Abstract
Recent discoveries of autophagy receptors, which specifically recognize different cellular cargo destined for degradation, have opened a new chapter in the autophagy field. Selective cargo recognition by autophagic machinery is important in the context of cellular homeostasis and survival. One of the crucial homeostasis events involving autophagy is the removal of damaged or excessive mitochondria through mitophagy. Future studies on mitochondrial receptors and proteins associated with mitochondrial clearance will help us better understand the role of mitophagy in normal physiological processes as well as in diverse pathological conditions.
Keywords: mitophagy, autophagy receptors, mitochondria, Nix, Bnip3, reticulocyte maturation
Introduction
Cellular homeostasis is established through biogenesis and degradation of proteins and organelles. Recently, research has been focused on removal of cellular components as well as their turnover with a special emphasis on autophagy, a sequestering process that involves the formation of double-membrane vesicles. Controlled and selective removal of damaged or redundant organelles via autophagy is involved in developmental processes and is physiologically important in certain disease states.1 Various types of selective autophagy can be categorized depending on the substrate, ranging from aggregated proteins, mitochondria or ribosomes to bacteria and are named accordingly aggrephagy, mitophagy, ribophagy or xenophagy.2,3 Cargo engulfment mechanisms are not well understood, but recent discoveries of numerous autophagic receptors and adaptors that uniquely recognize cargo species are emerging.2,3
The Role of Nix in Mitophagy
Mitochondria are one of the most vital organelles performing essential functions in the cell, from ATP production to calcium storage and cell death, and are indispensable for cellular homeostasis.4 Knowledge of the molecular mechanisms of mitochondrial recognition and removal by mitophagy has so far been limited. It is well known that during cellular development low energy states occur and excess mitochondria are not needed, and they may be the source of agents such as reactive oxygen species (ROS), cytochrome c and other apoptosis-inducing factors that when released could cause cell damage and death. In such a case, mitophagy is activated and occurs following induction or inhibition of apoptosis.5 Accumulation of damaged mitochondria is associated with neurodegenerative diseases, hypoxia and cancer, and it is important to comprehend how these abnormal conditions can be avoided. Moreover, partial or complete mitochondrial elimination during development and cell differentiation (e.g., during reticulocyte and lymphocyte maturation, and spermatogenesis) is likewise essential.
We have recently described Nix (also named Bnip3L) as mammalian mitophagy receptor important for selective removal of damaged mitochondria as well as complete removal of mitochondria during reticulocyte maturation.6 Nix-deficient mice develop anemia and severe reticulocytosis caused by inefficient loss of mitochondria. Although the basic autophagy machinery is functional in the absence of Nix, mitochonodria engulfment by autophagosomes is impaired.7,8 Therefore, we were able to demonstrate that Nix directly interacts with LC3/GABARAP proteins in an LIR (LC3 Interacting Region)-dependent manner where a single mutation in the LIR is sufficient to abolish recruitment of LC3/GABARAP to mitochondria. Analysis of programmed mitochondrial clearance in reticulocytes of Nix-deficient cells, where wild-type or LIR mutant Nix was introduced, revealed that the effect of the LIR mutation on Nix-mediated clearance is partial. This is similar to recent data where the yeast mitophagy receptor Atg32 was shown to interact with Atg8 in an LIR-dependent manner but this binding is also partial. For fully functional mitophagy in yeast, Atg32 needs a second binding partner, Atg11.9,10 Partial mitochondrial clearance in Nix−/− reticulocytes also suggests alternative mechanisms leading to mitophagy (Fig. 1). Further, Nix initiates mitophagy by preparing mitochondria for recognition by the autophagic machinery. When mitochondria get depolarized, Nix influences translocation of the E3 ligase Parkin to mitochondria in order to ubiquitinate mitochondrial proteins and, thus, mark mitochondria for degradation by autophagy.11,12 The importance of ubiquitin as a signal in autophagy has been demonstrated by identification of p62 and NBR1 as autophagy receptors that bind both ubiquitinated proteins, with their UBA domains and LC3/GABARAP, with LIR domains.13,14 In contrast, physiological localization of Nix to the mitochondrial outer membrane predisposes it to deliver mitochondria into autophagosomes without requiring the intermediate ubiquitin to label the cargo. However, ubiquitin, attached to other mitochondrial proteins, might be important for additional selection of damaged versus healthy mitochondria for autophagy.
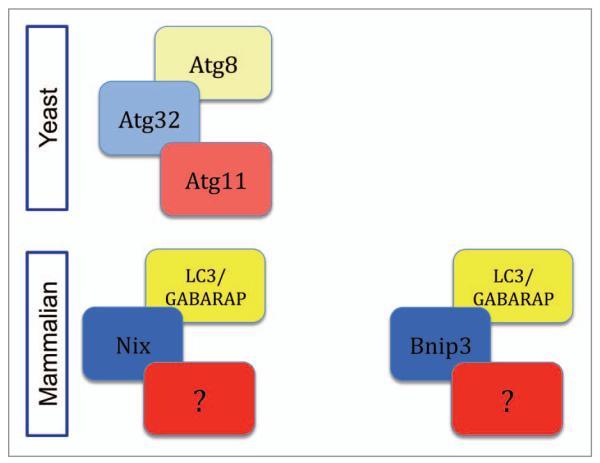
Figure 1.
Mitophagy receptors in yeast and mammals. The yeast mitophagy receptor Atg32 interacts with the Atg8 and Atg11 proteins in order to completely remove damaged mitochondria. An interaction between LC3/GABARAP proteins and the mitophagy receptors, Nix and Bnip3 is not sufficient for fully functional mitophagy in the mammalian system; an additional binding partner(s) needs to be defined.
Mitophagy Receptors in Development
Recent studies on Atg7−/− mice revealed the connection between general autophagy and mitophagy in red blood cell differentiation, and have suggested that the basic autophagic machinery is indispensable for programmed mitochondrial clearance. In contrast, mitochondrial mass and number in ex vivo hematopoietic lineage cells, such as myeloid and dendritic cells, are not affected by the loss of Atg7. The selective removal of mitochondria by autophagy during erythropoeisis is essential, and is a necessary developmental step. Degradation of other organelles in Atg7−/− erythroid cells, including the nucleus, ER and ribosomes is not affected.15 However, the Atg7−/− cells do not answer one of the key questions as to whether the depolarization of mitochondria is a cause or a consequence of mitochondria recruitment to autophagosomes. Because mitochondria in Nix−/− reticulocytes remain polarized, it has been hypothesized that Nix targets the mitohondria for mitophagy by triggering depolarization.7 However, the polarization state of mitochondria in Atg7−/− reticulocytes does not show mitochondrial depolarization, suggesting that Nix is not the only protein responsible for membrane depolarization.15 Therefore, current results are not giving a clear answer as to whether all mitochondria need to be depolarized for clearance by mitophagy. In contrast, it was proven that increased ROS levels lead to mitochondrial depolarization that signals translocation of the E3-ligase Parkin to mitochondria where it ubiquitinates mitochondrial proteins such as VDAC1 and mitofusins, and consequently obstructs the mitochondrial fission and fusion balance to finally trigger autophagy.16-18 Moreover, there is emerging evidence that other autophagy signals might be responsible for mitophagy. Another mitophagy receptor candidate is Bnip3, a Nix homolog previously shown to be involved in autophagic cell death that occurs independently of apoptosis.19 Hypoxia selectively induces Bnip3-dependent mitophagy through disruption of the interaction between Beclin 1 and Bcl-2.20 In addition, Nix was described to be a similar regulator of hypoxia-induced autophagy.21 Involvement of both proteins is important in hypoxia conditions where knockdown of both Bnip3 and Nix inhibits the pro-survival activation of hypoxia-induced mitophagy. In that regard, ectopic expression of both Bnip3 and Nix causes activation of mitophagy even in normoxia.21
Our preliminary results showed that Bnip3, similar to Nix, possess an LIR that strongly binds LC3/GABARAP proteins and its binding is abolished when a single mutation in the LIR is introduced (Fig. 2). Nix and Bnip3 could, consequently, act together and participate in mitophagy to finally result in cell rescue or differentiation. Furthermore, Nix and Bnip3 could be involved in mitochondrial removal through the same autophagic mechanism by binding LC3/GABARAP but in different normal or abnormal conditions. For example, Nix could be involved in complete removal of mitochondria during cellular differentiation (in reticulocytes or T-lymphocytes) and Bnip3 in conditions such as hypoxia. Both could be required when, in normal conditions and terminally differentiated cells, the population of damaged mitochondria needs to be removed to protect cells from further damage or death. Because p62 and NBR1 show redundancy while clearing aggregated ubiquitinated proteins, it is also possible that Nix and Bnip3 have overlapping functions as mitophagy receptors.
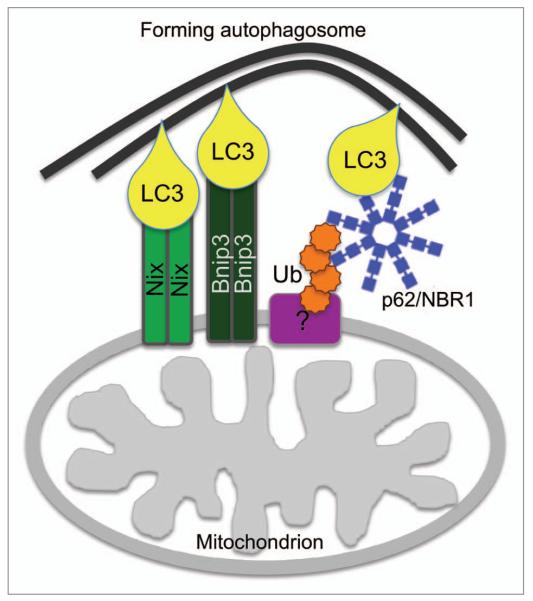
Figure 2.
The role of Nix, Bnip3 and ubiquitin in mitophagy. Mitochondrion-localized Nix and Bnip3 interact via their N-terminal LIR with LC3/GABARAP-conjugated to the newly forming autophagosome, thereby mediating tethering of the nascent phagophore to the target mitochondrion. Concomitant ubiquitination of a mitochondrion-localized substrate protein may, via the Ub-binding autophagy receptors p62 and NBR1, further increase tethering of mitochondrial and phagophore membranes.
Concluding Remarks
The role of Nix as a mitophagy receptor has extensively been studied in reticulocytes; however, other cellular systems, where mitochondria are physiologically removed due to developmental progression, like in spermatocytes or lens cells, needs to be additionally explored for both Nix and Bnip3. Moreover, future studies will enable better understanding of the mechanism by which the mitophagy receptors Nix and Bnip3 recognize mitochondria destined for elimination, and what are the upstream signals that label such mitochondria. What mediates full incorporation of mitochondria into autophagosomes remains to be further explored, but it is obvious that this process is tightly connected with regulation of mitochondrial quality.
Acknowledgements
We are grateful to Janos Terzic and Ivana Marinovic Terzic for comments and discussions. I.N. is supported by an EMBO Long term fellowship. Research in the I.D. laboratory is supported by the Deutsche Forschungsgemeinschaft, the Cluster of Excellence “Macromolecular Complexes” of the Goethe University Frankfurt (EXC115) and the European Research Council (ERC) grant agreement No. [250241-LineUb].
References
- 1.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–30. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirkin V, et al. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–69. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 3.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–41. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 4.Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–90. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xue L, Fletcher GC, Tolkovsky AM. Mitochondria are selectively eliminated from eukaryotic cells after blockade of caspases during apoptosis. Curr Biol. 2001;11:361–5. doi: 10.1016/s0960-9822(01)00100-2. [DOI] [PubMed] [Google Scholar]
- 6.Novak I, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandoval H, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–5. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schweers RL, et al. NIX is required for programmed Do mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104:19500–5. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanki T, et al. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Ding WX, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285:27879–90. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narendra D, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 14.Kirkin V, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–16. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 15.Mortensen M, et al. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci USA. 2010;107:832–7. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–31. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 17.Gegg ME, et al. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–70. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci USA. 2010;107:5018–23. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azad MB, et al. Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving BNIP3. Autophagy. 2008;4:195–204. doi: 10.4161/auto.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Bellot G, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–81. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]