, Gohil VM, Gu Z, Greenberg ML. Separation of yeast phospholipids using one-dimensional thin-layer chromatography. Anal Biochem. 2005;338:162–164. doi: 10.1016/j.ab.2004.11.020. [DOI] [PubMed] [Google Scholar]
- Vance JE, Tasseva G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbalip.2012.08.016. in press. [DOI] [PubMed] [Google Scholar]
- van den Brink-van der Laan E, Killian JA, de Kruijff B. Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim Biophys Acta. 2004;1666:275–288. doi: 10.1016/j.bbamem.2004.06.010. [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.