Abstract
In the past decade, the diversity of signals generated by the ubiquitin system has emerged as a dominant regulator of biological processes and propagation of information in the eukaryotic cell. A wealth of information has been gained about the crucial role of spatial and temporal regulation of ubiquitin species of different lengths and linkages in the nuclear factor-κB (NF-κB) pathway, endocytic trafficking, protein degradation and DNA repair. This spatiotemporal regulation is achieved through sophisticated mechanisms of compartmentalization and sequential series of ubiquitylation events and signal decoding, which control diverse biological processes not only in the cell but also during the development of tissues and entire organisms.
Ten years ago, we were only starting to suspect the leading part that the ubiquitin network was about to take in our understanding of cellular signalling. At that time, ubiquitylation was beginning to be acknowledged as more than simply a signal for protein degradation, and instead as a main regulator of biological events1. RING domains had been recognized as mediators of ubiquitin ligase activity, and differently linked ubiquitin chains were suspected to regulate distinct processes, as suggested by reports linking Lys63 ubiquitin chains to pro-inflammatory signalling pathways and DNA repair responses. Since then, the ubiquitin system has been found to be highly versatile, in terms of both the range of ubiquitin signals and the range of cellular responses that ubiquitin conjugation can trigger (BOX 1).
Box 1. The ubiquitin network — what a difference a decade makes.
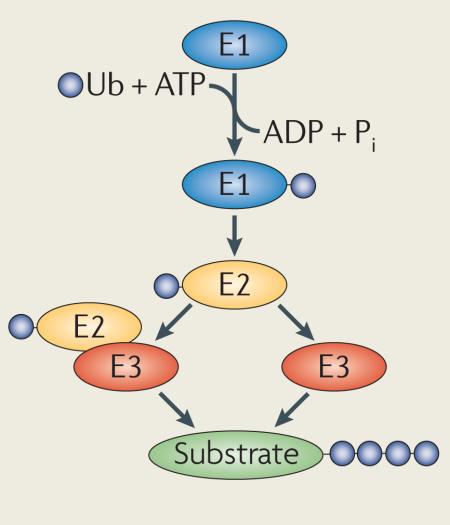
Ubiquitylation is mediated by the sequential activity of activating (E1), conjugating (E2) and ligating (E3) enzymes, resulting in the conjugation of monoubiquitin or polyubiquitin chains of different lengths and linkages to target proteins (see the figure). The past 10 years have shown great advancements in our understanding of ubiquitin signalling2,7,11,16,20,137.
2000
Eukaryotic genomes were thought to encode a single E1 enzyme, ≥20 E2 enzymes and <100 E3 enzymes.
RING fingers and HECT domains were associated with ubiquitylation.
Substrate specificity was mainly considered to be mediated by the E3 enzyme.
Information regarding ubiquitin-binding domains (UBDs) was limited, and only the ubiquitin-associated (UBA) domain and two putative UBD motifs in Rpn10 (S5A in mammals) had been described.
Only linkages involving Lys6, Lys11, Lys29, Lys48 and Lys63 were thought to exist.
The ubiquitin-like molecules recognized were NEDD8, small ubiquitin-related modifier 1 (SUMO1), SUMO2, SUMO3, ISG15, autophagy-related 8 (Atg8), Atg12 and Fub1.
2010
Eukaryotes are today estimated to have two E1 enzymes, ~40 E2 enzymes and >600 E3 enzymes.
Substrate specificity is thought to be achieved by the E2–E3 pair, together with surrounding domains, accessory molecules and post-translational modifications.
A large number of UBDs, as well as their preferences for binding surfaces and linkage specificities, has been identified.
Linkages are known to be formed on all internal Lys residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 and Lys63). Linear chains (by Gly76–Met1 conjugation) and atypical chains (heterologous, forked or mixed) have also been described.
The ubiquitin-like molecules recognized are NEDD8, SUMO1, SUMO2, SUMO3, ISG15, Atg8, Atg12, Fub1, ubiquitin-related modifier 1 (Urm1), ubiquitin-fold modifier (UFM) and FAT10.
Pi, inorganic phosphate; Ub, ubiquitin.
Ubiquitylation is achieved by the concerted action of activating (E1), conjugating (E2) and ligating (E3) enzymes, some of which possess elongating activities that support the generation of polyubiquitin chains2–6. Proteins can be modified through the conjugation of monoubiquitin or polyubiquitin chains of variable length on any of the seven Lys residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 or Lys63) or the amino-terminal Met (Met1) of the ubiquitin monomer. Ubiquitin chains can thus be connected by at least eight different homotypic linkages, as well as by a range of atypical chains (heterologous, forked or mixed)5,7,8. Occasionally, efficient protein polyubiquitylation requires additional conjugation factors, so-called E4 enzymes, that support the elongation of ubiquitin chains9 (reviewed in REF. 10). The myriad of ubiquitin signal species is recognized and decoded by specialized ubiquitin-binding domains (UBDs), which form transient, non-covalent interactions with either ubiquitin moieties or the linkage region in ubiquitin chains11. Strict control of ubiquitylation is accomplished by deubiquitylating enzymes (DUBs), a divergent family of isopeptidases that reverse ubiquitylation by removing conjugated ubiquitin tags12,13. Several recent reviews have elaborated on the principle mechanisms by which ubiquitylation is implicated in regulating different aspects of cellular physiology, including protein degradation, DNA repair, the ER-associated degradation (ERAD) pathway, receptor endocytosis, apoptosis and autophagy14–20.
In this Review, we focus on the spatiotemporal organization of ubiquitin signalling. We give a comprehensive overview of how spatial organization is achieved by the compartmentalization and distinct arrangements of proteins in the ubiquitin arsenal, from the structural basis of the molecular mechanisms to the signalling pathways that control their biological functions. Moreover, we illustrate how dynamic spatial and temporal changes in ubiquitin networks are used to coordinate processes in the cell, such as the nuclear factor-κB (NF-κB) signalling pathway and receptor endosomal sorting, as well as in cell and animal development.
Ubiquitylation in space and time
The abundance of ubiquitin in all cells and in most subcellular structures has forced the living cell to develop numerous strategies to properly organize the ubiquitin network in space and time. Interestingly, although ubiquitin does not seem to be found inside organelles such as the endoplasmic reticulum (ER), mitochondria21 or chloroplasts22, ubiquitin conjugation certainly seems to affect the function and homeostasis of all subcellular compartments. Several levels of specificity are achieved by the following: the selectivity for distinct target proteins that is displayed by individual E2–E3 pairs; the recognition of ubiquitin conjugate length and linkage by UBDs; and the apparent correlation between functional outcome and distinct ubiquitin species. However, these mechanisms do not fully explain how the ubiquitin system can specifically regulate thousands of cell ular functions23. Instead, fine-tuning of ubiquitylation depends on the dynamic changes and sequestration of components within the ubiquitin network, as well as on chaperones and scaffold proteins. Sequential complex assembly, based on multiple ubiquitin–UBD interactions that are regulated in space and time, enables the propagation of signalling events controlling the dynamics of receptor trafficking, the activation of kinases in the NF-κB pathway, synaptic plasticity and cell cycle progression.
Compartmentalization of the conjugation machinery
There is a growing body of evidence emphasizing essential roles for E2 enzymes and E3 enzymes in mediating substrate specificity as well as spatial and temporal control of ubiquitylation. Although some ubiquitin conjugation machinery proteins are predominantly expressed in certain tissues, for example the muscle-specific E3 ligase tripartite motif-containing 63 (TRIM63)24, many of the estimated ~40 E2 enzymes5 and >600 E3 enzymes4,25 show broad expression patterns and therefore require spatial organization at the subcellular level.
One of the most straightforward strategies to control protein localization is physical tethering to a specific organelle or subcellular membrane. Indeed, many E3 enzymes contain regions that are predicted to be transmembrane domains and to restrict them to vesicular structures. E2 and E3 subunits that are devoid of transmembrane domains can also associate with specific organelles by interacting with scaffold proteins or by forming multisubunit ubiquitin ligase complexes that include components which target them to specific subcellular locations.
Ubiquitin network proteins that are restricted to a specific subcellular compartment include the E2 enzymes ubiquitin-conjugating enzyme 6 (Ubc6) and Ubc7, which are associated with the ER and are involved in ERAD26. Ubc6 is tethered to the ER through its transmembrane domain, whereas Ubc7 (which is normally cytoplasmic) is recruited to the ER by the scaffold protein coupling of ubiquitin conjugation to ER degradation 1 (Cue1)27. Interestingly, in the absence of Cue1, Ubc7 is degraded through ubiquitin fusion degradation 4 (Ufd4)-dependent Lys48-linked ubiquitylation, a mechanism that elegantly ensures that Ubc7 acts only when properly localized to the ER28.
Multiple E3 ligases are specifically recruited to mitochondria29, and many of these use transmembrane domains to attach to the outer mitochondrial membrane (OMM), where they catalyse ubiquitylation in the cytoplasmic space surrounding the mitochondrion. Indeed, mitochondrial E3 ligases have many mitochondrion-related roles, such as the regulation of internal mitochondrial protein quality control30, mitochondrial trafficking and morphological dynamics during mitochondrial division25,31; however, they are also important for the integration of mitochondria within the cellular environment. For example, mitochondrial ubiquitin ligase activator of NF-κB25 (MULAN; also known as GIDE32) is an E3 ligase that is involved in mitochondrion-to-nucleus signalling25, regulation of cell growth and induction of caspase-dependent apoptotic cell death32.
In addition to proteins that constitutively localize to specific subcellular compartments, numerous E2–E3 pairs are targeted to their site of action only in response to specific physiological conditions and/or distinct post-translational modifications (BOX 2). For instance, members of the class III E2 family (UBCH6 (also known as UBE2E1), ubiquitin-conjugating enzyme E2 E2 (UBE2E2) and UBCM2 (also known as UBE2E3)) translocate to the nucleus following the attachment of ubiquitin to the Cys residue of their active site. This nuclear translocation is mediated by importin 11, which specifically recognizes ubiquitin-charged E2 enzymes33.
Box 2. Crosstalk between the ubiquitin network and other post-translational modifications.
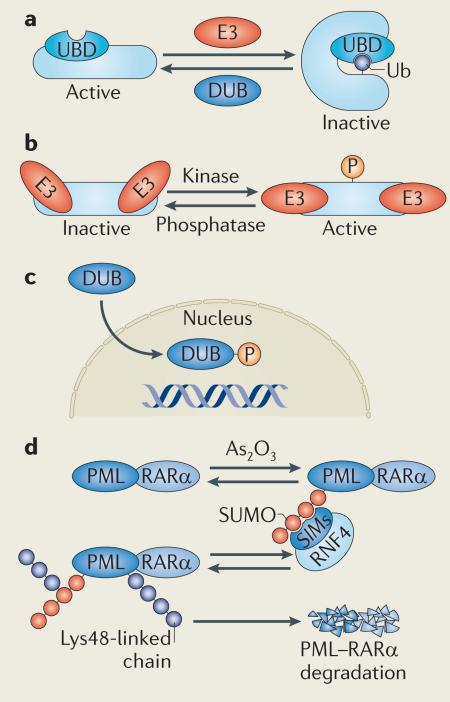
To regulate biological processes, the ubiquitin network is tightly integrated with other post-translational modifications, including phosphorylation, acetylation and conjugation by other ubiquitin-like molecules. Protein activity is often regulated by monoubiquitylation, leading to protein conformational changes (see the figure, part a). There is a significant interplay among different types of ubiquitin-like molecules with regard to enzymatic activity: for example, the ubiquitin-like modifier NEDD8 can be conjugated to cullin-based ubiquitin ligases and thereby induce a conformational change that facilitates ubiquitin transfer and stimulates ligase activity138. By contrast, sumoylation of ubiquitin-specific-processing protease 25 (USP25) inhibits its deubiquitylating enzyme (DUB) activity by steric hindrance139. Catalytic activity of the ubiquitin ligase machinery is also highly dependent on phosphorylation (see the figure, part b). For example, the activity of the E3 ligase ITCH is upregulated by Jun N-terminal kinase 1 (JNK1)-mediated phosphorylation140, whereas phosphorylation triggers nuclear translocation of the DUB ataxin 3 (REF. 141) (see the figure, part c).
Moreover, recent reports have brought new light to the role of different post-translational modifications in the N-end rule pathway142. Studies over the past decades indicate that certain amino-terminal residues, including Met, Ala, Val, Ser, Thr and Cys, are prone to acetylation and that this may function as a degradation signal (AcN-degrons) in the N-end rule pathway, promoting Doa10-dependent degradation in the proteasome143.
The significance of understanding the crosstalk between different post-translational modifications is clearly reinforced by recent advances made in the clinic. In patients with promyelocytic leukaemia (PML), treatment with arsenic trioxide (As2O3) has been found to induce polysumoylation of PML–retinoic acid receptor-α (RARα), the fusion protein that underlies the disease. Sumoylated PML–RARα is subsequently recognized and ubiquitylated with Lys48-linked chains by the small ubiquitin-related modifier (SUMO)-dependent E3 ligase RING finger 4 (RNF4), resulting in PML–RARα degradation and hence a therapeutic effect in PML patients144,145 (see the figure, part d).
SIMs, SUMO-interacting motifs; Ub, ubiquitin; UBD, ubiquitin-binding domain.
Another intriguing example of spatiotemporal regulation within ubiquitin networks is the time-dependent assembly of DNA damage response (DDR) complexes at specific nuclear foci in response to DNA damage, such as DNA double-strand breaks (DSBs). A key molecule during DDR complex assembly is the adaptor and nucleating protein mediator of DNA damage checkpoint 1 (MDC1). When phosphorylated by ataxia-telangiectasia mutated (ATM), MDC1 triggers the sequential recruitment of the E3 ligase RING finger 8 (RNF8; which, together with UBC13 (also known as UBE2N), ubiquitylates histone H2A and its variants), breast cancer type 1 susceptibility (BRCA1) and p53-binding protein 1 (53BP1) to DSBs (reviewed in REF. 34). To reinforce sustained ubiquitylation of histone H2A, an additional E3 ligase, RNF168, is subsequently recruited. This is mediated by RNF168’s two MIU (motif interacting with ubiquitin) domains and the recently identified UMI (UIM (ubiquitin-interacting motif)- and MIU-related) domain, which recognize ubiquitylated H2A. This results in local amplification of H2A modified by Lys63-linked polyubiquitin chains and the retention of 53BP1 and BRCA1 at DSBs (reviewed in REF. 16). Additional E3 ligases are also recruited to DSBs, including RAD18 (which promotes homologous recombination by directly binding the DNA recombinase RAD51C35) and the HECT E3 ligase HECT domain and RCC1-like domain-containing 2 (HERC2; which stabilizes RNF8–UBC13 to promote the amplification of Lys63-linked ubiquitin signals at DSBs36).
Timing is everything — dynamics through DUBs
Given that DUBs are highly involved in the timing of ubiquitin attachment and removal, the DUB branch of the ubiquitin network is also crucial for the spatiotemporal regulation of ubiquitylation. The close association of ubiquitin loading and removal is compellingly demonstrated by the identification of many E3–DUB pairs, in which two counteracting enzymes interact directly to fine-tune ubiquitin conjugation. The pairing of E3 ligases with their corresponding DUBs has also been suggested to be a regulatory mechanism to prevent autoubiquitylation of ligases and/or to induce proteasomal targeting of DUBs12.
Similarly to the ubiquitin-conjugation machinery, DUBs contain structural diversities that contribute to substrate specificity, subcellular localization and protein–protein interactions37. Of the estimated 95 DUBs in the human genome38, only ubiquitin-specific-processing protease 30 (USP30) 39 (anchored to the OMM) and USP19 (REF. 40) (anchored to the ER) have predicted transmembrane regions. However, again similarly to the ubiquitin-conjugation machinery, DUBs can be targeted to specific subcellular compartments by associating with proteins containing transmembrane domains. For example, ataxin 3 binds to the AAA-ATPase p97 (REF. 41), which recruits it to the cytoplasmic surface of the ER, where it directs the retrotranslocation of ubiquitylated ERAD substrates into the cytoplasm41.
An excellent example that illustrates the importance of DUBs in the temporal regulation of ubiquitin signalling is the assembly of DNA repair-competent complexes during DSB responses. At least seven DUBs have been reported to antagonize the activity of E3 ligases and enable dynamic changes in the DSB landscape. Among these, USP3 and USP16 directly deubiquitylate histone H2A42,43. By contrast, BRCA1–BRCA2-containing complex subunit 36 (BRCC36), which is recruited to RNF8–UBC13-synthesized Lys63-linked chains at DSBs as part of the RAP80 complex following irradiation44, functions as a general Lys63-specific DUB that counter acts the activity of the RNF8–UBC13 ubiquitin ligase complex by deubiquitylating γH2AX45. However, the molecular mechanisms underlying the temporal control of ligase activation followed by the precise involvement of BRCC36 in this signalling cascade remain elusive. Another key DUB is USP1, which modulates the ubiquity lation of proliferating cell nuclear antigen (PCNA)46 and the Fanconi anaemia proteins FANCD2 and FANCI47,48. Two recent surprises on the arena of DSB-associated DUBs are ubiquitin carboxy-terminal hydrolase 37 (UCH37; also known as UCHL5)49 and OTUB1 (REF. 50). UCH37, which has long been considered to be a proteasome-specific DUB51,52, was recently found to associate with the inositol-requiring 80 (INO80) chromatin-remodelling complex to catalyse nucleosome sliding. OTUB1 suppresses RNF168-dependent polyubiquitylation by binding to and inhibiting the E2 UBC13 independently of its catalytic activity50. It seems that in a physiological context, accumulation of OTUB1 in an E2 enzyme-enriched cell compartment might set the threshold for signalling initiation upon DNA damage50.
Ubiquitin recognition through UBDs
Following ubiquitin conjugation, cellular recognition and interpretation of ubiquitin signals are carried out by downstream effectors and scaffolds, which can also be spatially organized. The immediate decoders of ubiquitylation are proteins containing one or several UBDs (known as ubiquitin receptors), which often show specificity for ubiquitin length and linkage patterns (reviewed in REF. 11); these proteins commonly also have other functional domains. The low affinities reported for ubiquitin–UBD interactions (often in the micromolar range11,53), which for years have puzzled the research community, have led to the conclusion that the high specificity of ubiquitin–UBD interactions in physiological settings regularly relies on the local concentration of binding partners, the presence of additional binding interfaces and/or the formation of preformed signalling complexes.
Analogously to the ubiquitin-conjugation machinery and DUBs, ubiquitin receptors can be restricted to subcellular compartments by integral transmembrane anchors or domains that mediate membrane association by directly interacting with different phospholipids. This is a common strategy for proteins involved in endo cytosis and vesicle trafficking, which have the UBD UIM together with a phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5) P2)- and PtdIns(1,4,5) P3-binding epsin N-terminal homology (ENTH) domain (in the case of epsins) or a PtdIns3P-specific FYVE domain (in the case of hepatocyte growth factor-regulated Tyr kinase substrate (HRS) and signal-transducing adaptor molecule (STAM))18,54. Dynamic recruitment of ubiquitin receptors in response to physiological status is also common in the nucleus, where interactions with other nuclear proteins, as well as conjugation to ubiquitin and/or small ubiquitin-like modifier (SUMO), regulate step-by-step protein complex assembly on damaged chromatin16,55. One such example is the recruitment of translesion DNA polymerases to ubiquitylated PCNA, which is mediated by ubiquitin-binding motif (UBM) and ubiquitin-binding zinc-finger (UBZ) domains56, as well as MIU- and UMI-dependent chromatin targeting of RNF168 (REFS 57,58).
The functions of many of the >200 cellular ubiquitin receptors are mostly unknown11. Interesting combinations of UBDs with transmembrane domains, RNA recognition motifs and multiple protein–protein interaction modules indicate that there is much more to be discovered when it comes to spatial and temporal targeting of the ubiquitin network.
Functions of restricted ubiquitin signals
To demonstrate how stringent spatiotemporal regulation of ubiquitin networks is used to orchestrate cellular behaviour, we briefly discuss four well-characterized processes in which ubiquitylation has been studied in detail: the NF-κB signalling pathway; trafficking and endosomal sorting of transmembrane proteins; proteasomal degradation of damaged and unwanted proteins; and regulation of p53-mediated transcriptional activation by ubiquitin signals.
NF-κB — a prototype for ubiquitin compartmentalization
The past few years have brought a substantial amount of information concerning the integration of many ubiquitin signals in the NF-κB pathway (FIG. 1), which is important for proper regulation of inflammation, immune responses and cell survival59. Even though the activation of NF-κB (made up of p50 and p65) can originate from the stimulation of many cell-surface and intracellular receptors (such as tumour necrosis factor receptor 1 (TNFR1), interleukin-1 receptor type 1 (IL-1R1) or Toll-like receptor 4 (TLR4) and CD40), all pathways seem to converge at the stage of the inhibitor of NF-κB (IκB) kinase (IKK) complex. This master regulatory complex, composed of IKKα, IKKβ and NF-κB essential modulator (NEMO; also known as IKKγ), mediates the phosphorylation event that enables the Skp–cullin–F-box–βTrCP (SCFβTrCP)-induced Lys48-linked polyubiquitylation and subsequent degradation of the inhibitory IκB proteins. This ultimately allows NF-κB to translocate to the nucleus and activate transcription (reviewed in REFS 60,61).
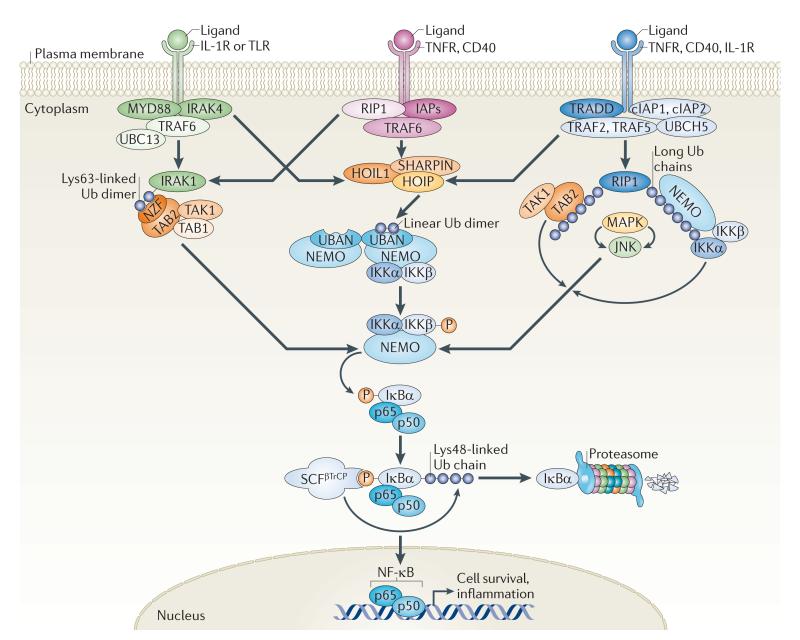
Figure 1. Ubiquitylation — bringing temporal and spatial order to the NF-κB pathway.
Activation of the nuclear factor-κB (NF-κB) pathway downstream of multiple inflammatory cell surface receptors, such as interleukin-1 receptor (IL-1R), Toll-like receptor (TLR), tumour necrosis factor receptor (TNFR) and CD40, triggers the recruitment and ubiquitylation of many molecules with scaffolding and catalytic activities. Recent evidence suggests that multiple ubiquitin species join forces to bring spatial and temporal organization to the different branches of the NF-κB pathway. Distinct ubiquitin signals seem to be spatially restricted close to specific ubiquitin-binding proteins such as TAK1-binding 2 (TAB2) and NF-κB essential modulator (NEMO; also known as IKKγ). TAB2 can bind Lys63-linked chains, which are produced by the E2 complex of ubiquitin-conjugating enzyme 13 (UBC13; also known as UBE2N) and UBE2 variant 1A (UEV1A) and E3 ligases such as TNFR-associated factor 6 (TRAF6). NEMO can bind linear diubiquitin chains, produced by the linear ubiquitin chain assembly complex (LUBAC), which is composed of HOIL1-interacting protein (HOIP), haeme-oxidized IRP2 ubiquitin ligase 1 (HOIL1; also known as RBCK1) and SHANK-associated RH domain-interacting protein (SHARPIN). NEMO can also bind longer ubiquitin chains with variable linkages, including Lys11, which is generated by the E2 UBCH5 and E3 ligase cellular inhibitor of apoptosis protein 1 (cIAP1)68. Regardless of which pathway is activated and by what type of ubiquitin chain, all scenarios result in the activation of the inhibitor of NF-κB (IκB) kinase (IKK) complex. Following activation, the IKKs phosphorylate the inhibitory IκBα, thus creating a binding site for the Skp–cullin–F-box–βTrCP (SCFβTrCP) ubiquitin E3 ligase complex. This triggers the Lys48-linked ubiquitylation and subsequent proteasomal degradation of IκBα. NF-κB (made up of p50 and p65) can then translocate to the nucleus and activate the transcription of genes promoting cell survival and inflammation. IRAK, IL-1R-associated kinase; JNK, Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; MYD88, myeloid differentiation primary response 88; NZF, neural zinc-finger factor; RIP1, receptor-interacting protein 1; TAK1, TGFβ-activated kinase 1; TRADD, TNFR1-associated DEATH domain; Ub, ubiquitin; UBAN, ubiquitin binding in ABIN and NEMO.
Different types of ubiquitin chains have been proposed to spatially and temporally control IKK complex activation in different branches of the NF-κB pathway7,8,61. Activation of the IKK complex depends on Lys63-linked ubiquitylation of numerous receptor-proximal substrates, including receptor-interacting protein 1 (RIP1), IL-1R-associated kinase (IRAK) and various TNFR-associated factors (TRAFs) in different settings, generated by the cooperative efforts of the Lys63-specific E2 complex UBC13–UBE2 variant 1A (UEV1A) together with a divergent set of ubiquitin E3 ligases (for example TRAFs, inhibitor of apoptosis proteins (IAPs) and Pellino)61. Lys63-linked chains, which function as docking sites for the ubiquitin-binding neural zinc-finger factor (NZF) domains of the TAK1-binding 2 (TAB2) and TAB3 adaptor proteins, have long been thought to be the dominant ubiquitin signal underlying activation of the IKKs61. Lys63-linked ubiquitin chains that are conjugated to adaptor proteins (RIP1, TRAFs and IRAKs) may indeed serve as platforms to recruit TAB2-bound TGFβ-activated kinase 1 (TAK1) and NEMO-associated IKK through the UBDs of TAB2 and NEMO, respectively61 (FIG. 1). This model is supported by precipitation experiments in which all of these proteins have been found in activated receptor complexes, together with several proteins decorated with Lys63-linked ubiquitin chains of unknown length. However, it remains unknown what determines the timing and specificity of kinase recruitment (TAB2, TAB3 and TAK1 should be recruited first, followed by NEMO–IKK) and how the directionality in signalling is accomplished in such complexes.
A possible explanation of this conundrum is offered by recent findings that ubiquitin chains coupled through Met1 (known as linear ubiquitin chains) can provide crucial specificity components upstream of IKK activation62–64. Linear ubiquitin chains are produced in cells by the linear ubiquitin chain assembly complex (LUBAC), which consists of the catalytic RING-between-RING domain protein HOIL1-interacting protein (HOIP) bound to two adaptor proteins, haeme-oxidized IRP2 ubiquitin ligase 1 (HOIL1; also known as RBCK1)62,64 and the recently identified SHANK-associated RH domain-interacting protein (SHARPIN)65–67. The current model indicates that the trimeric LUBAC is rapidly recruited from the cytosol to activated TNFR and CD40 receptor complexes by binding to substrates with Lys63- or Lys11-linked chains59,65,66,68. Following such recruitment, LUBAC can mediate linear ubiquitylation of NEMO, RIP1 and potentially other, unidentified components that are essential for activation of the NF-κB pathway62,64–67. Mechanistically, linear ubiquitin chains conjugated to NEMO molecules have been found to interact with the ubiquitin binding in ABIN and NEMO (UBAN) domain of neighbouring NEMO molecules63, leading to conformational changes in NEMO dimers that potentially can trigger activation of the IKKs63 (FIG. 1).
More recent data suggest that the length of ubiquitin chains may also be an important determinant in the activation of NF-κB23. Structural and biophysical data indicate that the isolated UBAN domain of NEMO preferentially binds linear diubiquitin, rather than Lys11-, Lys48- or Lys63-linked ubiquitin chains63,68–70. By contrast, when expressed together with the C-terminal zinc-finger of NEMO, the binding to longer Lys63-linked chains is increased71. Thus, it is likely that NEMO reads multiple ubiquitin chains using different mechanisms to control the dynamics and directionality of IKK complex activation (FIG. 1).
NF-κB inactivation is also temporally controlled. Two DUBs, A20 (also known as TNFAIP3) and CYLD, seem to be absolutely essential for NF-κB inactivation, as deletion or mutation of either of these causes uncontrolled inflammation and tumorigenesis (reviewed in REF. 72). A20 and CYLD have temporally distinct roles in inhibiting NF-κB at different stages during inflammation: CYLD restricts initial spontaneous activation of NF-κB, whereas A20 terminates inducible NF-κB signalling through a negative feedback loop. Although CYLD and A20 deubiquitylate similar substrates to impair NF-κB activation, their modes of action and spatiotemporal regulation differ significantly (reviewed in REF. 72). CYLD has been shown to bind NEMO directly and to display DUB activity towards both Lys63-linked and linear chains73. CYLD binding to NEMO is indeed required for its phosphorylation by the IKK complex, which in turn inhibits its DUB activity, leading to NF-κB activation74.
Although A20 is temporally regulated at the transcriptional level, it also binds to many interacting partners and is recruited to active receptor complexes to restrict NF-κB signalling. For example, following TNF stimulation, A20 interacts with the ubiquitin receptor TAX1-binding protein 1 (TAX1BP1) to form a complex with RIP1 (REF. 75). Together with TAX1BP1 and the E3 ligases ITCH and RNF11, A20 forms a ubiquitinediting complex that can inhibit the NF-κB cascade72. This is achieved by the removal of Lys63-linked chains from A20 substrates, such as TRAF6 and RIP1, followed by the addition of proteasome-targeting Lys48-linked chains76. Interestingly, although A20 preferentially cleaves Lys48-linked chains over Lys63-linked ones in vitro73,77, it preferentially removes Lys63-linked chains in cells, indicating that its specificity in vivo might be controlled by unknown determinants.
A20 can also temporally control NF-κB activation by restricting the interaction of the E3 ligases TRAF2, TRAF6 and cellular IAP 1 (cIAP1) with E2 enzymes78. Early after lipopolysaccharide stimulation, which activates TLRs, A20 disrupts the binding of TRAF2, TRAF6 and cIAP1 to the E2 enzymes UBC13 and UBCH5C, whereas at later time points A20 triggers Lys48-linked ubiquitylation and degradation of these enzymes78.
Ubiquitin controls trafficking and endosomal sorting
Plasma membrane proteins can be ubiquitylated by multiple monoubiquitins or oligoubiquitin chains, which leads to their internalization and subsequent endocytic sorting. This is known to occur not only for most receptor Tyr kinases but also for other membrane proteins54,79,80 (FIG. 2). The ubiquitylation of such cargoes is spatially regulated: it is initiated at the cell surface but can often be sustained on endosomal membranes by specific E3 ligases80,81. A set of endosomal ubiquitin receptors, including epidermal growth factor receptor substrate 15 (EPS15), epsin and the endosomal sorting complex required for transport (ESCRT) machinery, is responsible for orchestrating the directional flow and spatial sorting of ubiquitylated cargo along the endocytic route18,82. Each of the four ESCRTs (ESCRT-0, ESCRT-I, ESCRT-II and ESCRT-III) has at least three structural motifs that direct the spatial organization of the endosomal sorting machinery. These include domains that interact with membrane lipids, other ESCRTs and ubiquitin, although ESCRT-III lacks a UBD83.
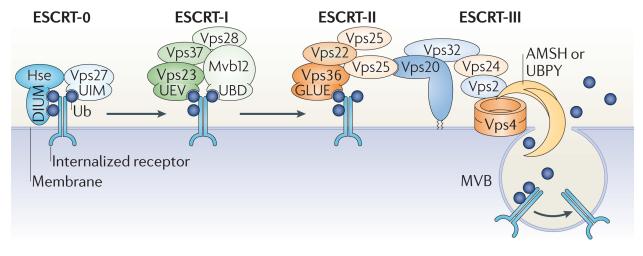
Figure 2. Ubiquitin-mediated coordination of receptor endocytosis.
The sequential recognition of ubiquitin signals by components of the endosomal sorting complexes required for transport (ESCRTs) is essential for orchestrating the transportation and sorting of internalized plasma membrane proteins along the endocytic pathway. Upon ligand binding, many plasma membrane proteins are marked by monoubiquitylation or Lys63-linked polyubiquitylation and internalized into early endosomes. At early endosomes, the internalized cargo is captured by ESCRT-0, which can simultaneously interact with three ubiquitin moieties through the diubiquitin motif (DIUM) of Hse (the yeast homologue of mammalian hepatocyte growth factor-regulated Tyr kinase substrate (HRS)) and the ubiquitin-interacting motif (UIM) of vacuolar protein sorting 27 (Vps27; the yeast homologue of signal-transducing adaptor molecule (STAM)). Following the ESCRT-0-mediated concentration of ubiquitylated cargo at early endosomes, ESCRT-I and ESCRT-II are sequentially recruited to the endosomal membrane by the ubiquitin-conjugating enzyme E2 variant (UEV) domain in Vps23 (the yeast homologue of tumour susceptibility gene 101 (TSG101)), the newly identified ubiquitin-binding domain (UBD) of MVB sorting factor 12 (Mvb12) and the GRAM-like ubiquitin-binding in EAP45 (GLUE) domain of Vps36, as well as by a direct interaction between the ESCRTs. Together with ESCRT-III, ESCRT-I and ESCRT-II facilitate the maturation of early endosomes into late endosomes, from which multivesicular bodies (MVBs) pinch-off to fuse with lysosomes, where the internalized cargo is ultimately degraded. In addition to the ESCRT components, several E3 ubiquitin ligases and deubiquitylating enzymes (DUBs) associate with the ESCRTs along the endocytic pathway, initially for directing the sorting events that separate cargo destined for either recycling or degradation and in the final stages to dissociate ubiquitin species from the internalized cargo before MVB formation. Dissociation of ubiquitin relies on the activity of the DUBs associated molecule with the SH3 domain of STAM (AMSH) and ubiquitin isopeptidase Y (UBPY), which, by directly interacting with ESCRT-III components, are suitably positioned at the membrane of late endosomes. Ub, ubiquitin.
In agreement with its role in physically capturing and concentrating ubiquitylated cargo at the endosomal membrane, the highest capacity to bind ubiquitin is displayed by ESCRT-0. This complex can simultaneously bind several ubiquitin moieties through the diubiquitin motif (DIUM) in HRS and UIM in STAM. It has been suggested that, after endocytosis has been initiated in this way, the accumulation of ubiquitin-decorated cargoes allows the subsequent recruitment of ESCRT-I and ESCRT-II. In yeast, ESCRT-I recognizes ubiquitin through the UEV domain in vacuolar protein sorting 23 (Vps23 ; the yeast homologue of mammalian tumour susceptibility gene 101 (TSG101)), together with a recently discovered UBD in MVB sorting factor 12 (Mvb12)84, whereas ESCRT-II relies on the GRAM-like ubiquitin-binding in EAP45 (GLUE) domain in Vps36 for ubiquitin binding. The recruitment of ESCRT-III and the machinery that finally carries out multivesicular body (MVB) formation and membrane scission (the AAA-ATPase Vps4) seems to be independent of ubiquitin signalling.
In addition to spatial accumulation of trafficking cargoes by the ESCRTs, temporal regulation of the entire process is ensured by the action of numerous ubiquitin-modulating proteins (E3 ligases and DUBs) that are localized to distinct parts of the endosomal compartment. For example, the DUBs associated molecule with the SH3 domain of STAM (AMSH) and ubiquitin isopeptidase Y (UBPY; also known as USP8, and termed Doa4 in yeast) are targeted to the early endosome by interacting with STAM and have important roles in the initial sorting of ubiquitylated cargo85. Furthermore, multiple E3 ligases and DUBs balance the ubiquitylation status of the internalized cargo to determine its fate, which is to be either recycled back to the membrane or degraded in the lysosome. In yeast, the level of Lys63-linked polyubiquitylation of membrane-bound substrates is controlled by the E3–DUB pair reverses SPT-phenotype 5 (Rsp5)–Ubp2, which is recruited to ESCRT-0 (containing Hse and Vps27, the yeast homologues of HRS and STAM)86,87. Indeed, Rsp5 activity is required for the lysosomal targeting of many plasma membrane proteins and was recently reported to be directly recruited to specific cell surface cargoes by arrestin-related trafficking adaptors (ARTs) in yeast88 through the ART PY motifs.
In the final stages of endocytosis, deubiquitylation is as important as the initial ubiquitylation event to complete the sorting of conjugates into MVBs. After an internalized cargo has finally been committed to degradation, removal of the conjugated ubiquitin species not only is essential for ubiquitin itself to be recycled but also allows the ESCRT machinery to dissociate from its cargo and to be reused in the cell. Such activities are carried out by specific DUBs, such as AMSH and UBPY, which are recruited to late endosomal compartments by directly associating with ESCRT-III or the scaffolding protein ALG2-interacting X (ALIX; also known as AIP1, and termed Bro1 in yeast) (reviewed in REF. 85).
Spatial control of substrate degradation
Even though the importance of ubiquitylation in protein degradation has long been appreciated, research during the past few years has challenged the traditional model assigning a linear correlation between ubiquitylation and proteasomal targeting by revealing a full battery of ubiquitin species that orchestrate protein removal in human cells.
Protein substrates, primarily conjugated by Lys48-linked ubiquitin chains (but possibly by all non-Lys63 linkages89) are routed to the proteolytic 26S protea some by several cooperating paths. Protein targeting to the proteasome can be direct, occurring through direct recognition of ubiquitin by the 19S regulatory particle subunits Rpn10 (S5A in mammals), which contains one to three UIMs depending on the species15, and Rpn13 (ADRM1 in mammals), which contains a newly identified UBD named Pru90,91. Alternatively, targeting can be indirect and, in that case, is mediated by proteasomal shuttle factors, such as Rad23, Dsk2 and DNA damage-inducible 1 (Ddi1) 92,93, which use their ubiquitin-associated domains (UBA domains) for binding ubiquitylated substrates and their N-terminal ubiquitin-like domains (UBL domains) for docking to the Rpn10, Rpn13 or Rpn1 (PSMD2 in mammals) subunit of the proteasomal base11,15. Shuttle factors have several important roles in the spatial organization of protein degradation, as they provide a mechanism for the capture and transport of distantly located ubiquitylated substrates to the proteasome. It has been suggested that they also stabilize the substrate–proteasome complex and protect ubiquitin chains from excessive extension and disassembly during the transfer to the proteasome. Interestingly, both Rad23 and Dsk2 are directly linked to the ubiquitin-conjugation machinery by interaction with Ufd2, an E4 enzyme that perfectly positions them close to ubiquitin chains immediately after conjugation, ensuring quick transport to the proteasome94.
When correctly positioned at the narrow entrance into the proteasomal core, proteins destined for destruction need to be deubiquitylated to ensure efficient degradation of the substrate and recycling of the ubiquitin tag. To accomplish a tightly regulated deubiquitylation, the proteasome is associated with at least three DUBs with complementary qualities. The JAMM (JAB1/MPN/MOV34 metalloenzyme)-type Rpn11 is an integral subunit of the regulatory particle lid, whereas Ubp6 (USP14 in mammals) and UCH37 (which is absent in budding yeast) are recruited and activated by direct binding to Rpn1 and ADRM1, respectively (reviewed in REF. 95). In a complementary manner, Rpn11 has a propensity to cleave ubiquitin chains with proximal specificity to displace the entire chain at once, whereas Ubp6 and UCH37 remove ubiquitin moieties from the distal tip of chains, resulting in progressive trimming of ubiquitylated substrates15. Functionally, these distinct activities are thought to fine-tune protein degradation, providing a mechanism to attenuate degradation of certain, possibly lightly or improperly ubiquitylated, substrates when needed. Adding another level of complexity to the dynamic control of substrate degradation, the HECT E3 and E4 ligase Hul5 was recently reported to extend existing ubiquitin chains and thereby counteract the chain-trimming activities of Ubp6 at proteasomes96, which indicates that we still do not have the complete picture of the dynamic regulation of substrate commitment to degradation.
Localized ubiquitin signalling also has a role in autophagy, which removes insoluble proteins and damaged or excess organelles from the cell20,97, but this is not discussed in this Review owing to space limitations.
Guarding the ‘guardian of the genome’
The stability, activity and localization of numerous proteins are regulated by various ubiquitin modifications, as well as other post-translational modifications (BOX 2). One intriguing example of such complex regulation is the tumour suppressor p53. Polyubiquitylation of p53 by the E3 ligase double-minute 2 (MDM2) leads to its proteasomal degradation98, but additional E3 ligases have also been found to modify p53 with monoubiquitin, Lys48-linked chains or Lys63-linked chains. This enables tight control of p53 levels, as well as changes in both its subcellular localization and its preference for a diverse panel of interaction partners (recently reviewed in REF. 99). Interestingly, when present at low levels, MDM2, as well as a selection of other E3 ligases100, can switch specificity and mediate p53 monoubiquitylation, which unmasks p53’s nuclear export signal, thereby leading to its nuclear export101,102. Cytosolic p53 directly binds the antiapoptotic proteins B cell lymphom a-extra large (BCL-XL) and BCL-2, thereby triggering the mitochondrial apoptotic pathway103, and was recently found to also repress autophagy in a cell cycle-dependent manner104. Moreover, p53 has been shown to be ubiquitylated by the ER-resident E3 ligase synoviolin in the vicinity of the ER, resulting in its degradation by the cytoplasmic ERAD pathway; however, the functional significance of this is unclear105.
Ubiquitylation in development
Spatial and temporal organization of the ubiquitin network has in recent years proved to orchestrate biological processes not only on the cellular level but also on the level of tissues and entire animals. Below, we discuss how differential modes of ubiquitylation at specific subcellular sites are used to control the development and functional output of specific cell types (in this case, neurons) and highlight the importance of spatiotemporally organized ubiquitylation during animal development.
Polarizing ubiquitin signals in neurons
A spatially controlled ubiquitin network is important to coordinate the polarized flow of information that is necessary for the life and functionality of a neuron, from early development and maintenance to modulation of plasticity and functional output. Together with protein synthesis, protein degradation provides a general mechanism to fine-tune protein availability and receptor function in synapses and to facilitate long-lasting storage of information by continuously remodelling the neuronal proteome106,107.
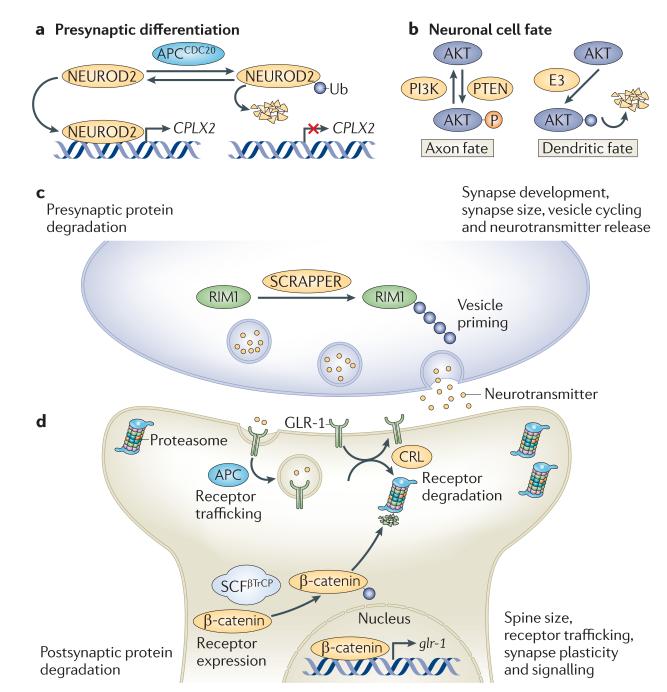
Every step of neuronal development, from early differentiation to fine-tuning of synapses, seems to be regulated by the ubiquitin network. For example, by joining forces with its two distinct co-activators, CDH1 and CDC20, the E3 ligase anaphase-promoting complex (APC) directs axon growth and patterning108, and controls presynaptic axonal differentiation109 by triggering the degradation of the transcription factor neurogenic differentiation factor 2 (NEUROD2) (FIG. 3a). Ubiquitin-mediated degradation is also a key event during the establishment of neuronal cell fate. Axonal and dendritic fates are distinguished by high and low levels of phosphoinositide 3-kinase (PI3K) signalling, respectively, and the distinctive activation of the PI3K pathway in axons has been found to depend on the spatially restricted proteasomal degradation of AKT in dendrites. This may be mediated by the specific recruitment of an unknown E3 ligase to dendritic tips, resulting in a local degradation of AKT in these structures110 (FIG. 3b).
Figure 3. Polarized functions of ubiquitylation in neurons.
Stringent spatial control of the ubiquitin network is crucial for many aspects in the life, death and functionality of the nervous system. a | The ubiquitin E3 ligase anaphase-promoting complex (APC)21 is considered to be a key regulator of presynaptic axonal differentiation. APC and its co-activator CD20 (APCCD20) induce the degradation of the proneuronal transcription factor neurogenic differentiation factor 2 (NEUROD2), which regulates the transcription of complexin II (CPLX2) and thereby suppresses presynaptic differentiation specifically at presynaptic sites. b | The spatially restricted ubiquitylation and subsequent proteasomal degradation of AKT in dendrites underlies the generation of neuronal polarity, during which axonal and dendritic fates are distinguished by high and low levels of phosphoinositide 3-kinase (PI3K) signalling, respectively. To further reinforce high PI3K activation in axons, phosphatase and tensin homologue (PTEN; which antagonizes PI3K signalling) was recently found to be reciprocally downregulated by NEDD4-mediated ubiquitylation in axonal growth cones. c,d | A spatially controlled ubiquitin–proteasome system network is essential for maintaining optimal protein levels and synaptic balance. At the presynaptic side, the turnover of proteins regulating vesicle priming and release, such as DUNC13 in Drosophila melanogaster and RAB3-interacting molecule 1 (RIM1) in mice, are strictly controlled by ubiquitylation. In mice, the E3 responsible for RIM1 ubiquitylation is SCRAPPER (c). In response to neuronal stimulation, active proteasomes are highly enriched in the postsynaptic compartment, where they are crucial for controlling spine size, receptor trafficking, synapse plasticity and signalling downstream of neurotransmitter receptors. In Caenorhabditis elegans, Glu receptor 1 (GLR-1) is regulated by ubiquitylation on many levels, including receptor internalization and trafficking (by the APC) and receptor degradation (by a cullin–RING ubiquitin ligase (CRL) complex made up of cullin 3 and the BTB-Kelch protein KEL-8). Moreover, GLR-1 is also transcriptionally controlled by WNT signalling, which in turn is regulated by the Skp–cullin–F-box–βTrCP (SCFβTrCP)-mediated ubiquitylation and degradation of β-catenin (d). Ub, ubiquitin.
It has long been known that growth cones contain an active ubiquitin–proteasome system and that inhibition of the proteasome hampers chemotropic-mediated growth cone turning in response to netrin 1, an extracellular axonal guidance molecule111, as well as synaptic function and plasticity112. In agreement, ubiquitylation is clearly involved in axon guidance, often by regulating the amount of guidance receptors at the plasma membrane. For example, the Drosophila melanogaster Roundabout protein (a repulsive axon guidance receptor that binds to Slit, a repellent secreted by midline glia113) is upregulated in commissural neurons by the E3 ligase NEDD4 after their compulsory crossing of the midline. This provides a switch in responsiveness from attractive to repellent midline cues, thereby inhibiting any further attempts to cross the midline114. Mechanistically, the surface levels of Roundabout depend on the sorting receptor Commisureless, which directs Roundabout from the synthetic to the late endocytic pathway, resulting in Roundabout degradation. Commisureless is specifically downregulated in a NEDD4-dependent manner in neurons that have crossed the midline, thus stimulating an enhanced sorting of Roundabout to the cell surface, where it acts to inhibit secondary attempts to cross the midline114,115.
In the mature nervous system, components of the ubiquitin–proteasome system localize near synapses107,112, where they control proper synaptic balance by maintaining optimal protein levels. On the presynaptic side, the levels of proteins, such as the synaptic vesicle priming proteins DUNC13 in D. melanogaster and RAB3-interacting molecule 1 (RIM1) in mice, are under the strict control of ubiquitylation (FIG. 3c), indicating an evolutionarily conserved link between neurotransmitter release and the ubiquitin–proteasome system116,117. More specifically, the mouse E3 ligase SCRAPPER is thought to polyubiquitylate RIM1 in an activity-dependent manner and thereby regulate neurotransmission strength, which is essential for proper synaptic fine-tuning during long-term potentiation117. On the postsynaptic side, ubiquitylation is highly important for regulating synaptic conductivity by controlling the number of neurotransmitter receptors that reside at postsynaptic membranes. In Caenorhabditis elegans, the AMPA-type Glu receptor 1 (GLR-1) has been reported to be negatively regulated in this way by multiple E3 ligases, including APC — which regulates its trafficking and recycling118 — and the E3 cullin–RING ubiquitin ligase complex containing the BTB-Kelch protein KEL-8 and cullin 3 (CUL-3) — which mediates its degradation in the proteasome119. In addition, GLR-1 is transcriptionally controlled by an SCF complex (SCFβTrCP) containing the F-box protein LIN-23, which promotes degradation of the C. elegans β-catenin homologue β-catenin- and armadillo-related 1 (BAR-1) and thereby regulates the transcription of WNT target genes, including GLR-1 (REF. 120) (FIG. 3c). Although the dynamics of GLR-1 regulation by ubiquitylation is not fully understood, it is clear that the ubiquitin–proteasome system is essential to ensure an appropriate adjustment of GLR-1 levels in response to synaptic requirements at different time points. In addition to receptor levels, activity-induced ubiquitylation of numerous postsynaptic density molecules, including postsynaptic density 95 (PSD95), SH3 and multiple ankyrin repeat domains protein (SHANK) and SAP90-and PSD95-associated protein (SAPAP; also known as GKAP)107, is crucial for the constitutively ongoing structural and functional adaptation of PSD structure and function in response to physiological needs.
Ubiquitin networking in animal development
Spatial restriction of the ubiquitin network is not only obvious within cells but also crucial in the context of entire organisms, in which developmental choices to a large extent depend on local activities of the ubiquitylation machinery.
In the early embryo, developmental decisions often rely on secreted signalling molecules, called morphogens, which form concentration gradients to orchestrate the induction of differential cell fates. Initially, morphogens, including members of the bone morphogenetic protein (BMP) family, establish the body plan of the animal. For example, in D. melanogaster the orthologue of BMP2 and BMP4, Decapentaplegic (DPP), is essential for dorsoventral axis formation (reviewed in REF. 121). Cells within the signalling range of DPP respond by activating a transcriptional programme mediated by the SMAD transcriptional activators MAD and MEDEA, which form a dimer and translocate to the nucleus121.
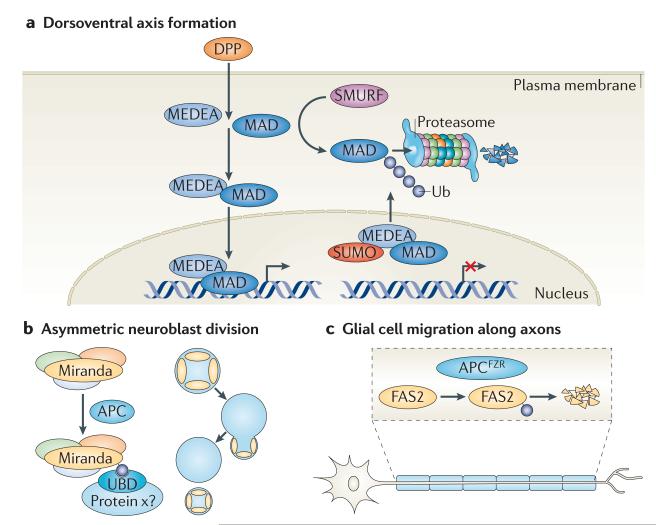
In addition to a restricted expression of DPP, recent data show that MAD and MEDEA are regulated by ubiquitylation and sumoylation, respectively (FIG. 4a). The SMAD ubiquitylation regulatory factor (SMURF) family of HECT E3 ubiquitin ligases targets MAD for proteasomal degradation during embryonic dorsoventral patterning122, whereas the nuclear conjugation of MEDEA by SUMO triggers its nuclear export and thereby negatively regulates its activity123; these events are essential for confining the signalling range of DPP.
Figure 4. Control of development by localized ubiquitylation events.
a | The developmental morphogen Decapentaplegic (DPP) is responsible for mediating dorsoventral polarity in Drosophila melanogaster. When bound to its receptor, DPP activates a transcriptional programme mediated by the SMAD transcriptional activators MAD and MEDEA, which translocate to the nucleus as dimers. The SMAD ubiquitylation regulatory factor (SMURF) family of HECT E3 ligases targets MAD for proteasomal degradation, and sumoylation of MEDEA in the nucleus triggers its nuclear export, thus negatively regulating its activity. b | Miranda is a multidomain scaffold protein that plays an important part in neuroblast asymmetric division in D. melanogaster. Its central domain binds cargo proteins, such as Prospero, Staufen and Brain tumour (BRAT). It has been shown that the localization of Miranda is regulated by the multidomain E3 ligase anaphase-promoting complex (APC), which probably directly monoubiquitylates Miranda, leading to its asymmetric localization and retention at the neuroblast cortex through binding to an unknown ubiquitin receptor. c | The migration of glial cells along motor axons in the D. melanogaster peripheral nervous system relies on the homophilic interaction between two distinct isoforms of the immunoglobulin superfamily cell adhesion molecule Fasciclin 2 (FAS2), which is expressed on the cell surface of motor neurons and glial cells, respectively. Recently, the ubiquitin E3 ligase APCFZR was reported to promote, in a graded manner, the ubiquitylation and subsequent degradation of FAS2 in motor neurons, thereby forming a gradient of cell surface-bound FAS2 along the length of the axon. This allows glial cells to migrate and encapsulate motor neuron axons as they grow. SUMO, small ubiquitin-related modifier; Ub, ubiquitin.
Spatial and temporal orchestration by the ubiquitin network is also important for the development of specific tissues and cell types. As already emphasized, ubiquitylation is an important determinant during development of the nervous system. In addition to its role in axonal differentiation, the APC has been reported to regulate the asymmetric distribution of Miranda and its associated cargo proteins (such as Prospero, Staufen and Brain tumour (BRAT)), which enables the asymmetric division of D. melanogaster neuroblasts into self-renewing neuroblasts and committed ganglion mother cells124. By monoubiquitylating Miranda, the APC can enable asymmetric localization of both Miranda and its cargo proteins, potentially by regulating its binding to ubiquitin receptors (FIG. 4b). At later stages, the APC locally induces endocytic internalization and downregulation of the adhesion molecule Fasciclin 2 (FAS2) in a graded manner along the axonal projections of motor neurons, thereby creating a subcellular gradient of adhesiveness that is thought to coordinate glial migration with axonal growth125 (FIG. 4c). Even though direct ubiquitylation of FAS2 has not been shown in D. melanogaster, its vertebrate homologue, neural cell adhesion molecule (NCAM), is known to be targeted for endocytosis by ubiquitylation126.
An alternative approach to impart spatial information through the ubiquitin network is applied during the terminal differentiation of D. melanogaster spermatids. This process, known as individualization, is controlled by the activity of a CUL3 E3 ligase complex in which the adaptor protein KLH10 targets Bruce, a member of the IAP family, for proteolytic degradation127. The degradation of Bruce is carried out in a graded manner, leading to a gradient of active caspases and consequent production of individualized sperm128. This gradient is generated by the recently identified KLH10–CUL3 E3 ligase inhibitor Scotti, which is expressed in a subcellular distal-to-proximal gradient in spermatids.
In addition to specific roles for distinct ubiquitin network components, the overall status of ubiquitylation and proteasomal degradation seems to be developmentally regulated. The levels of the polyubiquitin receptors RPN10 and RAD23 fluctuate between different developmental phases. This has been shown in D. melanogaster, in which high expression levels of these proteins during embryogenesis and pupal stages may reflect the intense tissue reorganizations that occur at these stages129. Interestingly, the composition of the proteasome has also been found to be closely linked to animal age. Indeed, ageing seems to perturb the assembly of the highly active 26S proteasome, favouring the formation of the weakly active 20S proteasome, an event that is associated with a marked decline in locomotor capacity and ATP levels130.
It therefore seems that there are many strategies to use the ubiquitin network during development. New findings, including the recent report that cellular responsiveness may be fine-tuned by opposing the activities of E3–DUB enzymes, as has been shown for the WNT signalling pathway in D. melanogaster131, indicate that we have so far only scratched the surface of how ubiquitylation is used to achieve spatial organization in development.
Conclusion and perspectives
Even though many of the basic principles of ubiquitylation are well understood, we are far from completely comprehending the full functional extent of ubiquitin-mediated events in specific biological processes. As the immediate access to free ubiquitin at any given location in the cell is a prerequisite for ubiquitylation, there is a need for spatial control of ubiquitin signalling. In this Review, we have discussed some examples in which ubiquitin signalling is spatially controlled; other examples, not discussed owing to space limitations, include regulation of parkin-mediated mitophagy by the protein kinase PTEN-induced putative kinase (PINK)132,133, and localization of ubiquitin signalling cascades regulating antiviral innate immunity in peroxisomes134.
Future studies will need to focus on increasing our understanding of the dynamics of the ubiquitin network in its physiological context and to clarify how diverse ubiquitin signals are generated and decoded by UBDs in the highly crowded micromilieu of the cell. We are now starting to grasp the link between specific E2–E3 pairs and the formation of distinct ubiquitin chains, but pinpointing which pairs are physiologically relevant and clarifying the importance of ubiquitin chain length in biological settings still requires further research. Related to this subject, it will also be interesting to dissect the mechanisms of ubiquitin editing and investigate whether this phenomenon is restricted to the proteasome and the NF-κB pathway or whether it is a general strategy used for the spatiotemporal regulation of ubiquitin signalling networks.
The ubiquitin system has been found to have integral roles in most cellular events and is therefore an important therapeutic target, for example for the development of treatments against cancer and neurodegenerative disorders, as well as inflammatory and infectious diseases135,136. A better understanding of the spatiotemporal regulation of the ubiquitin network may allow us to target such proteins at specific subcellular structures, leading to the development of more selective therapeutics and minimization of unwanted side effects136.
Acknowledgements
We apologize to all scientists whose important contributions were not referenced in this Review owing to limitations in the number of references. We are grateful to C. Joazeiro and H. Walczak for comments and discussions. C.G. is supported by the Swedish Foundation for Strategic Research (SSF). Research in the I.D. laboratory is supported by the Deutsche Forschungsgemeinschaft, the Cluster of Excellence ‘Macromolecular Complexes’ of the Goethe University Frankfurt (EXC115) and a European Research Council Advanced Grant.
Glossary
- RING
A zinc-binding protein–protein interaction motif, found in RING-type ubiquitin E3 ligases, that binds to the E2 ubiquitin thioester and thereby promotes ubiquitin transfer to substrate proteins.
- ER-associated degradation
A protein quality control pathway (mediated by p97–VCP–CDC48) in which misfolded or regulated proteins residing in the endoplasmic reticulum (ER) are translocated into the cytosol for proteasomal degradation.
- Ubiquitin-like molecule
A protein that structurally resembles ubiquitin and uses its own E1, E2 and E3 enzymes to conjugate to target proteins and to modify their properties. Examples are the small ubiquitin-like modifiers (SUMOs), NEDD8, ISG15, Fat10 and autophagy-related 8.
- N-end rule
A biological principle that relates the half-life of a cellular protein to the identity of its amino-terminal residue, in which N-terminal amino acids are recognized by specific E3 ubiquitin ligases (N-recognins).
- Breast cancer type 1 susceptibility (BRCA1)
An E3 ligase that catalyses multiple types of ubiquitin signals. It is often found in a heterodimeric RING complex with BRCA1-associated RING domain 1 and has an important role in homologous recombination repair of DNA double-strand breaks.
- Ubiquitin-interacting motif
An α-helical ubiquitin-binding domain that binds to the conserved hydrophobic patch that is centred around Ile44 in ubiquitin with an affinity in the range of ~100–400 μM.
- HECT
One of the major classes of ubiquitin ligases. HECT E3 ligases contain a domain with a catalytic Cys residue that forms a thioester intermediate during ubiquitin transfer to the substrate protein.
- Proliferating cell nuclear antigen (PCNA)
A ring-shaped molecule encircling DNA. It slides bidirectionally along DNA to constitutively monitor genomic integrity. Following DNA damage, ubiquitylation of PCNA is essential for the recruitment of damage-tolerant DNA polymerases, allowing translesion synthesis.
- F-box
A protein module of ~50 residues that is involved in mediating protein–protein interactions. F-box proteins commonly act as substrate recognition subunits in cullin–RING ubiquitin ligases.
- Receptor Tyr kinase
A membrane-bound protein Tyr kinase that often functions as a receptor for secreted hormones, growth factors and cytokines.
- Diubiquitin motif
A double-sided ubiquitin-binding domain, first identified in hepatocyte growth factor-regulated Tyr kinase substrate (HRS), that can simultaneously bind two ubiquitin moieties, both of which are required for the endocytic sorting function of HRS.
- GRAM-like ubiquitin-binding in EAP45
A ubiquitin-binding domain that folds into a split pleckstrin homology domain with a non-canonical lipid-binding pocket that interacts with phosphatidylinositol-3-phosphate.
- Multivesicular body (MVB)
An intermediate structure in the endosomal pathway that is formed when membrane portions bud into the lumen of late endosomes, forming intralumenal vesicles. MVB sorting is an essential event for the degradation of internalized cell surface proteins in the lysosome.
- Ubiquitin-associated domain
A short (~40 amino acids) sequence motif, first found in proteins associated with the ubiquitylation pathway, that mediates polyubiquitin binding.
- Ubiquitin-like domain (UBL domain)
A modular protein domain present in multiple cellular proteins. UBL domains structurally resemble ubiquitin by folding into the ubiquitin β-grasp superfold and can in general be recognized by ubiquitin-binding domains.
- JAMM (JAB1/MPN/MOV34 metalloenzyme)
A family of zinc metalloprotease deubiquitylating enzymes.
- Anaphase-promoting complex
A multifunctional ubiquitin ligase that, by pairing with its co-activators CDC20 and CDH1, specifically targets cell cycle proteins (among others) for degradation.
- Cullin–RING ubiquitin ligase (CRL)
A member of a large family of multisubunit E3 ligases, commonly comprising a cullin scaffold, a catalytic RING subunit, a substrate-recognition subunit (SRS), and, for most CRLs, an adaptor subunit linking the SRS to the complex.
- Postsynaptic density (PSD)
A structure in the postsynaptic membrane in which l-Glu neurotransmitter receptors are accumulated together with multiple adhesion, scaffold, cytoskeletal and signalling molecules. PSDs organize the postsynaptic signalling machinery, control synaptic plasticity and maintain synaptic homeostasis.
Footnotes
Competing interests statement The authors declare no competing financial interests.
FURTHER INFORMATION Ivan Dikic’s homepage: http://www.biochem2.com/default.html
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Weissman AM. Themes and variations on ubiquitylation. Nature Rev. Mol. Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Varshavsky A. Regulated protein degradation. Trends Biochem. Sci. 2005;30:283–286. doi: 10.1016/j.tibs.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 5.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nature Rev. Mol. Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nature Rev. Mol. Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of NF-κB activation. EMBO Rep. 2009;10:706–713. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koegl M, et al. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 10.Hoppe T. Multiubiquitylation by E4 enzymes: ‘one size’ doesn’t fit all. Trends Biochem. Sci. 2005;30:183–187. doi: 10.1016/j.tibs.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains — from structures to functions. Nature Rev. Mol. Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkinson KD. DUBs at a glance. J. Cell Sci. 2009;122:2325–2329. doi: 10.1242/jcs.041046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nature Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 14.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nature Rev. Mol. Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nature Rev. Mol. Cell Biol. 2010;11:479–489. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- 17.Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- 18.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 19.Wertz IE, Dixit VM. Regulation of death receptor signaling by the ubiquitin system. Cell Death Differ. 2010;17:14–24. doi: 10.1038/cdd.2009.168. [DOI] [PubMed] [Google Scholar]
- 20.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol. Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz AL, Ciechanover A, Brandt RA, Geuze HJ. Immunoelectron microscopic localization of ubiquitin in hepatoma cells. EMBO J. 1988;7:2961–2966. doi: 10.1002/j.1460-2075.1988.tb03158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beers EP, Moreno TN, Callis J. Subcellular localization of ubiquitin and ubiquitinated proteins in Arabidopsis thaliana. J. Biol. Chem. 1992;267:15432–15439. [PubMed] [Google Scholar]
- 23.Ikeda F, Crosetto N, Dikic I. What determines the specificity and outcomes of ubiquitin signaling? Cell. 2010;143:677–681. doi: 10.1016/j.cell.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 24.Bodine SC, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 25.Li W, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [Describes the identification of E3 ligase complexes involved in spatial ubiquitylation and degradation of proteins at the ER.] [DOI] [PubMed] [Google Scholar]
- 27.Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 28.Ravid T, Hochstrasser M. Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nature Cell Biol. 2007;9:422–427. doi: 10.1038/ncb1558. [DOI] [PubMed] [Google Scholar]
- 29.Neutzner A, Benard G, Youle RJ, Karbowski M. Role of the ubiquitin conjugation system in the maintenance of mitochondrial homeostasis. Ann. N. Y. Acad. Sci. 2008;1147:242–253. doi: 10.1196/annals.1427.012. [DOI] [PubMed] [Google Scholar]
- 30.Cohen MM, Leboucher GP, Livnat-Levanon N, Glickman MH, Weissman AM. Ubiquitin-proteasome-dependent degradation of a mitofusin, a critical regulator of mitochondrial fusion. Mol. Biol. Cell. 2008;19:2457–2464. doi: 10.1091/mbc.E08-02-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yonashiro R, et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang B, et al. GIDE is a mitochondrial E3 ubiquitin ligase that induces apoptosis and slows growth. Cell Res. 2008;18:900–910. doi: 10.1038/cr.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plafker SM, Plafker KS, Weissman AM, Macara IG. Ubiquitin charging of human class III ubiquitin-conjugating enzymes triggers their nuclear import. J. Cell Biol. 2004;167:649–659. doi: 10.1083/jcb.200406001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang J, et al. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nature Cell Biol. 2009;11:592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bekker-Jensen S, et al. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nature Cell Biol. 2010;12:80–86. doi: 10.1038/ncb2008. [DOI] [PubMed] [Google Scholar]
- 37.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [Systematic proteomic analysis of DUB interaction networks.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nijman SM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura N, Hirose S. Regulation of mitochondrial morphology by USP30, a deubiquitinating enzyme present in the mitochondrial outer membrane. Mol. Biol. Cell. 2008;19:1903–1911. doi: 10.1091/mbc.E07-11-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hassink GC, et al. The ER-resident ubiquitin-specific protease 19 participates in the UPR and rescues ERAD substrates. EMBO Rep. 2009;10:755–761. doi: 10.1038/embor.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong X, Pittman RN. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum. Mol. Genet. 2006;15:2409–2420. doi: 10.1093/hmg/ddl164. [DOI] [PubMed] [Google Scholar]
- 42.Nicassio F, et al. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 43.Joo HY, et al. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 44.Sobhian B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shao G, et al. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc. Natl Acad. Sci. USA. 2009;106:3166–3171. doi: 10.1073/pnas.0807485106. [References 43 and 44 show that spatial control of ubiquitin-editing mechanisms at the sites of DSBs is regulated by the ubiquitin-binding protein RAP80.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang TT, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nature Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 47.Nijman SM, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Sims AE, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nature Struct. Mol. Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 49.Yao T, et al. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol. Cell. 2008;31:909–917. doi: 10.1016/j.molcel.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakada S, et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature. 2010;466:941–946. doi: 10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
- 51.Yao T, et al. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nature Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- 52.Hamazaki J, et al. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006;25:4524–36. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winget JM, Mayor T. The diversity of ubiquitin recognition: hot spots and varied specificity. Mol. Cell. 2010;38:627–635. doi: 10.1016/j.molcel.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 54.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 55.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 56.Bienko M, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 57.Pinato S, Gatti M, Scandiuzzi C, Confalonieri S, Penengo L. UMI, a novel RNF168 ubiquitin binding domain involved in the DNA damage signaling pathway. Mol. Cell. Biol. 2011;31:118–126. doi: 10.1128/MCB.00818-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pinato S, et al. RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Mol. Biol. 2009;10:55. doi: 10.1186/1471-2199-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bianchi K, Meier P. A tangled web of ubiquitin chains: breaking news in TNF-R1 signaling. Mol. Cell. 2009;36:736–742. doi: 10.1016/j.molcel.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 60.Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 61.Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-κB regulatory pathways. Annu. Rev. Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 62.Haas TL, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [By using a proteomic approach, the authors describe spatial recruitment of the linear ubiquitin ligase complex LUBAC to the activated TNFR complexes.] [DOI] [PubMed] [Google Scholar]
- 63.Rahighi S, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [Describes the structure of the UBAN domain of NEMO in complex with linear diubiquitin, which explains the compartment-based specificity in activation of the IKK complex.] [DOI] [PubMed] [Google Scholar]
- 64.Tokunaga F, et al. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nature Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 65.Ikeda F, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gerlach B, et al. Linear ubiquitination prevents inflammation and regulates immune signaling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 67.Tokunaga F, et al. Sharpin is a component of the NF-κB activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [References 63–66 provide evidence for the spatiotemporal control of LUBAC (SHARPIN–HOIP–HOIL1) in ubiquitylating NEMO and mediating NF-κB activation in vivo. Deficiency in LUBAC functions leads to multi-organ inflammation and immune system defects.] [DOI] [PubMed] [Google Scholar]
- 68.Dynek JN, et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29:4198–4209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ivins FJ, et al. NEMO oligomerization and its ubiquitin-binding properties. Biochem. J. 2009;421:243–251. doi: 10.1042/BJ20090427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lo YC, et al. Structural basis for recognition of diubiquitins by NEMO. Mol. Cell. 2009;33:602–615. doi: 10.1016/j.molcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laplantine E, et al. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J. 2009;28:2885–2895. doi: 10.1038/emboj.2009.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harhaj EW, Dixit VM. Deubiquitinases in the regulation of NF-κB signaling. Cell Res. 2011;21:22–39. doi: 10.1038/cr.2010.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Komander D, et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reiley W, Zhang M, Wu X, Granger E, Sun SC. Regulation of the deubiquitinating enzyme CYLD by IκB kinase γ-dependent phosphorylation. Mol. Cell. Biol. 2005;25:3886–3895. doi: 10.1128/MCB.25.10.3886-3895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iha H, et al. Inflammatory cardiac valvulitis in TAX1BP1-deficient mice through selective NF-κB activation. EMBO J. 2008;27:629–41. doi: 10.1038/emboj.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 77.Lin SC, et al. Molecular basis for the unique deubiquitinating activity of the NF-κB inhibitor A20. J. Mol. Biol. 2008;376:526–540. doi: 10.1016/j.jmb.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shembade N, Ma A, Harhaj EW. Inhibition of NF-κB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Polo S, Di Fiore PP. Endocytosis conducts the cell signaling orchestra. Cell. 2006;124:897–900. doi: 10.1016/j.cell.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 81.Belgareh-Touze N, et al. Versatile role of the yeast ubiquitin ligase Rsp5p in intracellular trafficking. Biochem. Soc. Trans. 2008;36:791–796. doi: 10.1042/BST0360791. [DOI] [PubMed] [Google Scholar]
- 82.Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci. 2007;32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 83.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nature Rev. Mol. Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shields SB, et al. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J. Cell Biol. 2009;185:213–224. doi: 10.1083/jcb.200811130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Clague MJ, Urbe S. Endocytosis: the DUB version. Trends Cell Biol. 2006;16:551–559. doi: 10.1016/j.tcb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 86.Kee Y, Lyon N, Huibregtse JM. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J. 2005;24:2414–2424. doi: 10.1038/sj.emboj.7600710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ren J, Kee Y, Huibregtse JM, Piper RC. Hse1, a component of the yeast Hrs-STAM ubiquitin-sorting complex, associates with ubiquitin peptidases and a ligase to control sorting efficiency into multivesicular bodies. Mol. Biol. Cell. 2007;18:324–335. doi: 10.1091/mbc.E06-06-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–725. doi: 10.1016/j.cell.2008.09.025. [Describes spatial organization of the endocytic complexes at the cell surface by the family of arrestin-related proteins.] [DOI] [PubMed] [Google Scholar]
- 89.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [Quantitative proteomics identifies all non-Lys63 ubiquitin chains in proteasomal degradation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Husnjak K, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schreiner P, et al. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [References 89 and 90 identify a new UBD present in Rpn13, a ubiquitin receptor at the proteasome, that is implicated in binding and recruiting ubiquitylated proteins for degradation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Glickman MH, Raveh D. Proteasome plasticity. FEBS Lett. 2005;579:3214–3223. doi: 10.1016/j.febslet.2005.04.048. [DOI] [PubMed] [Google Scholar]
- 93.Grabbe C, Dikic I. Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins. Chem. Rev. 2009;109:1481–1494. doi: 10.1021/cr800413p. [DOI] [PubMed] [Google Scholar]
- 94.Richly H, et al. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 95.Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol. Cell. Proteomics. 2010 Sep;7 doi: 10.1074/mcp.R110.003871. (doi:10.1074/mcp.R110.003871) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Crosas B, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [Provides evidence for local editing of ubiquitin chains by opposing activities of E3 ligases and DUBs at the proteasome.] [DOI] [PubMed] [Google Scholar]
- 97.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nature Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 98.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 99.Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010;17:86–92. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li M, et al. Monoversus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 101.Stommel JM, et al. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lohrum MA, Woods DB, Ludwig RL, Balint E, Vousden KH. C-terminal ubiquitination of p53 contributes to nuclear export. Mol. Cell. Biol. 2001;21:8521–8532. doi: 10.1128/MCB.21.24.8521-8532.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mihara M, et al. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 104.Tasdemir E, et al. Regulation of autophagy by cytoplasmic p53. Nature Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yamasaki S, et al. Cytoplasmic destruction of p53 by the endoplasmic reticulum-resident ubiquitin ligase ‘Synoviolin’. EMBO J. 2007;26:113–122. doi: 10.1038/sj.emboj.7601490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cajigas IJ, Will T, Schuman EM. Protein homeostasis and synaptic plasticity. EMBO J. 2010;29:2746–2752. doi: 10.1038/emboj.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nature Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- 108.Konishi Y, Stegmuller J, Matsuda T, Bonni S, Bonni A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 2004;303:1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- 109.Yang Y, et al. A Cdc20-APC ubiquitin signaling pathway regulates presynaptic differentiation. Science. 2009;326:575–578. doi: 10.1126/science.1177087. [Shows that the E3 ubiquitin ligase APCCDC20 is important for proper synapse formation in the developing neurons in the brain, possibly through the degradation of brain-enriched transcription factor NEUROD2.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yan D, Guo L, Wang Y. Requirement of dendritic Akt degradation by the ubiquitin-proteasome system for neuronal polarity. J. Cell Biol. 2006;174:415–424. doi: 10.1083/jcb.200511028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32:1013–1026. doi: 10.1016/s0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- 112.Patrick GN, Bingol B, Weld HA, Schuman EM. Ubiquitin-mediated proteasome activity is required for agonist-induced endocytosis of GluRs. Curr. Biol. 2003;13:2073–2081. doi: 10.1016/j.cub.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 113.Simpson JH, Kidd T, Bland KS, Goodman CS. Short-range and long-range guidance by Slit and its Robo receptors. Robo and Robo2 play distinct roles in midline guidance. Neuron. 2000;28:753–766. doi: 10.1016/s0896-6273(00)00151-3. [DOI] [PubMed] [Google Scholar]
- 114.Myat A, et al. Drosophila Nedd4, a ubiquitin ligase, is recruited by Commissureless to control cell surface levels of the roundabout receptor. Neuron. 2002;35:447–459. doi: 10.1016/s0896-6273(02)00795-x. [DOI] [PubMed] [Google Scholar]
- 115.Keleman K, et al. Comm sorts Robo to control axon guidance at the Drosophila midline. Cell. 2002;110:415–427. doi: 10.1016/s0092-8674(02)00901-7. [DOI] [PubMed] [Google Scholar]
- 116.Speese SD, Trotta N, Rodesch CK, Aravamudan B, Broadie K. The ubiquitin proteasome system acutely regulates presynaptic protein turnover and synaptic efficacy. Curr. Biol. 2003;13:899–910. doi: 10.1016/s0960-9822(03)00338-5. [DOI] [PubMed] [Google Scholar]
- 117.Yao I, et al. SCRAPPER-dependent ubiquitination of active zone protein RIM1 regulates synaptic vesicle release. Cell. 2007;130:943–957. doi: 10.1016/j.cell.2007.06.052. [Reports the identification of the synaptic E3 ligase SCRAPPER as a regulator of proteasome-mediated degradation of RIM1 required for synaptic tuning.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Juo P, Kaplan JM. The anaphase-promoting complex regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Curr. Biol. 2004;14:2057–2062. doi: 10.1016/j.cub.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 119.Schaefer H, Rongo C. KEL-8 is a substrate receptor for CUL3-dependent ubiquitin ligase that regulates synaptic glutamate receptor turnover. Mol. Biol. Cell. 2006;17:1250–1260. doi: 10.1091/mbc.E05-08-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dreier L, Burbea M, Kaplan JM. LIN-23-mediated degradation of β-catenin regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Neuron. 2005;46:51–64. doi: 10.1016/j.neuron.2004.12.058. [DOI] [PubMed] [Google Scholar]
- 121.O’Connor MB, Umulis D, Othmer HG, Blair SS. Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development. 2006;133:183–193. doi: 10.1242/dev.02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Podos SD, Hanson KK, Wang YC, Ferguson EL. The DSmurf ubiquitin-protein ligase restricts BMP signaling spatially and temporally during Drosophila embryogenesis. Dev. Cell. 2001;1:567–578. doi: 10.1016/s1534-5807(01)00057-0. [DOI] [PubMed] [Google Scholar]
- 123.Miles WO, et al. Medea SUMOylation restricts the signaling range of the Dpp morphogen in the Drosophila embryo. Genes Dev. 2008;22:2578–2590. doi: 10.1101/gad.494808. [References 121 and 122 describe the role of ubiquitylation and sumoylation in the regulation of spatial and temporal organization of signalling pathways during embryo development.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Slack C, Overton PM, Tuxworth RI, Chia W. Asymmetric localisation of Miranda and its cargo proteins during neuroblast division requires the anaphase-promoting complex/cyclosome. Development. 2007;134:3781–3787. doi: 10.1242/dev.010900. [DOI] [PubMed] [Google Scholar]
- 125.Silies M, Klambt C. APC/CFzr/Cdh1-dependent regulation of cell adhesion controls glial migration in the Drosophila PNS. Nature Neurosci. 2010;13:1357–1364. doi: 10.1038/nn.2656. [DOI] [PubMed] [Google Scholar]
- 126.Diestel S, Schaefer D, Cremer H, Schmitz B. NCAM is ubiquitylated, endocytosed and recycled in neurons. J. Cell Sci. 2007;120:4035–4049. doi: 10.1242/jcs.019729. [DOI] [PubMed] [Google Scholar]
- 127.Arama E, Bader M, Rieckhof GE, Steller H. A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol. 2007;5:e251. doi: 10.1371/journal.pbio.0050251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kaplan Y, Gibbs-Bar L, Kalifa Y, Feinstein-Rotkopf Y, Arama E. Gradients of a ubiquitin E3 ligase inhibitor and a caspase inhibitor determine differentiation or death in spermatids. Dev. Cell. 2010;19:160–173. doi: 10.1016/j.devcel.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 129.Lipinszki Z, et al. Developmental-stage-specific regulation of the polyubiquitin receptors in Drosophila melanogaster. J. Cell Sci. 2009;122:3083–3092. doi: 10.1242/jcs.049049. [DOI] [PubMed] [Google Scholar]
- 130.Vernace VA, Arnaud L, Schmidt-Glenewinkel T, Figueiredo-Pereira ME. Aging perturbs 26S proteasome assembly in Drosophila melanogaster. FASEB J. 2007;21:2672–82. doi: 10.1096/fj.06-6751com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mukai A, et al. Balanced ubiquitylation and deubiquitylation of Frizzled regulate cellular responsiveness to Wg/Wnt. EMBO J. 2010;29:2114–2125. doi: 10.1038/emboj.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tanaka A, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62 SQSTM1. Nature Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 134.Dixit E, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [Describes the local organization of signalling complexes at the peroxisomal membranes that are essential for antiviral responses.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cohen P, Tcherpakov M. Will the ubiquitin system furnish as many drug targets as protein kinases? Cell. 2010;143:686–693. doi: 10.1016/j.cell.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 136.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 137.Varshavsky A. The ubiquitin system. Trends Biochem. Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 138.Rabut G, Peter M. Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell. 2008;30:610–619. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 140.Gallagher E, Gao M, Liu YC, Karin M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc. Natl Acad. Sci. USA. 2006;103:1717–1722. doi: 10.1073/pnas.0510664103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pastori V, et al. CK2 and GSK3 phosphorylation on S29 controls wild-type ATXN3 nuclear uptake. Biochim. Biophys. Acta. 2010;1802:583–592. doi: 10.1016/j.bbadis.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 142.Varshavsky A. The N-end rule. Cell. 1992;69:725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]
- 143.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lallemand-Breitenbach V, et al. Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ ubiquitin-mediated pathway. Nature Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 145.Tatham MH, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nature Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [References 143 and 144 provide evidence for regulated ubiquitylation of the promyelocytic leukaemia–retinoic acid receptor-α complex upon recruitment of E3 ligase RNF4 to SUMO chains. This pathway is used as an anti-cancer treatment mechanism through treatment with arsenic trioxide.] [DOI] [PubMed] [Google Scholar]