Abstract
This protocol describes a single molecule pull-down (SiMPull) assay for analyzing physiological protein complexes. The assay combines the conventional pull-down assay with single molecule total internal reflection fluorescence microscopy, and allows probing single macromolecular complexes directly from cell or tissue extracts. In this method, antibodies against the protein of interest are immobilized on a passivated microscope slide. When cell extracts are applied, the surface-tethered antibody captures the protein together with its physiological interaction partners. After washing away the unbound components, single molecule fluorescence microscopy is used to probe the pulled down proteins. Captured proteins are visualized through genetically encoded fluorescent protein tags or through antibody labeling. This ultra-sensitive assay requires at least 10-fold less reagents, is significantly faster and provides quantitative data compared to western blot analysis. Furthermore, SiMPull can distinguish between multiple association states of the same protein. SiMPull is generally applicable to proteins from a variety of cellular contexts and to endogenous proteins. Starting with the cell extracts and passivated slides, the assay requires 1.5 – 2.5 hours for data acquisition and analysis.
Keywords: protein-protein interaction, single molecule, immunoprecipitation, co-immunoprecipitation pull-down
Introduction
Nearly all vital cellular processes are performed by coordinated action of proteins acting in multimeric assemblies or complexes1. Cellular processes are also dynamically regulated by signaling networks, wherein interaction between biomolecules link physical or chemical stimuli to the effector molecules2, 3. Thus, analysis of protein interactions is central for understanding cell function and regulation. One of the most widely used techniques for studying interacting proteins is the pull-down or the co-immunoprecipitation assay4, 5. In the classical pull-down assay, the protein of interest or bait is selectively isolated from cell or tissue extracts. The physiological binding partners of the bait protein, or the prey proteins, co-purify with the bait protein. The identity of the prey proteins is determined using western blot or mass spectrometry.
Many proteins participate in different types of complexes and exhibit diverse functionality. Though instrumental in discovery of new binding partners, the data from ensemble pull-down assays represents an average of protein complexes. The true composition of physiological complexes is not easily revealed due to multiplicity in protein interactions5. Furthermore, no information about the stoichiometry of protein interactions is obtained. Similarly, one can determine the pair-wise interacting partners using yeast two hybrid6 or the complementation assays7, but the physiological assembly of these interactions is not deciphered. Single molecule experiments can provide additional insight about molecular architecture of protein assemblies8, but have been typically limited to reconstituted complexes with purified proteins9–12.
We recently described a single molecule pull-down13 or SiMPull assay that combines the classical pull-down assay with single molecule fluorescence imaging, and enables direct visualization of cellular protein complexes at the single molecule level. In this paper, we describe the detailed procedure for the SiMPull assay. The assay requires a single molecule total internal reflection fluorescence (TIRF) microscope, and can be performed using the same reagents as required for western blot analysis. In one incarnation, an antibody against a bait protein is immobilized on a polymer-passivated flow chamber. Cell extracts are made to flow through the chamber so that the bait protein is captured. If the bait is in complex with other proteins or nucleic acids, the antibody will capture these additional biomolecules. After washing out unbound proteins, multi-color fluorescence imaging with single fluorophore sensitivity is used to analyze the composition of the protein complexes. We describe the assay for pull-down using biotin labeled primary or secondary antibodies. The pulled down proteins are visualized through fluorescent protein-fused chimeras or through immunofluorescence labeling, as depicted in Fig. 1
Fig 1.
Schematic for single molecule pull-down. Microscope slides and coverslips are passivated with PEG, doped with biotin-PEG. Antibodies against the bait protein are immobilized using NeutrAvidin. When cell extract is added to the chamber, the surface immobilized antibody captures the bait protein, together with the prey. Other cellular components do not bind and are washed away. When the prey protein bears a fluorescent protein tag, it can be directly visualized using a single molecule fluorescence microscope. Alternatively, the pulled down complexes can be labeled via immuno-labeling for prey protein detection.
Several advantages stem from the single molecule imaging of cellular protein complexes. SiMPull can discriminate between multiple association states of a protein. It provides quantitative data on bait and prey protein population. SiMPull also allows us to determine the stoichiometry of the complexes by using photobleaching step analysis14. The prepared complexes may also be used for biochemical analysis of their activities at the single molecule level13, 15, 16. Thus, SiMPull can be used as a preparatory tool to study the functional activity of protein complexes that are not accessible through recombinant methods. In addition, SiMPull promises improvement by orders of magnitude in cost, time, and sensitivity over the conventional western blot. The method is generally applicable to a wide variety of cellular contexts and can be tailored for any alternative pull-down or fluorophore labeling schemes. Though we describe the assay for detecting prey proteins using fluorescent protein tags or using antibodies, other suitable specific labeling schemes should be compatible with the assay. Of particular note, is the recent demonstration of specific labeling of proteins in cell lysates15.
However, unlike western blot analysis, this method does not separate biomolecules based on their size, and relies solely on fluorophore-based detection. Hence, appropriate controls are necessary for correct interpretation of the data. In its current form, SiMPull is applicable when one already knows the anticipated binding partners. Lysates are typically diluted to obtain sufficiently low protein concentration for single molecule imaging. Hence, weak interactions with dissociation rate constants > 0.01 s−1 may not be accessible to the method13.
Experimental design
Surface passivation and construction of flow chambers
The key aspect of the pull-down assays is the selective immobilization of the protein of interest or bait on a solid matrix, which will bring along its interaction partners. The bait recruiting surfaces must specifically bind to the protein of interest while rejecting the non-specific adsorption of biomolecules that exist in cell extracts. We achieve this using methoxy polyethylene glycol (mPEG) coated microscope slide and cover slip. The PEG coating substantially reduces the binding of proteins to the glass surfaces17, 18 as demonstrated in Fig. 2. A small amount (~2%) of biotinylated PEG is added during the slide preparation: this functionalizes the surface and allows specific immobilization of biotinylated biomolecules using an avidin linker. Flow chambers are constructed using a passivated slide and coverslip for rapid and convenient exchange of contents (Fig. 3). A detailed protocol for surface passivation with mPEG and construction of flow chambers is also described in previous publications19, 20.
Fig. 2.
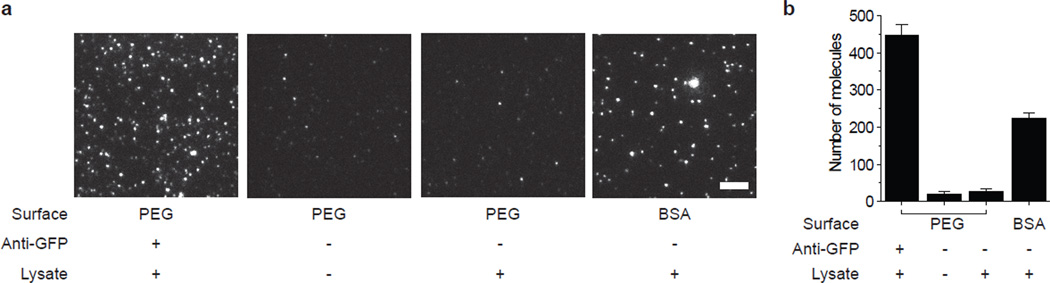
PEG passivation hinders non-specific protein adsorption. (a) Typical TIRF image of YFP-tagged protein pulled down using polyclonal anti-GFP antibody from the lysate (a, left). Non-specific binding of the protein to the PEG passivated surface (a, second from right) is comparable to the blank (a, second from left). Significant non-specific adsorption of YFP is observed when the same amount of lysate is added to surface passivated with 1 mg ml−1 BSA (a, right). Scale bar is 5 µm. (b) Bar graph with average number of fluorophores per image. Error bars represent standard deviation of the mean across > 20 images.
Fig 3.
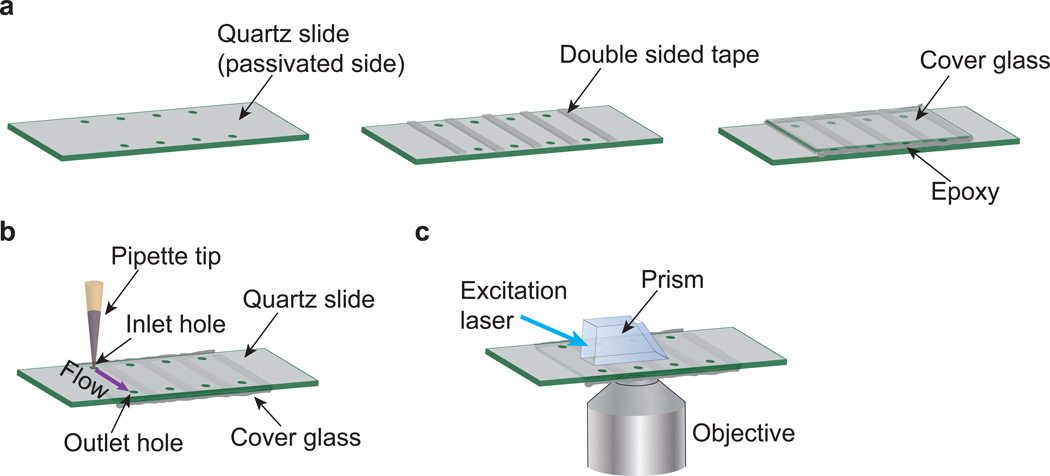
Preparation of flow chambers. (a) To assemble the flow chamber, place the quartz slide with the passivated side facing up. Put double-sided tape between the successive holes to create channels. Place the cover glass on top of the tape, and seal the edges with epoxy. (b) The holes on the slides are used for flowing solutions through the chamber using a pipette. (c) The prism is placed on top of the slide to create an evanescent excitation field at the quartz-aqueous buffer interface.
Sample preparation
SiMPull can be performed using the same samples (purified proteins, tissue or cell lysates) as used for a conventional pull-down analysis. Extracts are typically prepared by lysing cultured cells or tissues with detergents. We have tested a variety of detergents and cell types for SiMPull analysis13. A protocol for lysis of cultured cells is included in Reagent Setup and for preparation of animal tissue extracts is provided as a Supplementary Method. Similar to conventional pull-down assays, it is critical to use non-denaturing conditions in order to preserve the physiological interactions that may occur. We avoid SDS and other strong ionic detergents for lysis as they can potentially denature the immobilized antibodies or disassemble protein complexes.
Antibody immobilization
The selective capture of the bait protein is achieved using surface immobilized antibodies. The biotin-doped surfaces are saturated with NeutrAvidin, and biotinylated antibody against the bait protein is immobilized at a concentration of 10–20 nM. This should yield a surface density of about 20–40 antibody molecules per µm2.
Often times, it is difficult to label the primary antibody with biotin. Many primary antibodies are supplied either as sera or in buffer with BSA as stabilizer and are hence, not suitable for labeling reactions. For immobilizing unlabeled antibodies, we use a biotinylated secondary antibody on the surface that can recruit the primary antibody against the bait protein. Secondary antibodies against one species can often cross-react with other species unless they have been specifically adsorbed to minimize cross-reactivity. For assays involving multiple antibodies, we use cross-adsorbed and affinity purified antibodies.
Single molecule pull-down
For imaging single molecules, the protein of interest must be immobilized at sufficiently low density such that the bait molecules are well separated on the slide surface. Cell lysates typically require dilution to obtain this low density. As the concentration of the protein of interest in the lysates is not known a priori, and also varies across preparations, it is difficult to predict the appropriate dilution factor for the lysate. To this end, we use an iterative approach: we choose a suitable starting point and then titrate the concentration of lysate to obtain 0.1 – 0.2 molecules-µm−2 imaging area upon 20 min incubation on the antibody coated surface. In this regime, the density of antibody molecules on surface is at least 100-fold higher than the concentration of immobilized bait/prey molecules and hence, the protein capture is not limited by the availability of binding sites. Fig. 4 illustrates how the optimal dilution factor is determined for pull-down of yellow fluorescent protein (YFP) fused protein.
Fig. 4.
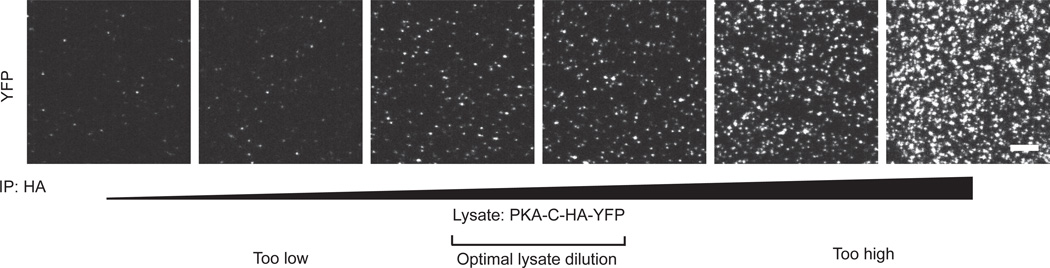
Determination of lysate dilution factor for protein kinase A tagged with YFP and HA tags. The lysate concentration is serially increased starting with a 5,000-fold dilution (second from left), to a 250-fold dilution (right). Image in left depicts background. The optimal fluorophore density for resolving single fluorophores is 0.1 – 0.2 fluorophores per µm2 as depicted in the center two images. Scale bar is 5 µm.
Fluorophore labeling
We describe two approaches for fluorescence detection of pulled-down proteins. First, the proteins of interest can be expressed as fluorescent protein-fused chimera. Proteins expressed as fluorescent protein fusions can be directly visualized under TIRF microscope with single molecule sensitivity. This approach ensures a one-to-one labeling of the protein, and hence, can be used for determining the stoichiometry of the protein in the complex8, 14. Enhanced yellow fluorescent protein is our probe of choice owing to its superior photophysical properties8 as compared against other GFP and RFP variants.
Alternatively, one can detect the prey protein using an immunofluorescence labeling scheme. This approach involves an antibody against the prey protein that is orthogonal to the bait-capturing antibody and a corresponding secondary antibody labeled with fluorophores. We demonstrate this scheme through pull-down of endogenous protein complexes from mouse brain tissue (Fig. 5). Caution should be taken when using multiple antibodies for pull-down or detection as secondary antibodies can cross-react with antibodies from other animals and lead to false positives.
Fig. 5.
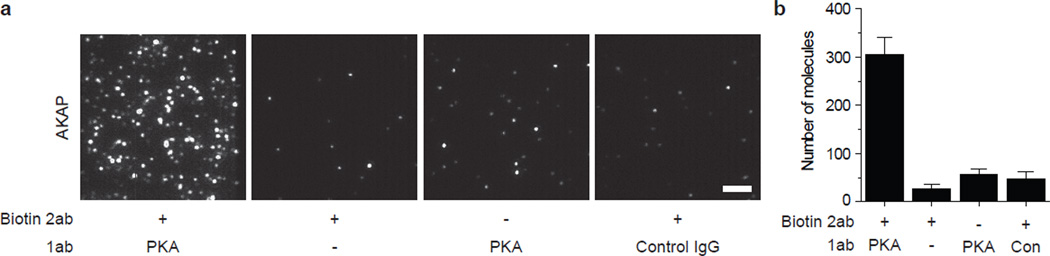
PKA-AKAP pull-down. PKA antibody is immobilized through a biotinylated secondary antibody. A 10-fold dilution of mouse brain extract is added. The pulled down AKAP protein is probed using an orthogonal antibody and a corresponding secondary antibody labeled with Alexa647 (a, left). Control experiments show a 5-fold reduced secondary antibody binding. Scale bar is 5 µm. (b) Bar graph with average number of Alexa647 spots per image. Error bars represent standard deviation of the mean across > 20 images. 2ab, secondary antibody; 1ab, primary antibody.
Single molecule microscopy
We use a prism type TIRF microscope equipped with an electron-multiplying charge-coupled device (EM-CCD) for single molecule imaging. The TIR illumination creates an evanescent field of excitation light that extends only 100–200 nm from the surface, and hence, almost exclusively excites the fluorophores tethered to the surface. This significantly reduces the background from fluorophores in solution, and the technique has been widely applied for single molecule microscopy19. A comprehensive guide for the construction of TIRF microscope for single molecule fluorescence imaging has been described earlier17, 21. Multi-color imaging is achieved using different and spectrally separated fluorophores, and corresponding excitation sources and emission filters. We describe the assay using a prism-type TIRF microscope, though in principle, it should be possible to obtain similar results using an objective-type TIRF setup.
Control experiments
The quality of passivation plays an important role in determining specificity of pull-down. It is important to check the quality of PEG passivation for each preparation of slides, as described below (Steps 14–15). Though the PEG surfaces substantially reduce the non-specific binding, non-specific binding would occur at protein concentrations > 100 nM. Also, antibodies may bind non-specifically to pulled down proteins or may cross react, leading to false positives. Control experiments with suitable control antibodies and control lysates are essential to verify that the detected fluorescence arises from the anticipated protein complexes. We typically perform control experiments by replacing each of the capture or detection antibodies with corresponding control antibodies and using control lysates without bait or prey protein expression. Similar to the conventional pull-down assays, additional controls, for example, mutations that prohibit binding between the bait and the prey proteins, may be performed, depending on the context.
Materials
Reagents
Methanol (Fisher Scientific, cat. no. A412-4) Caution: Methanol is flammable; liquid and vapors are toxic. Wear mask, gloves and chemical safety goggles while handling.
Acetone (Fisher Scientific, cat. no. A18-4) Caution: Acetone is flammable and eye and skin irritant. Wear gloves and chemical safety goggles while handling.
KOH pellets (Fisher Scientific, cat. no. P250-3) Caution: KOH is corrosive and eye and skin irritant. Use under a chemical fume hood.
Acetic acid, Glacial (Fisher Scientific, cat. no. A38-500) Caution: Acetic acid causes severe eye and skin burns. Handle with a glass pipette. Wear gloves and chemical safety goggles while handling.
Aminosilane (N-(2-Aminoethyl)-3-Aminopropyltrimethoxysilane), (United Chemical Technologies, cat. no. A0700) Caution: Aminosilane is potentially toxic and skin and eye irritant. Wear gloves and chemical safety goggles while handling.
Methoxy PEG (Laysan Bio. Inc., cat. no. mPEG-Succinimidyl Valerate, MW 5,000)
Biotin-PEG (Laysan Bio. Inc., cat. no. Biotin-PEG-SVA, MW 5,000)
Sodium bicarbonate (Fisher Scientific, cat. no. S233)
Double sided tape (3M)
Epoxy (Devcon, cat. no. 14250)
Tris-HCl (Fisher Scientific, cat. no. BP153-1)
NaCl (Fisher Scientific, cat. no. S641-212)
EDTA (Sigma, cat. no E9884-1KG)
Bovine serum albumin (BSA) (New England Biolabs, cat. no. B9001S)
NeutrAvidin (Pierce, cat. no. 31000)
Fluorophore labeled protein for testing quality of PEG passivation
Antibodies: Biotinylated anti-Flag (Sigma, cat. no. F9291), Biotinylated anti-rabbit (Santa Cruz Biotech., cat. no. SC2089), Biotinylated anti-GFP (Rockland Immunochemicals, cat. no. 600-106-215), Anti-goat (Rockland Immunochemicals, cat. no. 605-701-125), Rabbit anti-PKARII (Santa Cruz Biotech., cat. no. SC909), Anti-AKAP150 (Santa Cruz Biotech., cat. no. SC6445) Rabbit pre-immune IgG (Cell Sciences, cat. no. NRI01)
Cell culture media (Dulbecco's modified eagle medium, Hyclone, cat. no. SH30284 or equivalent, supplemented with serum if appropriate)
Transfection reagent (Lipofactamine 2000, Invitrogen, cat. no. 11668019)
NP-40 (United States Biological, cat. no. N3500)
Equipment
Rotary drill (Dremel, model no. 395)
Drill bits (0.75 mm) (Kingsley North Inc., cat. no. 1-0500-100)
Quartz slides (G. Finkenbeiner Inc., 1" × 3" × 1 mm thick)
Coverslips (24 × 40 mm, VWR International, cat. no. 48393230)
Slide holders (Fisher Scientific cat. no. 08-817)
Bath sonicator (Bransonic tabletop ultrasonic cleaner)
Propane torch Caution: Propane is highly flammable.
Single molecule TIRF microscope:
Inverted fluorescence microscope (Olympus cat. no. IX70),
1.2 NA water immersion objective (Olympus cat. no. UPLAPO60XW)
Pellin-Broca prism (EKSPLA, cat. no. 325-3206)
EM-CCD detector (Andor Technologies, cat. no. iXon DV 887-BI)
Laser shutter (Uniblitz electronic shutter, Vincent Associates, cat. no. LS6T2)
Shutter driver (Vincent Associates, cat. no. VMM-D1)
Mirrors to align lasers (Thorlabs cat. no. BB1-E02)
Polarizing beam splitter (Thorlabs, cat. no. PBS3)
XYZ micrometer translation stage (Newport Corp., cat. no. 462-XYZ)
Excitation and emission focusing lenses
Excitation lasers 488 nm (Coherent Inc, cat. no. Sapphire 488 LP-050), 568 nm (Coherent Inc., cat. no. Sapphire 568-50), 633nm (HeNe Laser, Newport Corp. cat. no. R-30995).
Emission filters: YFP (Chroma Technology, cat. no. HQ 535/30m), mCherry (Semrock Inc., FF01-607/36-25), Alexa647 (Chroma Technology, cat. no. 640DCLP).
Reagent setup
T50: 10 mM Tris-HCl pH 8.0, 50 mM NaCl, can be stored at room temperature, 22–25°C for up to 1 month.
T50-BSA: 10 mM Tris-HCl pH 8.0, 50 mM NaCl, 0.1 mg ml−1 BSA, can be stored at 4°C for up to 1 month.
Lysis buffer: 10 mM Tris pH 7.5, 1% vol/vol NP-40, 150 mM NaCl, 1 mM EDTA, with protease inhibitors). It is recommended to prepare fresh lysis buffer for each use.
Sample preparation: A method for lysate preparation from tissues is provided as a Supplementary Method. Here, we describe the method used for lysate preparation from transfected cells. The method can be tailored for different lysis conditions or cell types. HEK293 cells are cultured and maintained in Dulbecco’s modified eagle medium supplemented with antibiotics and 10% vol/vol fetal bovine serum. Transfect the cells with suitable transfection reagent following manufacturer’s protocol. For lysis, wash cells twice with PBS and add lysis buffer. Incubate at 4°C for 30 min. Pre-clean the lysate by centrifuging at 14,000g for 20 min at 4°C. It is recommended to use fresh lysates, within ~12 h after preparation.
Equipment setup
TIRF microscope: We use a custom-built prism-type TIRF microscope for single molecule imaging. An inverted microscope is adapted to hold a trapezoid fused-silica prism on top of the flow chamber. The excitation laser beam is directed towards the objective through the prism at an incidence angle greater than the critical angle (68°). The prism is index matched with the flow chamber using immersion oil, such that the evanescent excitation field is created at the quartz-aqueous buffer interface. A 60× water immersion objective (NA = 1.2) is used to collect the fluorescence signal. The scattered light is rejected using suitable filters/dichroic mirrors. The emitted light is imaged onto a 512 × 512 pixel EM-CCD. The final pixel size in the imaging plane is ~146 nm. The excitation laser intensity is modulated using a half-wave plate and a polarizing beam splitting cube to achieve 0.5 – 3 µW-µm−2 at the quartz-aqueous buffer interface, so as to obtain a 5–10 fold signal above noise.
Software: Fluorescence signal is recorded using custom software written in Visual C++. The software acquires movies as series of frames at specified time-resolution and depending on the camera used. Single molecule time trajectories are extracted from the movies using scripts written in IDL. The program creates an averaged image of the first 10-frames, and identifies single molecules as intensity maxima greater than a predetermined threshold. The algorithm also fits the point-spread function to a Gaussian to avoid including multiple molecules or aggregations into the analysis. Next, the local background is subtracted and intensities from 7 by 7 pixels surrounding the peak are added to obtain intensities of single molecules. The single molecule time trajectories and mean fluorophore count per movie are analyzed using MATLAB codes. All programs are available from the authors upon request.
Procedure
PEG passivation of microscope slides Timing: 6 – 8 h
-
1
Drill two holes in quartz slide, about 0.75 mm in diameter, 3–4 mm away from the edge (Fig. 3a). This is to create inlet and outlet for the flow channel. Rinse the slide, and place the slide and a coverslip in the slide holder. We typically make 4–5 flow channels per slide.
-
2
Rinse twice and bath sonicate the slide and the coverslip in MilliQ water for 10 min. Repeat the process with methanol followed by acetone, to remove any organic residue from the surfaces.
Caution: Methanol is flammable; liquid and vapors are toxic. Acetone is flammable and eye and skin irritant. Wear mask, gloves and chemical safety goggles while handling.
-
3
Sonicate in 1 M KOH for 20 min, rinse with MilliQ water. KOH treatment activates the surface for silane functionalization (Steps 5–7).
Caution: KOH is corrosive and eye and skin irritant. Use under a chemical fume hood.
-
4
Burn the slides for about 1 min and the coverslips for 1–2 sec with a propane torch to dry off any surface moisture. Place the slide and coverslip in a dry slide holder.
Caution: Propane is highly flammable.
-
5
Mix 95 ml methanol with 5 ml of acetic acid in a conical flask. Add 1 ml aminosilane, mix and immediately pour this solution in the slide holder with slide and coverslip. Incubate in dark for 10 min at room temperature.
Critical step: Aminosilane is photosensitive and hydrolyzes rapidly in water. Store it under nitrogen in dark.
Caution: Aminosilane is potentially toxic and skin and eye irritant. Acetic acid causes severe eye and skin burns. Wear appropriate safety equipment while handling.
-
6
Bath sonicate the slide and coverslip for 1 min and then incubate for another 10 min at room temperature.
-
7
Wash the slide and coverslip with methanol and water for 1–2 min each. Dry and place them in a humidified box.
-
8
Weigh 16 mg of mPEG with 0.3 mg biotin-PEG per slide/coverslip pair. Dissolve in 70 µl freshly prepared sodium bicarbonate buffer (10 mM sodium bicarbonate, pH 8.5). Mix well, and spin down for 30 s at 10,000g at room temperature to remove bubbles.
Critical step: The passivation on the coverslip surface is not required when using prism type TIRF microscope, but is recommended to prevent sample loss.
Critical step: The half-life of succinimidyl valerate PEG in pH 8.5 buffer is only ~10 min. After adding the buffer to PEG, move to Step 9 as soon as possible.
-
9
Apply this solution to the slide surface and sandwich it immediately with the coverslip. Store in dark for 3–4 h in the humidified boxes at room temperature.
-
10
Wash with copious amount of water, blow dry with nitrogen and store the slide and coverslip in vacuum at −20°C in dark. We use a food grade vacuum sealer for sealing the slides in vacuum.
Pause point: The slides can be stored for up to 2 weeks under these conditions.
Construction of flow chambers Timing ~30 min
-
11
Thaw the slides at room temperature for 10 min.
-
12
Sandwich a double-sided tape between the slide and cover slip excluding a ~5 mm channel with holes (Fig. 3a). Ensure that the tape sticks on both surfaces.
-
13
Seal the edges with epoxy and allow it to dry for 10 min. Volume of the flow channel is ~20 µl. Prepare additional chambers for the control experiments as required.
Testing quality of PEG slides Timing ~30 min
-
14
Flow 100 µl of T50 buffer in the flow channel (Fig. 3b), and image it under TIRF microscope. Acquire 10 short movies, at suitable time resolution, and determine the average number of fluorescent molecules per unit imaging area. This is the background fluorescence arising likely from impurities during surface preparation. Typically, the observed background fluorescence is < 0.02 molecules-µm−2 under our experimental conditions.
Troubleshooting
-
15
Test the slides for the quality of passivation. Flow 100 µl of a 10 nM fluorophore labeled protein to the flow chamber; incubate for 10 min and wash by flowing 200 µl T50 twice to remove unbound protein. Image under the TIRF microscope, and determine average number of non-specifically bound molecules. A good passivation should yield < 0.01 molecules-µm−2 non-specifically adsorbed molecules above background spot count (as determined in Step 14).
Troubleshooting
Immobilizing antibody against bait protein Timing 30 min – 1 h
-
16
Prepare a 0.2 mg/ml solution of NeutrAvidin in T50 buffer. Add 70 µl of this solution to the flow chamber. Incubate for 5 min. All incubations are performed at room temperature, unless specified.
-
17
Wash excess NeutrAvidin by flowing in 200 µl T50 twice.
-
18
For immobilizing biotinylated primary antibody against bait protein, follow option (A). For immobilizing unlabeled bait antibody via a biotinylated secondary antibody, follow option (B).
Critical step: The sensitivity and performance of the assay depends on the affinity of the antibody, and accessibility of the epitope. Like any antibody-based assay, it may be required to test different antibodies for this application.- Option A: immobilizing biotinylated primary antibody against bait protein
- Dilute the biotinylated antibody to a working concentration around 10–20 nM in T50-BSA.
- Add 100 µl of this solution to the chamber and incubate for 10 min.
- Rinse twice with 200 µl T50-BSA.
- Option B: using biotinylated secondary antibody to immobilize the primary antibody against the bait protein
- Flow 100 µl of 20–40 nM of biotin labeled appropriate secondary antibody and incubate for 10 min.
- Wash twice with T50-BSA.
- Flow 100 µl of 10–20 nM unlabeled primary antibody against the bait protein. Incubate for 10–20 min.
- Wash twice with T50-BSA.
Pull-down of proteins from cell lysates Timing ~30 min
-
19
Flow in 100 µl of an appropriate dilution of cell or tissue lysate on the antibody coated chambers. Dilutions are typically made in the lysis buffer without detergent, or in T50-BSA. Incubate for 10–20 min and then flush out the unbound extract. If the prey protein bears a fluorescent protein tag, proceed directly to Step 22 for imaging. Otherwise, proceed to Step 20 to fluorescently label the prey protein using antibodies.
Critical step: Expression levels of proteins vary significantly depending on the protein being studied and across preparations. Hence, this dilution factor needs to be determined for each experiment (or sample preparation). As the concentration of the protein of interest in the crude cell extracts is difficult to estimate, we typically start with a 2,000-fold dilution of cell extract for overexpressed proteins or a 20-fold dilution of cell extract for endogenous proteins, where the lysate is prepared from 103–104 cells µl−1. The concentration of protein is titrated to obtain 0.1 – 0.2 fluorophores-µm−2 as determined in Step 22.
Immunofluorescence labeling of pulled down protein Timing ~30 min
-
20
Incubate the pulled down protein with 100 µl of 5–10 nM antibody against the prey protein for 10–20 min. Wash twice with T50-BSA.
Critical step: When using a secondary antibody for detection, the primary antibody against bait and prey proteins must be from different organisms.
-
21
Add 100 µl 1–2 nM of fluorophore labeled secondary antibody against the prey protein. Incubate for 10 min and flush out the unbound antibody.
Critical step: The secondary antibody against the prey protein can potentially bind to the antibodies against the bait protein. Appropriate controls are recommended to rule out false positives.
Single molecule imaging and spot counting Timing 10 min – 1 h
-
22
Image the slide under a prism type TIRF microscope. Acquire 20 or more short movies (~20 frames each) depending on the statistics desired. Analyze the movies to determine the mean number of fluorophores per unit imaging area.
Critical step: Fluorescent proteins have a short fluorescence photobleaching lifetime, typically a few seconds under our excitation scheme. Turn off the excitation shutter when not imaging. When imaging organic dyes, through immunofluorescence labeling scheme, one can use appropriate oxygen scavenging systems to prolong the fluorescence lifetime of the dyes.
Troubleshooting
-
23
Titrate the concentration of cell lysate depending on the observed surface density of prey molecules to obtain 0.1 – 0.2 molecules-µm−2 and repeat steps 16–22 as necessary.
Critical step: Typically, there is some fluorescent background from the surface arising due to impurities. The concentration of immobilized molecules due to specific binding should be kept at least 5 to 10-fold above this background. Under our imaging conditions, the background is ~0.01 molecules-µm−2. For applications involving co-localization of multiple fluorophores and for stoichiometry determination via photobleaching analysis, the density of molecules should be kept between 0.05 – 0.15 molecules-µm−2.
Troubleshooting
-
24
Once the optimal concentration of the lysate is determined, perform appropriate control experiments by repeating steps 16–22 with a suitable control antibody (say, control IgG) and with control lysates lacking the bait-prey interaction.
Troubleshooting
Timing
Steps 1 – 10, PEG passivation of microscope slides: 6 – 8 h
Steps 11 – 13, Construction of flow chambers: ~30 min
Steps 14 – 15, Testing quality of PEG slides: ~30 min
Steps 16 – 18, Immobilizing antibody against bait protein: 30 min – 1 h
Steps 19, Pull-down of proteins from cell lysates: ~30 min
Steps 20 – 21, Immunofluorescence labeling of pulled down protein: ~30 min
Steps 22 – 24, Single molecule imaging and spot counting Timing 10 min – 1 h
Troubleshooting
Troubleshooting advice can be found in Table 1.
Table 1.
Troubleshooting guide.
| Step | Problem | Possible cause | Solution |
|---|---|---|---|
| 14 | High background fluorescence without protein immobilization | Surface impurities during slide preparation. For example, aminosilane or PEG may be contaminated. | Make new PEG slides, using fresh reagents. |
| 15 | Non-specific binding of proteins | The PEG passivation is not good or has deteriorated | Make new PEG slides. |
| 22 | No or few bait protein detected | Low sample concentration in the lysate. | Increase the concentration of lysate added to the flow chamber. |
| 22 | No bait protein detected | Antibodies do not recognize the protein | Try a different antibody against a different epitope on the bait protein |
| 23 | No expected prey protein detected | Antibodies do not recognize the protein | Try a different antibody against a different epitope on the prey protein |
| 24 | The specific binding yields only a 2–3 fold higher binding as compared to the control channel | High non-specific binding of antibodies against prey | Optimize the concentration of cell lysate and/or antibodies Wash with more stringent buffer conditions (high salt or a buffer with detergent) to remove non-specifically adsorbed proteins. |
| 24 | The control channel has same number of fluorophores as the sample channel | Surface passivation is not good | Make new PEG slides. |
Anticipated results
The PEG passivated surfaces resist non-specific protein adsorption. Fig. 2 depicts a typical TIRF image of surface immobilized YFP molecules, pulled down from cell lysate. When the same amount of lysate was added to a PEG passivated surface without antibodies, it yielded only ~5 fluorescent spots on average above the background in a 2500 µm2 imaging area. On the other hand, a surface passivated with BSA, yielded substantial non-specific protein binding (Fig. 2). Prior to the experiments, the slides should be checked for quality of passivation by flowing in ~10 nM of labeled protein.
The lysate concentration is titrated to obtain an optimal fluorescent spot count above background. Protein kinase A (PKA) tagged with YFP and HA tags is pulled down with antibodies against HA tag, and visualized using YFP fluorescence (Fig. 4). As we increased the lysate concentration, the number of YFP molecules observed increased. The optimal density for single molecule imaging is 0.1 – 0.2 fluorophores-µm−2 imaging area, as depicted in Fig 4. At higher concentrations, it is not possible to resolve single molecules.
We demonstrate the immunofluorescence labeling strategy through pull down of endogenous PKA-AKAP complexes from mouse brain extracts. We immobilized 20 nM biotinylated donkey-anti-rabbit antibody followed by 10 nM rabbit-anti-PKA antibody. To these surfaces, we then added a 10-fold dilution of whole brain extract. A detailed protocol for preparation of tissue samples is included as a Supplementary Method. The pulled down AKAPs are labeled with goat-anti-AKAP antibody (10 nM) followed by 2 nM secondary antibody against goat labeled with Alexa647. Control experiments excluding the anti-PKA antibody (Fig. 5a, second from left), excluding biotinylated secondary antibody against PKA (Fig. 5a, second from right) and with a control rabbit IgG (Fig. 5a, right) are performed to verify that the observed fluorescence arises from specific binding of prey antibodies to AKAP. Additional controls, by knocking out PKA or AKAP may be performed to confirm the interaction.
Supplementary Material
Acknowledgments
We thank Biswarathan Ramani, Yuji Ishitsuka and Kaushik Ragunathan for help with developing the protocol. This work was funded by NIH grants (AI083025, GM065367 to T.H.; HL082846 to Y.K.X.). Additional support was provided by NSF grants (0646550, 0822613 to T.H.). T.H. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Author contributions:
A.J., Y.K.X. and T.H. designed the research. R.L. prepared cell extracts. A.J. conducted single molecule experiments and analyzed the data. A.J., Y.K.X. and T.H. wrote the paper.
Competing financial interests:
The authors declare that they have no competing financial interests.
Supplementary Method: Immunoprecipitation of endogenous proteins from mouse tissues
References
- 1.Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 2.Barrios-Rodiles M, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 3.Yamada T, Bork P. Evolution of biomolecular networks: lessons from metabolic and protein interactions. Nat Rev Mol Cell Biol. 2009;10:791–803. doi: 10.1038/nrm2787. [DOI] [PubMed] [Google Scholar]
- 4.Puig O, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 5.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 6.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 7.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reyes-Lamothe Rodrigo JSD, Leake MC. Stoichiometry and Architecture of Active DNA Replication Machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Zhou D, Browne H, Balasubramanian S, Klenerman D. Molecule by molecule direct and quantitative counting of antibody-protein complexes in solution. Anal Chem. 2004;76:4446–4451. doi: 10.1021/ac049512c. [DOI] [PubMed] [Google Scholar]
- 10.Taguchi H, Ueno T, Tadakuma H, Yoshida M, Funatsu T. Single-molecule observation of protein-protein interactions in the chaperonin system. Nat Biotechnol. 2001;19:861–865. doi: 10.1038/nbt0901-861. [DOI] [PubMed] [Google Scholar]
- 11.Kapanidis AN, et al. Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc Natl Acad Sci U S A. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blosser TR, Yang JG, Stone MD, Narlikar GJ, Zhuang X. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022–1027. doi: 10.1038/nature08627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain A, et al. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoskins AA, et al. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeom KH, et al. Single-molecule approach to immunoprecipitated protein complexes: insights into miRNA uridylation. EMBO Rep. 2011;12:690–696. doi: 10.1038/embor.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleminger G, Solomon B, Wolf T, Hadas E. Effect of polyethylene glycol on the non-specific adsorption of proteins to Eupergit C and agarose. J Chromatogr. 1990;510:271–279. doi: 10.1016/s0021-9673(01)93761-6. [DOI] [PubMed] [Google Scholar]
- 19.Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu Rev Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 20.Rothenberg E, Ha T. Single-molecule FRET analysis of helicase functions. Methods Mol Biol. 2010;587:29–43. doi: 10.1007/978-1-60327-355-8_3. [DOI] [PubMed] [Google Scholar]
- 21.Selvin PR, Ha T. Single-molecule techniques : a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.