SUMMARY
Aggregation of misfolded proteins and the associated loss of neurons are considered as a hallmark of numerous neurodegenerative diseases. Optineurin is present in protein inclusions observed in various neurodegenerative diseases including amyotrophic lateral sclerosis (ALS), Huntington’s disease, Alzheimer’s disease, Parkinson’s disease, Creutzfeld-Jacob disease and Pick’s disease. Optineurin deletion mutations have also been described in ALS patients. However, the role of optineurin in mechanisms of protein aggregation remains unclear. In this report, we demonstrate that optineurin recognized various protein aggregates via its C-terminal coiled-coil domain in a ubiquitin-independent manner. We also show that optineurin depletion significantly increase protein aggregation in HeLa cells and morpholino-silencing of the optineurin ortholog in zebrafish causes the motor axonopathy phenotype similar to a zebrafish model of ALS. A more severe phenotype is observed when optineurin is depleted in zebrafish carrying ALS mutations. Furthermore, TANK1 binding kinase 1 (TBK1) is co-localized with optineurin on protein aggregates and is important in clearance of protein aggregates through the autophagy-lysosome pathway. TBK1 phosphorylates optineurin at position Ser-177 and regulates its ability to interact with autophagy modifiers. This study provides evidence for a ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates as well as additional relevance for TBK1 as an upstream regulator of the autophagic pathway.
Keywords: amyotrophic lateral sclerosis, Huntington disease, huntingtin, optineurin, phosphorylation, SOD1, TBK1, ubiquitin
INTRODUCTION
Accumulation of misfolded proteins and the associated loss of neurons are considered as a hallmark of many neurodegenerative diseases such as Parkinson’s disease, Alzheimer’s disease, Huntington’s disease (HD), amyotrophic lateral sclerosis (ALS), Creutzfeld-Jacobs disease, Pick’s disease as well as several types of spinocerebellar ataxias (Aguzzi and O’Connor, 2010). Two main proteolytic systems are responsible for the degradation of proteins in the cell; the ubiquitin-proteasome system (UPS) and the autophagy-lysosomal pathway. Misfolded and/or damaged proteins are usually successfully degraded by the UPS. If the amount of proteins targeted to the UPS surpasses the degradation efficiency rate of the proteasome, subsequent oligomerization and accumulation of proteins activate autophagy (Tyedmers et al., 2010). The autophagy-lysosomal degradation machinery can also, to some extent, clear protein aggregates (Korolchuk et al., 2010).
Macroautophagy (hereafter referred to as autophagy) is an intracellular process responsible for the degradation of protein aggregates, long-lived proteins, defective organelles and intracellular pathogens (Dikic et al., 2010; Chen and Klionsky, 2011; Mizushima et al., 2011). During autophagy, a double-membrane vesicle (the autophagosome) engulfs the cargo and subsequently fuses with the lysosome which degrades the cargo from the autophagosome. Over the last few years it has been shown that autophagic membranes can selectively target cargo via ubiquitin-like microtubule-associated protein light chain 3 modifiers [MAP1(LC3)] and gamma-aminobutyrate receptor associated protein (GABARAP), which act as receptor molecules (Kirkin et al., 2009a; Kraft et al., 2010). For example, autophagy of protein aggregates is coordinated by specific autophagy adaptor proteins p62 and NBR1, which simultaneously bind to polyubiquitinated protein aggregates and LC3 (Pankiv et al., 2007; Kirkin et al., 2009b). Recently, other autophagy adaptor proteins were reported, such as Alfy, Nix, NDP52, optineurin and FYCO1, all of which have been demonstrated to play a critical role in autophagy of protein aggregates (Filimonenko et al., 2010), defective mitochondria (Novak et al., 2010) and bacteria Salmonella enterica (Thurston et al., 2009; Wild et al., 2011) or in the general movement of autophagosomes by binding to microtubules (Pankiv et al., 2010), respectively.
Optineurin (OPTN) is a 577 amino acid protein of versatile functions which interacts with a variety of proteins involved in membrane and vesicle trafficking (Rab8, huntingtin, myosin VI), cellular morphogenesis, NF-κB regulation, signal transduction, transcription regulation and cellular division control (Hattula and Peranen, 2000; Anborgh et al., 2005; Sahlender et al., 2005; Zhu et al., 2007; del Toro et al., 2009; Kachaner et al., 2012). Mutations in OPTN gene have been associated with normal-tension glaucoma, primary open-angle glaucoma (Sarfarazi and Rezaie, 2003; Chalasani et al., 2009), Paget’s disease (Albagha et al., 2010; Chung et al., 2010) and ALS (Maruyama et al., 2010; Del Bo et al., 2011; van Blitterswijk et al., 2012). Recently, OPTN was characterized as an autophagy adaptor protein which regulates selective autophagy of ubiquitin-coated cytosolic Salmonella enterica. This OPTN function depends on the phosphorylation of its LC3-interacting motif by the protein kinase TANK-binding kinase 1 (TBK1) (Wild et al., 2011). OPTN has been shown to co-localize with protein inclusions in ALS (Maruyama et al., 2010), HD (Schwab et al., 2012) and many other neurodegenerative diseases (Osawa et al., 2011). However, little is known about the role of OPTN in neurodegenerative disorders, particularly in respect to the autophagy-mediated degradation of misfolded protein inclusions.
In this report, we use different aggregation-prone protein models: short fragment of the protein huntingtin carrying an extended polyglutamine (polyQ) mutation (htt ex1 Q103) and superoxide dismutase 1 (SOD1) protein carrying point mutations at position Gly-93 (SOD1 G93C or SOD1 G93A), commonly used as models of HD and ALS, respectively (Witan et al., 2008; Filimonenko et al., 2010). We show that OPTN recognizes htt ex1 Q103 and SOD1 G93C aggregates through its C-terminal coiled-coil domain in a ubiquitin-independent manner and actively participates in the degradation of both htt ex1 Q103 and SOD1 G93C inclusions. Furthermore, we provide evidence for the hypothesis that the degradation of htt ex1 Q103 and SOD1 G93C aggregates is regulated by OPTN via phosphorylation of OPTN Ser-177 by TBK1. Finally, OPTN depletion in wild-type zebrafish causes a motor axonopathy phenotype similar to the phenotype reported in a zebrafish model of ALS generated by overexpression of mutant form of SOD1.
RESULTS
OPTN interacts with aggregating proteins involved in ALS and HD
Recently, mutations in the gene encoding OPTN have been associated with some familial and sporadic forms of ALS. OPTN is frequently detected in protein inclusions in neurons of ALS and HD patients (Maruyama et al., 2010; Schwab et al., 2012). To address the possible interaction of OPTN with protein aggregates involved in neurodegenerative pathologies, we transiently transfected HeLa cells with GFP-SOD1 G93C mutant. This mutation has been identified in ALS patients and is responsible for familial cases of ALS (Gaudette et al., 2000). We also used HeLa cells stably transfected with CFP-htt ex1 Q103 as a well-known model of HD (Filimonenko et al., 2010). The interaction between OPTN and SOD1 G93C or htt ex1 Q103 aggregates was studied using co-immunostaining, immunoprecipitation and binding experiments.
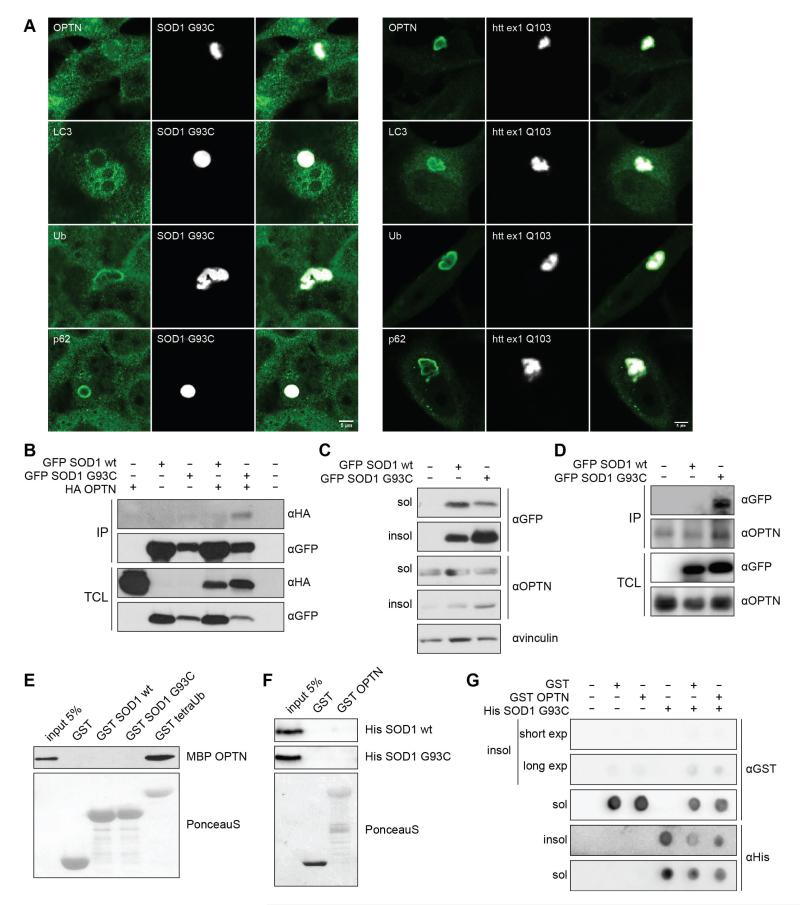
Confocal analyses indicated that OPTN readily co-localized with both SOD1 G93C and htt ex1 Q103 inclusions (Fig. 1A, upper panels). Similarly, we observed a co-localization of ubiquitin, p62 and LC3 with SOD1 G93C and htt ex1 Q103 aggregates (Fig. 1A). To further confirm OPTN interaction with SOD1 G93C inclusions, we co-expressed wild-type SOD1 (SOD1 wt) or mutant SOD1 G93C with a wild-type HA-OPTN (HA-OPTN wt) in HeLa cells. Wild-type HA-OPTN co-immunoprecipitated only with the aggregation-prone SOD1 G93C mutant and not with wild-type SOD1 (Fig. 1B). Moreover, fractionation experiments, separating soluble and insoluble fractions of the cells, revealed that a significant amount of endogenous OPTN was found in the insoluble fraction of HeLa cells expressing GFP-SOD G93C while the portion of insoluble OPTN detected in cells transfected with the empty vector or with the wild-type GFP-SOD1 was found to be less prominent (Fig. 1C). Co-immunoprecipitation experiments of soluble GFP-SOD1 G93C with endogenous OPTN were performed to determine if OPTN interacted with the soluble monomeric and oligomeric forms of mutant GFP-SOD1 G93C present in the soluble fraction of HeLa cells. Mutant GFP-SOD1 G93C but not wild-type SOD1 co-immunoprecipitated with endogenous OPTN from the soluble fraction (Fig. 1D). To test if the interaction of OPTN with SOD1 G93C mutant was direct, we carried out a GST pull-down binding assay with either purified MBP-OPTN and GST-SOD1 G93C or GST-OPTN and His-SOD1 G93C. Neither purified MBP-OPTN nor GST-OPTN interacted with mutant GST-SOD1 G93C or His-SOD1 G93C, respectively (Fig. 1E, F). Finally we tested whether OPTN directly interacted with aggregated mutant SOD1 G93C using a dot blot assay. We induced aggregation of His-SOD1 G93C in vitro (Yerbury et al., 2012) and mature aggregates were incubated with purified GST-OPTN. OPTN did not bind to mature SOD1 G93C aggregates. The results obtained suggest that OPTN indirectly recognized soluble monomeric or oligomeric forms of mutant SOD1 G93C but also the aggregated form of SOD1 (Fig. 1B-G).
Fig. 1. OPTN interacts with aggregating proteins involved in ALS and HD.
(A) Endogenous OPTN, LC3, ubiquitin (Ub) and p62 co-localized with GFP-SOD1 G93C aggregates in transiently transfected HeLa cells for 48 hours (left side) and with CFP-htt ex1 Q103 aggregates in stable transfected HeLa cells (right side). Scale bar, 5 μm. (B) Co-immunoprecipitation experiment from HeLa cells co-transfected with wild-type GFP-SOD1 or GFP-SOD1 G93C and HA-OPTN. Total cell lysates (TCL) were subjected to immunoprecipitation using anti-GFP antibody and then probed with anti-HA antibody. (C) Immunoblot analysis of cellular fractions using antibodies against vinculin, GFP and OPTN. HeLa cells transfected with empty vector, wild-type GFP-SOD1 and GFP-SOD1 G93C were lysed in NP40 lysis buffer 48 hours post transfection and lysates were separated in soluble and insoluble fraction. OPTN showed enrichment in insoluble fractions together with SOD1 G93C aggregates. (D) Co-immunoprecipitation experiment from HeLa cells transfected with wild-type GFP-SOD1 or mutant GFP-SOD1 G93C. Total cell lysates (TCL) were subjected to immunoprecipitation using anti-OPTN antibody and then probed with anti-GFP antibody. (E) GST pull-down assay of purified wild-type MBP-OPTN using GST, wild-type GST-SOD1 or G93C mutant. Wild-type MBP-OPTN did not interact with either wild-type GST-SOD1 or G93C mutant. (F) GST pull-down assay of purified wild-type His-SOD1 or G93C mutant using GST and wild-type GST-OPTN. Both wild-type His-SOD1 and G93C mutant did not interact with wild-type GST-OPTN. (G) Aggregated His-SOD1 G93C mutant was incubated with GST or GST-OPTN at 37°C for 1 hour. Soluble and insoluble fraction were separated and spotted onto nitrocellulose membrane. Membranes were blotted with either anti-His or anti-GST antibody.
The C-terminal coiled-coil domain of OPTN is responsible for binding to protein aggregates in a ubiquitin-independent manner
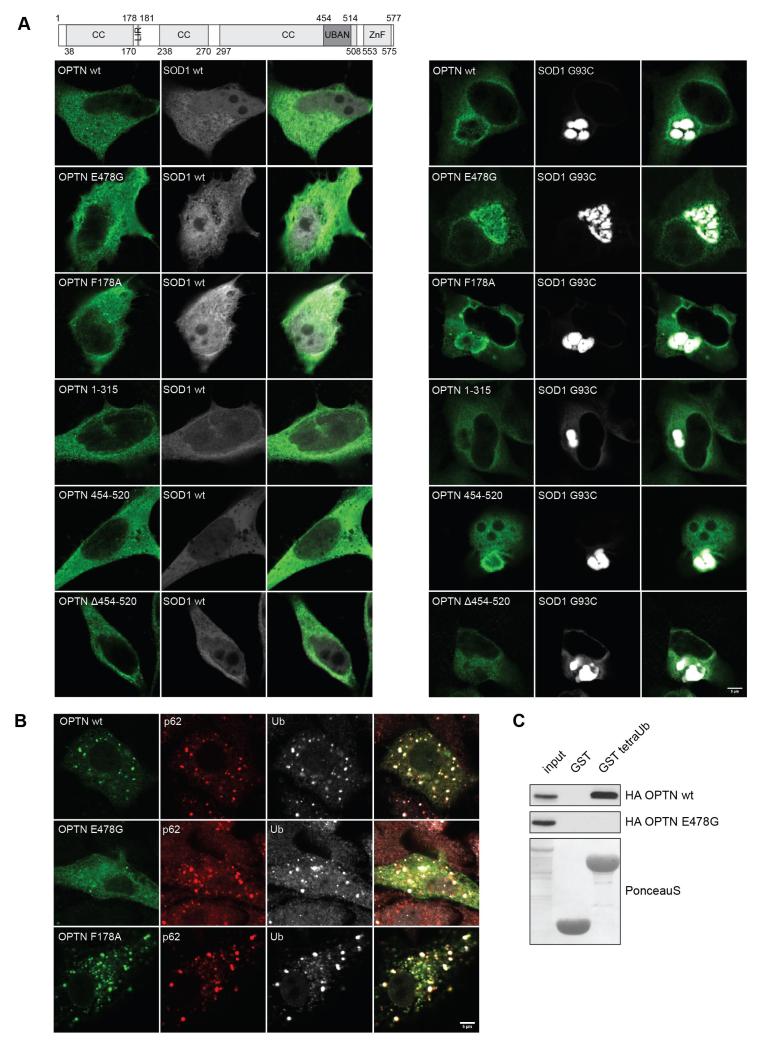
Next, we mapped the domain of OPTN responsible for the interaction with aggregation-prone proteins. We co-expressed various fragments of OPTN with GFP-SOD1 G93C in HeLa cells. The N-terminal fragment of OPTN (1-315) did not co-localize with GFP-SOD1 G93C aggregates (Fig. 2A, right panel), while the C-terminal fragment (297-577) co-localized with the inclusions (data not shown). Further fragmentation of the C-terminal part of OPTN showed that the isolated coiled-coil domain (amino acids 454-520) interacted with GFP-SOD1 G93C protein aggregates while a deletion of the same domain abolished this interaction (Fig. 2A, right panel), suggesting that the C-terminal coiled-coil domain is critical for OPTN binding to GFP-SOD1 G93C protein inclusions. Co-expression of the aforementioned fragments of OPTN with wild-type SOD1 demonstrated that OPTN did not form aggregates in the absence of mutant SOD1 G93C (Fig. 2A, left panel).
Fig. 2. The C-terminal coiled-coil domain of OPTN is responsible for binding to proteins aggregates in a ubiquitin-independent manner.
(A) Schematic representation of OPTN domain structure. Numbers indicate the amino acids. Transient co-transfection with the indicated fragments of HA-OPTN and wild-type GFP-SOD1 (left panel) or GFP-SOD1 G93C (right panel) in HeLa cells was performed. Wild-type HA-OPTN, HA-OPTN F178A, E478G and 454-520 localized to GFP-SOD1 G93C aggregates. Expression of neither HA-OPTN 1-315 nor Δ454-520 led to co-localization with aggregates. Scale bar, 5 μm. (B) HeLa cells transfected with wild-type GFP-OPTN, GFP-OPTN F178A and GFP-OPTN E478G were treated 24 hours post transfection with puromycin for 2 hours, fixed and stained with anti-ubiqutin and anti-p62 antibodies. Wild-type GFP-OPTN and GFP-OPTN F178A showed localization with puromycin-induced aggregates while GFP-OPTN E478G showed lack of co-localization. Scale bar, 5 μm. (C) GST pull-down assay of wild-type HA-OPTN and HA-OPTN E478G using GST and GST-tetraUb. Wild-type HA-OPTN interacted with tetraUb while HA-OPTN E478G showed lack of interaction with tetraUb.
OPTN contains the UBAN domain, an ubiquitin binding domain present in various proteins such as NEMO and ABIN-3 (Wagner et al., 2008; Husnjak and Dikic, 2012). It was previously shown that the E478G mutation in UBAN domain of OPTN abolished OPTN binding to ubiquitin (Wild et al., 2011). We observed that OPTN E478G mutant was still able to co-localize with GFP-SOD1 G93C protein aggregates, suggesting that OPTN interacts with protein aggregates in a ubiquitin-independent manner. Moreover, the LC3-interacting motif (LIR) mutant, OPTN F178A, unable to bind LC3/GABARAP modifiers, had no influence on OPTN localization to GFP-SOD1 G93C protein aggregates. We obtained similar results when we expressed the aforementioned OPTN fragments in stably transfected CFP-htt ex1 Q103 HeLa cells (data not shown), confirming that OPTN recognizes protein aggregates through its C-terminal coiled-coil domain in a ubiquitin-independent manner.
We also investigated co-localization of OPTN with puromycin-induced ubiquitin and p62 positive aggregates (Pankiv et al., 2007) by monitoring the localization of wild-type HA-OPTN, HA-OPTN E478G and HA-OPTN F178A in HeLa cells treated with puromycin for 2 hours (Fig. 2B). We observed that wild-type HA-OPTN and HA-OPTN F178A co-localized with protein inclusions, but HA-OPTN E478G co-localization could not be detected. The results obtained suggest that OPTN interacts with puromycin induced ubiquitin and p62 positive aggregates through the UBAN domain in a ubiquitin-dependent manner, similarly to its interaction with Salmonella enterica (Wild et al., 2011). The GST pull-down binding assay confirmed that wild-type HA-OPTN binds ubiquitin chains, while the OPTN E478G mutation abolished this binding (Fig. 2C), as previously described (Wild et al., 2011).
OPTN depletion increases protein aggregation
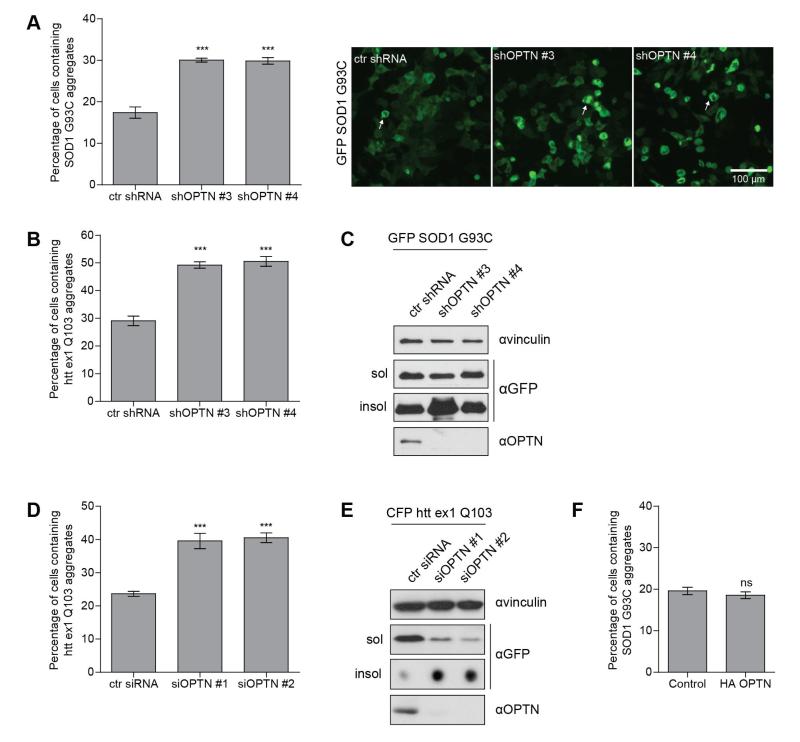
Following the confirmation of an indirect interaction between OPTN and protein aggregates, we investigated the functional relevance of OPTN in protein aggregation. For this purpose, we established HeLa cell lines stably expressing a control non-targeting shRNA (ctrl shRNA) or shRNAs directed against OPTN (shOPTN #3 and shOPTN #4). We transiently transfected OPTN knock-down and shRNA control cells with GFP-SOD1 G93C or GFP-htt ex1 Q103 for 48 hours and quantified the aggregates of GFP-SOD1 G93C and GFP-htt ex1 Q103 (representative pictures for GFP-SOD1 G93C shown in Fig. 3A). OPTN depletion induced a significant increase of GFP-SOD1 G93C or htt ex1 Q103 aggregates (Fig. 3A, B). Aggregation observed by immunofluorescence was further confirmed by investigation of the soluble and insoluble fractions. Immunoblot analysis showed an increase of insoluble GFP-SOD1 G93C in HeLa cells depleted of OPTN (Fig. 3C). Similarly, transient depletion of OPTN using two different siRNA targeting OPTN (siOPTN #1 and siOPTN #2) for 48 hours in HeLa cells stably expressing CFP-htt ex1 Q103 led to an increased number of cells containing aggregates as well as insoluble CFP-htt ex1 Q103 compared to control cells expressing OPTN (Fig. 3D, E). Inversely, overexpression of HA-OPTN had no significant effect on the number of GFP-SOD1 G93C (Fig. 3F) or GFP-htt ex1 Q103 inclusions (data not shown). The results obtained indicate that the lack of OPTN increases the accumulation of GFP-SOD1 G93C and GFP-htt ex1 Q103 protein aggregates.
Fig. 3. OPTN depletion increases protein aggregation.
(A) Quantification of transfected cells containing GFP-SOD1 G93A aggregates in ctrl shRNA and two different shRNA OPTN-depleted HeLa cells transiently transfected with GFP-SOD1 G93C for 48 hours, *** P<0.001. Representative images of each group are shown. Arrows show typical transfected cells containing GFP-SOD1 G93C aggregates. Scale bar, 100 μm. (B) Quantification of transfected cells containing GFP-htt ex1 Q103 aggregates in ctrl shRNA and two different shRNA OPTN-depleted HeLa cells, *** P<0.001. (C) Immunoblot analysis of cellular fractions using antibodies against vinculin, GFP and OPTN. HeLa cells transfected with GFP-SOD1 G93C in ctrl shRNA and two different shRNA OPTN-depleted HeLa cells were lysed in NP40 lysis buffer 48 hours post transfection and lysates were separated in soluble and insoluble fractions. The quantity of GFP-SOD1 G93C increased in the insoluble fraction in shRNA OPTN-depleted HeLa cells. (D) Quantification of aggregates in HeLa cells stably expressing CFP-htt ex1 Q103 transiently transfected with ctrl siRNA or two different siRNAs targeting OPTN for 48 hours, *** P<0.001. (E) Immunoblot analysis of cellular fractions using antibodies against vinculin, GFP and OPTN. HeLa cells stably expressing CFP-htt ex1 Q103 and transfected with ctrl siRNA or two different siRNAs targeting OPTN were lysed in NP40 lysis buffer 48 hours post transfection and lysates were separated in soluble and insoluble fractions. The soluble fraction was analyzed by SDS-PAGE gels and insoluble fraction was applied on a dot-blot apparatus. The amount of CFP-htt ex1 Q103 in the insoluble fraction increased in HeLa cells transfected with siRNA targeting OPTN. (F) Quantification of transfected cells containing GFP-SOD1 G93C aggregates in HeLa cells transiently co-transfected with the empty vector (control) or HA-OPTN for 48 hours, ns – not significant. All data are shown as mean values ± s.e.m.
OPTN modulates protein aggregation via autophagy
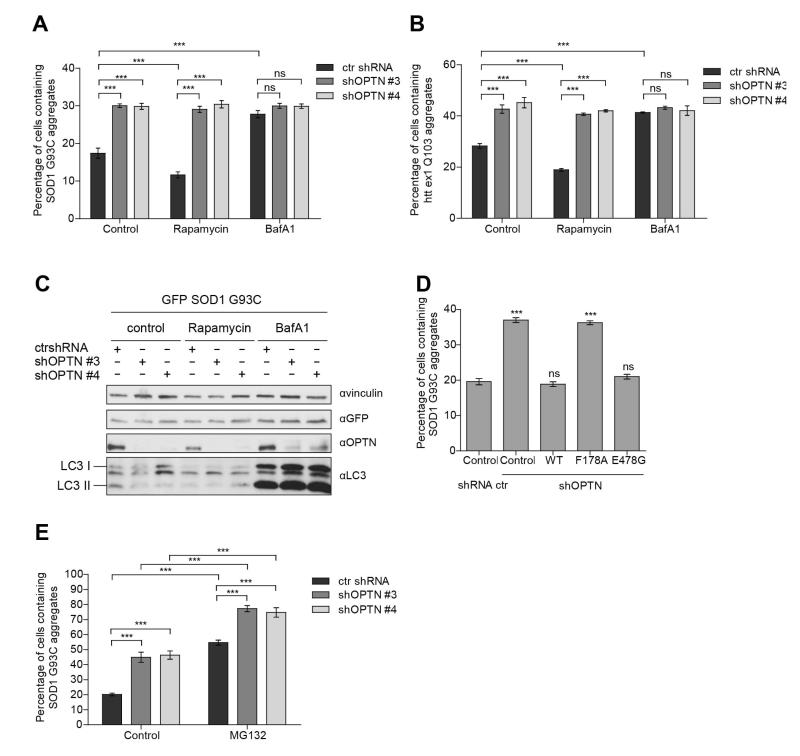
In light of the recently established role of OPTN in selective autophagy of ubiquitin-coated Salmonella enterica (Wild et al., 2011) we investigated the role of OPTN in the autophagy clearance of protein aggregates. Stable ctrl shRNA and shOPTN HeLa cells were transfected with GFP-SOD1 G93C or GFP-htt ex1 Q103 and treated with DMSO (control), rapamycin (autophagy activator) and bafilomycin A1 (autophagy inhibitor). A significant decrease in GFP-SOD1 G93C and GFP-htt ex1 Q103 aggregates in ctrl shRNA HeLa cells treated with rapamycin was observed (Fig. 4A, B). In contrast, treatment with bafilomycin A1 caused a 2-fold increase of the number of GFP-SOD1 G93C and GFP-htt ex1 Q103 inclusions (Fig. 4A, B), which was comparable with the augmentation of protein aggregates induced by OPTN depletion in shOPTN HeLa cells. In OPTN-depleted HeLa cells rapamycin and bafilomycin A1 had no effect on the quantity of cells with GFP-SOD1 G93C and GFP-htt ex1 Q103 protein inclusions. Autophagy activation or inhibition was confirmed by assessing the levels of LC3-I/LC3-II in total cell lysates (Fig. 4C). In order to demonstrate a specific role of OPTN in the aggregation phenotype observed in cellular models we performed a reconstitution of OPTN-depleted shOPTN HeLa cells with wild-type OPTN, OPTN F178A (LC3-binding-deficient OPTN) and OPTN E478G (ubiquitin-binding-deficient OPTN). Both wild-type and OPTN E478G resulted in a decrease in aggregation of GFP-SOD1 G93C to levels similar to ctrl shRNA HeLa cells (Fig. 4D). However, OPTN F178A was not able to rescue the levels of GFP-SOD1 G93C (Fig. 4D). The results observed indicate that OPTN regulates GFP-SOD1 G93C and GFP-htt ex1 Q103 aggregation through autophagy via binding to LC3.
Fig. 4. OPTN modulates protein aggregation via autophagy.
(A, B) Quantification of transfected cells containing GFP-SOD1 G93C (A) or GFP-htt ex1 Q103 (B) aggregates in HeLa cells transiently transfected with control shRNA or two different shRNA silencing OPTN (shOPTN) and treated with DMSO (control), 2 μM rapamycin for 48 hours or 200 nM bafilomycin A1 (BafA1) for 24 hours, *** P<0.001, ns – not significant. (C) Immunoblot analysis using antibodies against vinculin, GFP, OPTN and LC3. Cells were lysed in RIPA buffer and analyzed on SDS PAGE gel. LC3 levels confirmed autophagy activation by rapamycin and autophagy inhibition by bafilomycin A1. (D) Quantification of transfected cells containing GFP-SOD1 G93C aggregates in control shRNA or two different shOPTN HeLa cells and treated with DMSO (control) for 48 hours or 10 μM MG132 for 24 hours, *** P<0.001, ns – not significant. (E) Quantification of transfected cells containing GFP-SOD1 G93C aggregates in OPTN-depleted HeLa cells co-transfected with GFP-SOD1 G93C and the empty vector or various shRNA-resistant OPTN constructs – wild-type OPTN, OPTN F178A and OPTN E478G. Wild-type OPTN and OPTN E478G mutant rescued OPTN-depleted HeLa cells while OPTN F178A failed to rescue it. *** P<0.001, ns – not significant. All data are shown as mean values ± s.e.m.
Next, we treated ctrl shRNA and shOPTN HeLa cells transfected with GFP-SOD1 G93C and GFP-htt ex1 Q103 with the proteasomal inhibitor MG132 for 24 hours (Fig. 4E). Inhibition of proteasomal degradation significantly increased the number of cells with protein aggregates in ctrl shRNA HeLa cells but also in OPTN depleted HeLa cells, indicating that the effect of OPTN on protein aggregation is independent of ubiquitin-proteasome system.
TBK1 phosphorylates OPTN on protein aggregates
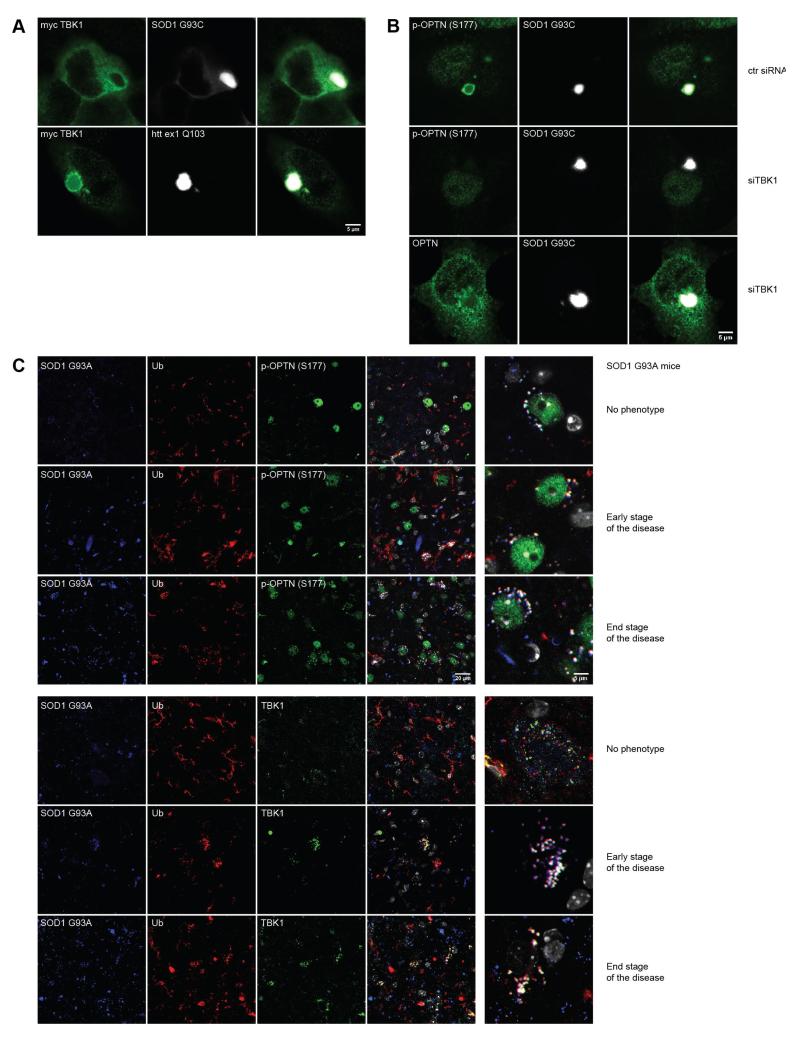
As the targeting of protein aggregates to the autophagy-lysosome pathway requires the LIR motif of OPTN, we investigated whether TBK1-mediated phosphorylation of Ser-177, adjacent to the LIR motif, is involved in the regulation of the interaction of OPTN with protein inclusions (Wild et al., 2011). We showed that TBK1 co-localized with protein aggregates in HeLa cells transiently co-transfected with myc-TBK1 and GFP-SOD1 G93C or GFP-htt ex1 Q103 (Fig. 5A). OPTN phosphorylated at the Ser-177 (phospho S177 OPTN) co-localized with GFP-SOD1 G93C inclusions. As no phospho S177 OPTN was detected on GFP-SOD1 G93C aggregates in HeLa cells co-transfected with TBK1-targeted siRNA, we hypothesized that phosphorylation of OPTN on protein aggregates is mediated by TBK1 (Fig. 5B).
Fig. 5. TBK1 phosphorylates OPTN on protein aggregates.
(A) Co-transfection of myc-TBK1 and GFP-SOD1 G93C or GFP-htt ex1 Q103 for 48 hours in HeLa cells showed localization of myc-TBK1 on GFP-SOD1 G93C and GFP-htt ex1 Q103 aggregates. Scale bar, 5 μm. (B) Phospho S177 OPTN [p-OPTN (S177)] localized to GFP-SOD1 G93C aggregates in HeLa cells co-transfected with control siRNA while co-transfection with siTBK1 abolished phospho S177 OPTN co-localization with GFP-SOD1 G93C inclusions. Endogenous OPTN localized to GFP-SOD1 G93C aggregates in HeLa cells co-transfected with siRNA targeting TBK1. Scale bar, 5 μm. (C) Immunohistochemical staining of the spinal cord of 5 month (no phenotype, asymptomatic), 7 month (early stage of the disease) and 10-month-old (end stage of the disease) transgenic mouse expressing human SOD1 G93A showed that SOD1 G93A proteins aggregates were immunopositive for TBK1 and phospho S177 OPTN in motor neurons. Gray – DAPI staining, scale bars, 20 μm and 5 μm.
Additionally, we analyzed spinal cord sections of transgenic mice expressing human mutant SOD1 G93A (Gurney et al., 1994) at three different stages of the pathology. We examined 5 month-old (no phenotype), 7 month-old (early-stage of the disease) and 10 month-old (end-stage of the disease) human mutant SOD1 G93A transgenic mice. We confirmed the co-localization of phospho S177 OPTN and TBK1 with SOD1 G93A protein aggregates (Fig. 5C) at all phenotype stages investigated even in asymptomatic 5 month-old SOD1 G93A mice where only sparse SOD1 aggregates were observed (Gurney et al., 1994). Occasionally, ubiquitinated SOD1 aggregates were not co-stained with phospho S177 OPTN or TBK1 (Fig. 5C). These results demonstrate a co-localization of TBK1 and phospho S177 OPTN with SOD1 G93A aggregates in this mouse model of ALS, suggesting that OPTN targeting of SOD1 G93A aggregates to the autophagy-lysosome pathway is regulated by phosphorylation of OPTN by TBK1.
TBK1 plays a role in autophagic degradation of protein aggregates
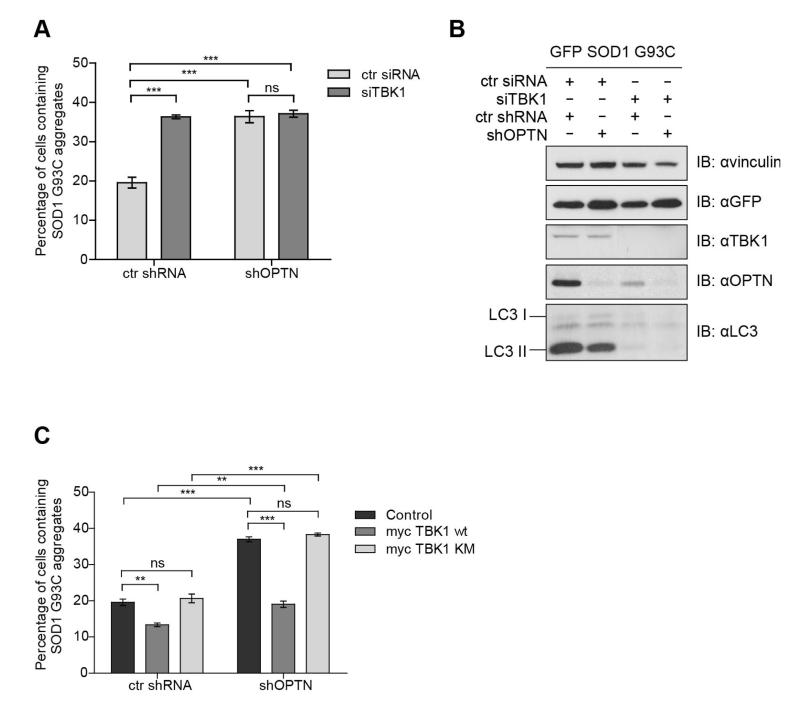
We further investigated the role of TBK1 in autophagic degradation of protein aggregates. Depletion of TBK1 in ctrl shRNA HeLa cells induced an increase in GFP-SOD1 G93C aggregation similar to the increase observed after silencing of OPTN. In contrast, TBK1 silencing had no additional effect in shOPTN HeLa cells (Fig. 6A). These results indicate a common pathway of modulation of protein aggregation shared between TBK1 and OPTN. The silencing of TBK1 induced decreased levels of the lipidated form of LC3 (LC3-II), representing a decreased incorporation of LC3 into autophagosomal membranes and a decreased autophagic activity (Fig. 6B).
Fig. 6. TBK1 plays a role in autophagy degradation of protein aggregates.
(A) Quantification of transfected cells containing GFP-SOD1 G93C aggregates transiently co-transfected with control siRNA or siRNA targeting TBK1 in control shRNA or OPTN-depleted HeLa cells, *** P<0.001, ns – not significant. (B) Immunoblot analysis using antibodies against vinculin, GFP, TBK1, OPTN and LC3. Control shRNA or OPTN-depleted HeLa cells co-transfected with GFP-SOD1 G93C and control siRNA or siRNA targeting TBK1 for 48 hours were lysed in RIPA buffer and run on SDS PAGE gel. (C) Quantification of transfected cells containing GFP-SOD1 G93C aggregates in shRNA control or OPTN-depleted HeLa cells transiently co-transfected with empty vector as control, wild-type myc-TBK1 and myc-TBK1 kinase mutant K38A (TBK1 KM). *** P<0.001, ** P<0.01, ns – not significant. All data are shown as mean values ± s.e.m.
Overexpression of myc-TBK1 in ctrl shRNA HeLa cells decreased GFP-SOD1 G93C aggregation (Fig. 6C) in a way comparable to rapamycin-treated cells (Fig. 4A). Interestingly, the overexpression of myc-TBK1 in shOPTN HeLa cell partially decreased GFP-SOD1 G93C aggregation, indicating potential activation of autophagic degradation of GFP-SOD1 G93C by TBK1 in the absence of OPTN. In contrast, the kinase-inactive TBK1 mutant K38A had no impact on GFP-SOD1 G93C aggregation (Fig. 6C), indicating that the kinase activity of TBK1 could be responsible for its effect on protein aggregation. The results obtained suggest an important role of TBK1 phosphorylation of OPTN and potentially other proteins involved in degradation of protein aggregates via the autophagy-lysosome pathway.
OPTN depletion in zebrafish (Danio rerio) causes motor axonopathy and increases motor axonopathy induced by mutant SOD1 overexpression
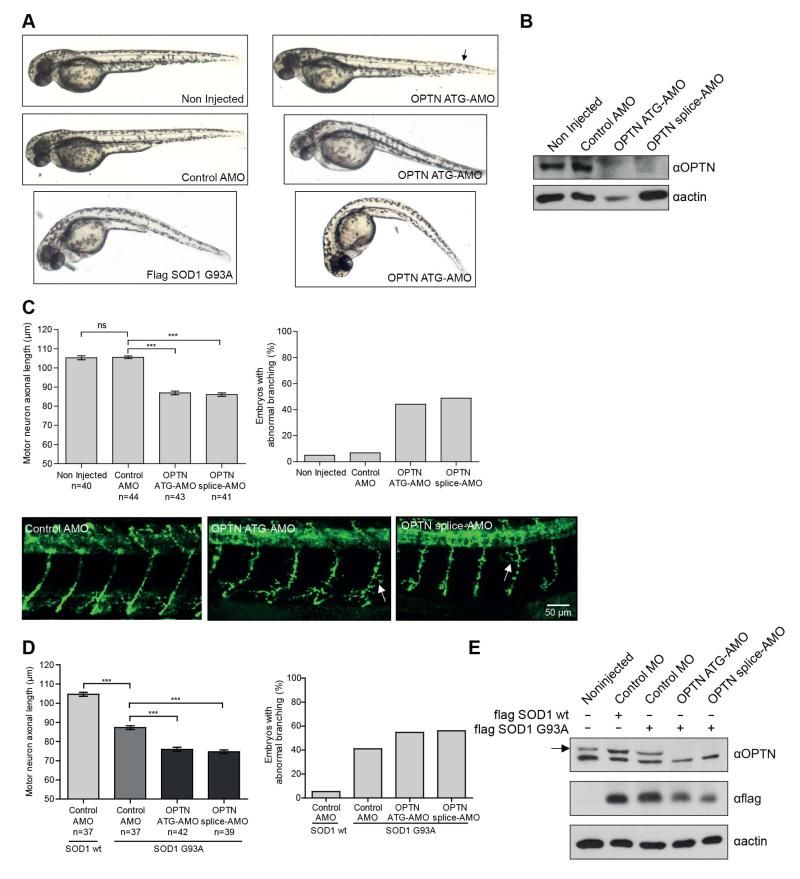
In order to investigate the effects of OPTN depletion on protein aggregation in vivo, we employed a well-validated zebrafish model of ALS overexpressing the SOD1 G93A mutant characterized by the development of a specific and dose-dependent axonopathy (Lemmens et al., 2007; Laird et al., 2010; Xi et al., 2011). In order to deplete OPTN levels in zebrafish, two different antisense morpholino oligonucleotides (AMO) targeting OPTN were used and the zebrafish were analysed 48 hours post-fertilization. Both AMOs induced an efficient silencing of OPTN and depletion of OPTN in wild-type zebrafish resulted in a specific phenotype characterized by embryos with curved tails (Fig. 7A, B) and delayed and/or no ability to swim away after touch (data not shown). This phenotype was also observed in zebrafish overexpressing SOD1 G93A without silencing of OPTN (Fig. 7A). We also observed shorter and abnormally branched motor axons measured by labeling embryos with the synaptic vesicle marker, SV2 (Lemmens et al., 2007). Knockdown of OPTN led to a decrease in motor axon length and an increase in abnormal motor axon branching (Fig. 7C). These data indicate that OPTN depletion causes motor axonopathy in zebrafish embryos. Similarly, co-injection of OPTN AMO and mRNA coding for flag-SOD1 G93A produced a more pronounced decrease in motor axon length in zebrafish embryos as well as an increase in abnormal motor axon branching (Fig. 7D, E).
Fig. 7. OPTN depletion in zebrafish causes motor axonopathy and increases motor axonopathy induced by mutant SOD1 overexpression.
(A) Zebrafish injected with OPTN specific translation blocking (OPTN ATG-AMO) and splice blocking (OPTN splice-AMO) morpholino showed phenotype similar to the one observed in zebrafish overexpressing SOD1 G93A 48h post fertilization in comparison to non-injected zebrafish or injected with control morpholino (control AMO). (B) Immunoblot analysis of protein extracted from zebrafish using antibodies against OPTN and actin showed efficiency of OPTN silencing. (C) Quantification of motor axon length (μm) and branching (%) in zebrafish injected with two different OPTN AMO revealed shorter motor neuron axons and increase premature branching in comparison to non-injected zebrafish or zebrafish injected with control AMO. Abnormal branching is expressed as percentage of embryos showing premature branching in two or more axons, *** P<0.001, ns – not significant. Representative images of motor neuron axons 48 hours post fertilization from each group are shown. Motor neurons are labeled with SV2 antibody. Arrow indicates premature branched axon. Scale bar, 50 μm. (D) Quantification of motor axon length (μm) and branching (%) in zebrafish with overexpressed flag-SOD1 G93A and control AMO showed a decrease in motor neurons length and increase in premature branching when compared to zebrafish with overexpressed wild-type flag-SOD1 and control AMO. Zebrafish with overexpressed flag-SOD1 G93A and OPTN AMO showed additional decrease in motor neurons length and additional increase in premature branching, *** P<0.001, ns – not significant. (E) Immunoblot analysis of protein extracts from zebrafish with RIPA buffer using antibodies against flag, OPTN and actin shows efficiency of OPTN silencing and flag-SOD1 expression. All data are shown as mean values ± s.e.m.
DISCUSSION
OPTN has been reported to co-localize with protein inclusions in a wide range of neurodegenerative diseases such as ALS (Maruyama et al., 2010), HD (Schwab et al., 2012), Alzheimer’s disease, Parkinson’s disease, Creutzfeldt-Jakob disease and Pick disease (Osawa et al., 2011). Moreover, recent studies described OPTN deletion mutations in ALS patients (Maruyama et al., 2010; Del Bo et al., 2011; Sugihara et al., 2011; Iida et al., 2012; Tumer et al., 2012; van Blitterswijk et al., 2012). Despite the existing observations, little is known about the potential function of OPTN in neurodegenerative diseases. Our data suggest an important role for OPTN in protein aggregation, accumulation and degradation. We also provide insights into the regulation of OPTN function in autophagic degradation of protein aggregates involved in the pathogenesis of ALS and HD.
In this work, we report on the co-localization of OPTN, SOD1 and htt protein aggregates as previously described for other proteins involved in autophagy such as ubiquitin, p62 and LC3 (Fig. 1) (Filimonenko et al., 2010). We demonstrate that the interaction of OPTN with aggregation-prone proteins is indirect and occurred already with soluble monomeric or oligomeric forms of aggregating proteins (Fig. 1). The identity of the protein that functions as an adaptor between OPTN and aggregating protein remains unknown. A potential candidate is ubiquitin. Indeed, OPTN is an autophagy cargo receptor with the ability to bind LC3 through an LC3-interacting motif (LIR) and ubiquitin through ubiquitin-binding domain (UBAN). The importance of the UBAN domain in autophagic clearance of cytosolic Salmonella enterica has been shown (Wild et al., 2011). Here we show that OPTN interacts with p62 bodies through its UBAN domain recognizing polyubiquitin chains. However, OPTN interaction with SOD1 and htt aggregates is ubiquitin-independent. The mutation in the UBAN domain (E478G) that completely abolishes OPTN binding to polyubiquitin chains (Wild et al., 2011) had no impact on OPTN interaction with SOD1 aggregates (Fig. 2). It is possible that ubiquitin is not the signal/motif of protein aggregates recognized by OPTN to be recruited to the autophagy machinery, as ubiquitination might not be a universal marker of protein aggregates. Indeed, ubiquitin-negative inclusions were found in ALS patients (Allen et al., 2002; Osawa et al., 2011). It was also shown that p62 interaction with SOD1 protein aggregates (Gal et al., 2009) as well as with a non-ubiquitinated substrate STAT5A_ΔE18, an aggregation-prone isoform of STAT5A (Watanabe and Tanaka, 2011), could be independent of ubiquitin signals. Moreover, as recently shown co-chaperone BAG3 mediated aggresome targeting and selective autophagy of SOD1 aggregates do not depend on substrate ubiquitination (Gamerdinger et al., 2011). OPTN interacts with SOD1 and htt aggregates via its C-terminal coiled-coil domain (454-520 amino acids). However, future studies will have to determine which specific residues of OPTN are involved in its interaction with protein inclusions and which protein is recognized by OPTN on protein inclusions.
Furthermore, OPTN depletion in HeLa cells resulted in a significant increase in aggregation of SOD1 G93C or htt ex1 Q103, suggesting a role of OPTN in the degradation of aggregation-prone proteins (Fig. 3). OPTN was modulating SOD1 G93C or htt ex1 Q103 degradation by the autophagy-lysosome system. This effect of OPTN was regulated by phosphorylation of Ser-177 by TBK1, necessary for OPTN binding to LC3, following recruitment of the autophagy machinery during the degradation of Salmonella enterica (Wild et al., 2011). Even though non-phosphorylated OPTN was detected in protein inclusions, phosphorylation of OPTN by TBK1 is a rate-limiting step in autophagic degradation of protein aggregates. Therefore, we hypothesize that OPTN is first recruited to the protein aggregates following recognition and phosphorylation by TBK1, allowing the subsequent recruitment of LC3 and the autophagy machinery.
Interestingly, we also showed that even in the absence of OPTN, overexpression of TBK1 could still decrease the number of cells containing SOD1 G93C aggregates (Fig. 6C). This observation suggests that in addition to OPTN, TBK1 potentially could phosphorylate other proteins involved in the degradation of protein aggregates. Potential candidates might be other previously identified autophagy receptors such as p62, Alfy and NBR1. As TBK1 depletion caused decreased incorporation of LC3 into autophagosomal membranes and a decreased autophagic activity, potential candidates could also be components of autophagy initiation ULK1-dependent complex or members of the PI3K complex involved in vesicle nucleation (Behrends et al., 2010).
Our cellular observations were confirmed in vivo as we co-localized TBK1 and phosphorylated OPTN with SOD1 G93A protein aggregates in the spinal cord of a murine model of ALS (Gurney et al., 1994). Most of the SOD1 protein inclusions were imunoreactive for phosphorylated OPTN and TBK1 as well as for ubiquitin even at the non-symptomatic stage when the spinal cord contained almost no inclusions and the SOD1 G93A mice had not developed any phenotype. This observation concurs with our in vitro results showing OPTN interaction with soluble monomeric or oligomeric mutant SOD1 G93C. There was a slight discrepancy between ubiquitin and phospho S177 OPTN staining, as some ubiquitinated aggregates were not co-stained with phospho S177 OPTN or TBK1. Recently, it was shown that OPTN was absent or rare in ALS patients (Hortobagyi et al., 2011), which might be in agreement with our observation that some SOD1 aggregates are not recognized by phospho S177 OPTN or TBK1 in the mutant SOD1 G93A mouse. However, we also demonstrated that ubiquitin is not the signal recognized by OPTN on the protein aggregates and therefore not surprisingly some ubiquitinated SOD1 aggregates were not co-stained with phospho S177 OPTN or TBK1 in the mutant SOD1 G93A mouse. This might be an in vivo confirmation that the presence of ubiquitin is not attracting OPTN to the protein aggregates. On the other hand, it might also indicate that the phospho S177 OPTN/TBK1-negative aggregates are lacking the protein recognized by OPTN on aggregation-prone proteins. Further studies are needed to identify the adaptor protein between OPTN and protein inclusions as its presence might facilitate the degradation of aggregation-prone proteins. Although controversial results have been obtained on the presence of OPTN in protein aggregates in different neurodegenerative diseases (Deng et al., 2011; Ito et al., 2011; Schwab et al., 2012; Hortobagyi et al., 2011) the localization of phospho S177 OPTN or TBK1 in inclusions in human cases of neurodegenerative diseases still remains to be determined.
We also investigated the effect of OPTN depletion in zebrafish and showed that silencing of OPTN in wild-type zebrafish induced a phenotype similar to the one displayed by a zebrafish model of ALS overexpressing SOD1 G93A. The fact that OPTN deletion caused an ALS motor axonopathy phenotype in wild-type zebrafish is further supported by the description of OPTN deletion mutations in ALS patients (Maruyama et al., 2010; Del Bo et al., 2011; Sugihara et al., 2011; Iida et al., 2012; Tumer et al., 2012; van Blitterswijk et al., 2012). Furthermore, OPTN depletion in zebrafish expressing flag-SOD1 G93A provoked an even more severe motor axonopathy phenotype. This observation of an additive effect of OPTN deletion and SOD1 G93A overexpression might suggest that the pathological mechanisms involved in OPTN and SOD1 G93A are complementary. However, as the severity of the motor axonopathy phenotype in ALS zebrafish model overexpressing SOD1 G93A is dose dependent (Lemmens et al., 2007), OPTN depletion might cause a lack of autophagic clearance and a subsequent accumulation of SOD1 G93A aggregates. We have showed that OPTN is an important player in autophagy clearance of misfolded and aggregation-prone proteins and taking this into consideration, neurodegenerative pathology caused by mutation or complete deletion of OPTN in zebrafish or in human ALS cases can be explained as a dysfunctional autophagy clearance of misfolded and aggregation-prone proteins. However, in ALS patients without OPTN mutation OPTN is recruited to the protein aggregates as an attempt to degrade the aggregation-prone proteins but an increase stimulation of this potentially protective pathway might be needed to avoid the accumulation of proteins in the nervous system.
In conclusion, we provided evidence that OPTN regulates protein aggregation through autophagy pathway by recognizing protein aggregates via the C-terminal coiled-coil domain and by recruiting LC3 to protein aggregates via its LIR motif. Furthermore, TBK1 regulates protein aggregation by phosphorylating OPTN and also various other proteins involved in the regulation of autophagy. These findings support a potential protective role of OPTN and TBK1 in neurodegenerative disorders associated with intracellular protein aggregation.
MATERIALS AND METHODS
Plasmids, reagents and cell lines
pcDNA3.1-HA-OPTN (wild-type, F178A and E478G), pMAL-c2x-OPTN, pGEX4T1-OPTN, pGEX4T1-tetraUb and pcDNA3.1-myc-TBK1 (wild-type, K38A) plasmids were described previously (Ikeda et al., 2007; Wild et al., 2011). Homodimer GFP-SOD1 G93C was subcloned in pcDNA3.1 vector from vector pEGFP-N1-SOD1 G93C/G93C previously described (Witan et al., 2008). pcDNA3-GFP-htt ex1 Q103 was a kind gift from D. Krainc (Institute for Neurodegenerative Disease, Harvard Medical School, USA). Plasmids encoding flag-SOD1 (wild-type and G93A) were kindly provided by R. Takahashi (Department of Neurology, Kyoto University Hospital and Graduate School of Medicine, Japan). HA-OPTN 1-315 and HA-OPTN 454-520 were subcloned into BamHI-XhoI sites of the pcDNA3.1 vector. HA-OPTN Δ454-520 was generated by in vitro mutagenesis (Stratagene). Plasmids encoding wild-type His-SOD1 and G93C mutant or GST-SOD1 and G93C mutant were subcloned into BamH1 site of the pRSET vector or pGEX4T1, respectively. All constructs were verified by sequencing before use. GeneJuice® (Merck) and Lipofectamine™ RNAiMAX were used for mammalian cells transfection. GFP-Trap® was purchased from ChromoTek, puromycin, MG132 and rapamycin from Sigma-Aldrich, bafilomycin A1 from LC Laboratories, Protein G Agarose from Roche, Glutathione-Sepharose 4B from GE Healthcare, Amylose resin from New England Biolabs, Ni NTA Agarose from Qiagen.
Cell lines and transfection of siRNA and plasmids
HeLa cells were obtained from ATCC. Stable HeLa cells expressing CFP-htt ex1 Q103 were kindly provided by A. Simonsen (Institute of Basic Medical Sciences, University of Oslo, Norway) (Filimonenko et al., 2010). HeLa cell lines stably expressing a control non-targeting shRNA (ctrl shRNA) or shRNAs directed against OPTN (shOPTN #3 and shOPTN #4) were previously described (Wild et al., 2011). All cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin/streptomycin at 37°C, 5% CO2. For siRNA experiments, HeLa cells were transfected with siRNA using Lipofectamine™ RNAiMAX in reverse transfection according to the manufacturer’s protocol. After 24 hours cells were transfected with plasmids using GeneJuice and all analyses were performed 48 hours later. Sequences of siRNA used in this study: Non-Targeting siCONTROL #2 from Thermo Fischer, siOPTN #1 5′-GCACGGCAUUGUCUAAAUA-3′, siOPTN #2 5′-GAAGUUUACUGUUCUGAUU-3′, siTBK1 5′-GACAGAAGUUGUGAUCACA-3′. A final concentration of 10 (ctrl siRNA, siTBK1, siOPTN #2) or 20 nM (siOPTN #1) of siRNAs was used for silencing.
Antibodies
The following antibodies were used in the study: rabbit polyclonal anti-OPTN (Abcam, ab23666), mouse monoclonal anti-GFP (Roche), mouse monoclonal anti-ubiquitin (FK2, Enzo Life Science), mouse monoclonal anti-FLAG M2 (Sigma-Aldrich), mouse monoclonal anti-actin (Sigma-Aldrich), mouse monoclonal anti-vinculin (Sigma-Aldrich), mouse monoclonal anti-myc (Santa Cruz), rabbit polyclonal anti-LC3 (MBL, PM036), rabbit polyclonal anti-LC3B (a kind gift from Z. Elazar, the Weizmann Institute of Science, Israel), mouse monoclonal anti-synaptic vesicle 2 (DSHB), rabbit polyclonal anti-TBK1 (Cell Signaling), rabbit polyclonal anti-p62 (Enzo Life Sciences), mouse monoclonal anti-HA (Covance), mouse monoclonal anti-MBP (Sigma), mouse monoclonal anti-GST (Santa Cruz), mouse monoclonal anti-His (Novagen). Rabbit polyclonal anti-SOD1 (Enzo) was conjugated to DyLight 650 using the DyLight antibody labeling kit (Thermo scientific). The phospho S177 antibody was generated by immunoGLobe® as previously described (Wild et al., 2011). Secondary horseradish peroxidase (HRP)-conjugated goat anti-mouse and goat anti-rabbit IgGs antibodies were purchased from Santa Cruz. Cy3, Cy5 and Alexa 488 conjugated secondary antibodies were purchased from Jackson Immuno Reasearch.
Immunoprecipitation, binding assay and dot blot assay
HeLa cells were transfected with the indicated plasmids using GeneJuice according to the manufacturer’s instructions. After 24 (Fig. 1D) or 48 hours (Fig. 1B, 2C), cells were lysed in lysis buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 0.5% NP40, pH 7.4) supplemented with Complete protease inhibitor cocktail (Roche). Lysates were incubated with GST fusion protein bound beads for 4 hours in binding assay or were incubated either with 3 μl of anti-GFP-coupled beads for 2 hours or 5 μl anti-OPTN antibody for 4 hours followed by 1 h incubation with Protein G agarose in immunoprecipitation assay. The washed beads were subjected to SDS-PAGE gels for immunoblot analysis. GST, MBP or His fusion proteins were expressed in E. coli BL21 cells in LB medium and purified using glutathione sepharose, amylose resin or Ni-NTA agarose, respectively. MBP-tagged proteins were eluted from the amylose resin by 10 mM maltose, GST-tagged proteins with 20 mM reduced glutathione and His-tagged proteins with 200 mM imidazole. Supernatant containing eluted MBP or His-tagged proteins were used for subsequent GST pull-down assays with GST-proteins bound to beads. GST pull-down assays for recombinant protein-protein interactions were performed in 1× incubation buffer (20 mM Tris-HCl, pH8.4; 150 mM NaCl; 2.5 mM CaCl2; 10% glycerol, 1% Triton X-100, 1 mM DTT) for 4 hours at 4°C. Beads were washed 3 times with incubation buffer and subjected to SDS-PAGE gels for immunoblot analysis.
Dot blot assay was performed as previously described (Yerbury et al., 2012). Purified His-SOD1 G93C mutant (20 μM) was incubated in 10 mM potassium phosphate buffer, pH 7.4 containing 5 mM EDTA and 10 mM DTT at 37°C for 20 hours for inducing in vitro aggregation. Aggregated His-SOD1 G93C (20 μM) was incubated in absence or presence of GST-OPTN (20 μM) for 1 hour at 37°C. All samples were centrifuged for 30 min at 4°C, soluble fraction were separated and pellet containing insoluble fraction was washed three times with potassium phosphate buffer. Soluble and insoluble fractions were spotted onto a nitrocellulose membrane and membrane was blotted with either His or GST antibody.
Immunofluorescence
HeLa cells were grown on glass coverslips, transfected with indicated plasmids and 48 hours post transfection were fixed with 2% paraformaldehyde (PFA) for 20 min. Subsequently, cells were permeabilized with 0.2% Triton X-100 solution in PBS at room temperature for 10 min. All cells were blocked in PBS containing 5% BSA at 4°C overnight. Primary and secondary antibodies were diluted in the blocking solution and incubated for 1h at room temperature. Washes were performed in 0.1% Tween 20 in PBS. The coverslips were mounted with aqueous mounting medium (Mowiol) containing DAPI (Molecular Probes). Images were acquired by the LSM 510 META laser-scanning microscope (Zeiss) and processed using ImageJ software.
Transgenic Mice and tissue preparation
Transgenic C57/B6 mice expressing human mutant SOD1 G93A (Gurney et al., 1994) were kept in the local animal facility under a 12 hour light cycle and food and drinking water ad libitum. Mice were housed, anesthetized and sacrificed according to the European and German guidelines for the care and use of laboratory animals. The animals were sacrificed at 5 month-old (no phenotype), 7 month-old (early-stage of the disease) and 10 month-old (end-stage of the disease i.e. when the mice were not able to right themselves up after 10 sec when put to the side) human mutant SOD1 G93A transgenic mice (Gurney et al., 1994). Animals were perfused transcardially with 4% PFA. The lumbar spinal cord was removed and post-fixed overnight. The tissue was then incubated in 20% sucrose in phosphate buffer overnight for cryoprotection. 15 μm spinal cord sections were cut on a cryostat and immunostaining was performed on floating sections. Briefly, spinal cord sections were permeabilized with 0.5% Triton X-100 in PBS and then washed with 0.1% Triton X-100 in PBS (PBST). After a 2 hour blocking with PBST containing 3% BSA, the sections were incubated with the primary antibodies directed against ubiquitin and TBK1 or phospho S177 OPTN at 4°C for 48 hours. Sections were then washed and incubated for 2 h with the secondary antibodies. After washing, the sections were incubated overnight at 4°C with the Cy5-conjugated anti-SOD1 antibody. Finally, sections were washed, mounted and observed under the LSM 510 META laser-scanning microscope (Zeiss).
Cell fractionation and filter trap assay
Cells were lysed in NP40 lysis buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 0.5% NP40, pH 7.4 supplemented with Complete protease inhibitor cocktail). After centrifugation (10 min, 5000 g) the supernatant was collected and constituted the soluble fraction. The pellet was resuspended in NP40 buffer containing Bensonaze® (Merck) and 2% SDS which represented the insoluble fraction. The soluble and insoluble fractions were analyzed by SDS-PAGE. The insoluble fraction was applied on dot-blot apparatus. Total cell lysate was obtained by lysing cells in RIPA buffer (50 mM Tris, 150 mM NaCl, 0.5% Na-Deoxycholate, 1% SDS, 1% Triton-X 100, pH 7.4) supplemented with Complete protease inhibitor cocktail at room temperature for 15 min.
Quantification of aggregate formation and statistics
Cells were analyzed on a Leica CTR7000 HS epifluorescence microscope. A minimum of 500 transfected cells were counted for each condition. The experiments were performed in triplicate and repeated twice. An example of cells containing SOD1 G93A aggregates is shown in figure 3A. Statistical significance was determined by Student’s t-test for comparison between two samples. To compare three or more ungrouped or grouped samples were used respectively one-way ANOVA or two-way ANOVA followed by Bonferroni multiple comparison test as post-hoc analysis. All statistical analysis was performed using Graph PadPrism (Prism Software INC, USA).
Zebrafish (Danio rerio)
The zebrafish were maintained, mated, and raised according to established procedures (Westerfield, 2000). The Tuebingen wild-type strain was used for all experiments. Morpholinos used in this study were purchased from Gene Tools (Philomath, USA): Standard control morpholino 5′-CCTCTTACCTCAGTTACAATTTATA)-3′, translation blocking morpholino targeted to start codon (OPTN ATG-AMO) 5′-CGATGATCCAGATGCCATGCTTTCT-3′ and splice blocking morpholino exon2-intron2 (OPTN splice-AMO) 5′-AAATTTCTCTCACCTCAGCTCCACT-3′. In vitro transcription of wild-type flag-SOD1 and flag-SOD1 G93A was performed with the MessageMachine T7 Kit (Ambion). Morpholinos were diluted in ultrapure water with 0.05% phenol red and injected into 2-8 cell stage zebrafish embryo as previously described (Laird and Robberecht, 2011). Dose-dependent curves of AMO and mRNA toxicity were performed and 1 nL of 0.5 mM AMOs (ctrl AMO, OPTN splice-AMO) or 0.4 mM OPTN ATG-AMO and 200 pg wild-type flag-SOD1 and flag SOD1 G93A mRNA were injected to minimize morpholino or cDNA-induced developmental delay and toxicity. After injections, embryos were stored in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) at 28°C. At 48 hours post fertilization, zebrafish embryos were fixed with 4% PFA in PBS and immunostained using SV2 antibody in order to visualize the motor neurons. Bright field images were acquired using a Leica MZ165 dissecting microscope and fluorescent images were acquired using a LSM 510 META laser-scanning microscope (Zeiss). Motor neuron length was measured in ImageJ and the average measure of the first ten motor axons after the yolk sac was calculated in each embryo. Embryos were marked as embryos with abnormal branching when two or more axons per embryo showed premature branching. Zebrafish lysates were prepared in RIPA lysis buffer and analyzed by immunoblotting.
Acknowledgment
We are thankful to Ivana Novak, Doris Popovic and Dinko Relkovic for critically reading the manuscript. We would like to thank the members of the Dikic Lab for discussions and constructive comments. We thank A. Simonsen for the HeLa cells expressing CFP-htt ex1 Q25/Q103, D. Krainc for GFP-htt ex1 Q25/Q103 plasmids, R. Takahashi for flag-SOD1 constructs, Z. Elazar for LC3B antibody and J. Puizina (Faculty of Science, University of Split) for the use of her microscopy facility. This work was supported by the Cluster of Excellence “Macromolecular Complexes” of the Goethe University Frankfurt (EXC115), the LOEWE funded OSF network and Gene and Cell Therapy Center, and the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013) / ERC grant agreement n° [250241-LineUb] to ID. JK is supported by City of Split scholarship and Boehringer Ingelheim Fonds Travel Grant. BJ is supported by the Deutsche Forschungsgemeinschaft (Exzellenzcluster 147 “Cardio-Pulmonary Systems”). CB is supported by grant from the Fritz-and-Hildegard-Berg-Foundation of the Stifterverband.
REFERENCES
- Aguzzi A, O’Connor T. Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nat Rev Drug Discov. 2010;9:237–248. doi: 10.1038/nrd3050. [DOI] [PubMed] [Google Scholar]
- Albagha OM, Visconti MR, Alonso N, Langston AL, Cundy T, Dargie R, Dunlop MG, Fraser WD, Hooper MJ, Isaia G, et al. Genome-wide association study identifies variants at CSF1, OPTN and TNFRSF11A as genetic risk factors for Paget’s disease of bone. Nature genetics. 2010;42:520–524. doi: 10.1038/ng.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen B, Ingram E, Takao M, Smith MJ, Jakes R, Virdee K, Yoshida H, Holzer M, Craxton M, Emson PC, et al. Abundant tau filaments and nonapoptotic neurodegeneration in transgenic mice expressing human P301S tau protein. J Neurosci. 2002;22:9340–9351. doi: 10.1523/JNEUROSCI.22-21-09340.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anborgh PH, Godin C, Pampillo M, Dhami GK, Dale LB, Cregan SP, Truant R, Ferguson SS. Inhibition of metabotropic glutamate receptor signaling by the huntingtin-binding protein optineurin. The Journal of biological chemistry. 2005;280:34840–34848. doi: 10.1074/jbc.M504508200. [DOI] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalasani ML, Swarup G, Balasubramanian D. Optineurin and its mutants: molecules associated with some forms of glaucoma. Ophthalmic Res. 2009;42:176–184. doi: 10.1159/000232400. [DOI] [PubMed] [Google Scholar]
- Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. Journal of cell science. 2011;124:161–170. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung PY, Beyens G, Boonen S, Papapoulos S, Geusens P, Karperien M, Vanhoenacker F, Verbruggen L, Fransen E, Van Offel J, et al. The majority of the genetic risk for Paget’s disease of bone is explained by genetic variants close to the CSF1, OPTN, TM7SF4, and TNFRSF11A genes. Human genetics. 2010;128:615–626. doi: 10.1007/s00439-010-0888-2. [DOI] [PubMed] [Google Scholar]
- Del Bo R, Tiloca C, Pensato V, Corrado L, Ratti A, Ticozzi N, Corti S, Castellotti B, Mazzini L, Soraru G, et al. Novel optineurin mutations in patients with familial and sporadic amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2011;82:1239–1243. doi: 10.1136/jnnp.2011.242313. [DOI] [PubMed] [Google Scholar]
- del Toro D, Alberch J, Lazaro-Dieguez F, Martin-Ibanez R, Xifro X, Egea G, Canals JM. Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus. Mol Biol Cell. 2009;20:1478–1492. doi: 10.1091/mbc.E08-07-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Bigio EH, Zhai H, Fecto F, Ajroud K, Shi Y, Yan J, Mishra M, Ajroud-Driss S, Heller S, et al. Differential involvement of optineurin in amyotrophic lateral sclerosis with or without SOD1 mutations. Archives of neurology. 2011;68:1057–1061. doi: 10.1001/archneurol.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, Johansen T, Kirkin V. Selective autophagy in cancer development and therapy. Cancer Res. 2010;70:3431–3434. doi: 10.1158/0008-5472.CAN-09-4027. [DOI] [PubMed] [Google Scholar]
- Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Molecular cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal J, Strom AL, Kwinter DM, Kilty R, Zhang J, Shi P, Fu W, Wooten MW, Zhu H. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem. 2009;111:1062–1073. doi: 10.1111/j.1471-4159.2009.06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Kaya AM, Wolfrum U, Clement AM, Behl C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO reports. 2011;12:149–156. doi: 10.1038/embor.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudette M, Hirano M, Siddique T. Current status of SOD1 mutations in familial amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1:83–89. doi: 10.1080/14660820050515377. [DOI] [PubMed] [Google Scholar]
- Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- Hattula K, Peranen J. FIP-2, a coiled-coil protein, links Huntingtin to Rab8 and modulates cellular morphogenesis. Curr Biol. 2000;10:1603–1606. doi: 10.1016/s0960-9822(00)00864-2. [DOI] [PubMed] [Google Scholar]
- Hortobagyi T, Troakes C, Nishimura AL, Vance C, van Swieten JC, Seelaar H, King A, Al-Sarraj S, Rogelj B, Shaw CE. Optineurin inclusions occur in a minority of TDP-43 positive ALS and FTLD-TDP cases and are rarely observed in other neurodegenerative disorders. Acta neuropathologica. 2011;121:519–527. doi: 10.1007/s00401-011-0813-3. [DOI] [PubMed] [Google Scholar]
- Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annual review of biochemistry. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- Iida A, Hosono N, Sano M, Kamei T, Oshima S, Tokuda T, Nakajima M, Kubo M, Nakamura Y, Ikegawa S. Novel deletion mutations of OPTN in amyotrophic lateral sclerosis in Japanese. Neurobiol Aging. 2012;33:1843, e1819–1824. doi: 10.1016/j.neurobiolaging.2011.12.037. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Hecker CM, Rozenknop A, Nordmeier RD, Rogov V, Hofmann K, Akira S, Dotsch V, Dikic I. Involvement of the ubiquitin-like domain of TBK1/IKK-i kinases in regulation of IFN-inducible genes. The EMBO journal. 2007;26:3451–3462. doi: 10.1038/sj.emboj.7601773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fujita K, Nakamura M, Wate R, Kaneko S, Sasaki S, Yamane K, Suzuki N, Aoki M, Shibata N, et al. Optineurin is co-localized with FUS in basophilic inclusions of ALS with FUS mutation and in basophilic inclusion body disease. Acta neuropathologica. 2011;121:555–557. doi: 10.1007/s00401-011-0809-z. [DOI] [PubMed] [Google Scholar]
- Kachaner D, Filipe J, Laplantine E, Bauch A, Bennett KL, Superti-Furga G, Israel A, Weil R. Plk1-dependent phosphorylation of optineurin provides a negative feedback mechanism for mitotic progression. Molecular cell. 2012;45:553–566. doi: 10.1016/j.molcel.2011.12.030. [DOI] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Molecular cell. 2009a;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Molecular cell. 2009b;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS letters. 2010;584:1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nature cell biology. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- Laird AS, Robberecht W. Modeling neurodegenerative diseases in zebrafish embryos. Methods Mol Biol. 2011;793:167–184. doi: 10.1007/978-1-61779-328-8_11. [DOI] [PubMed] [Google Scholar]
- Laird AS, Van Hoecke A, De Muynck L, Timmers M, Van den Bosch L, Van Damme P, Robberecht W. Progranulin is neurotrophic in vivo and protects against a mutant TDP-43 induced axonopathy. PLoS One. 2010;5:e13368. doi: 10.1371/journal.pone.0013368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmens R, Van Hoecke A, Hersmus N, Geelen V, D’Hollander I, Thijs V, Van Den Bosch L, Carmeliet P, Robberecht W. Overexpression of mutant superoxide dismutase 1 causes a motor axonopathy in the zebrafish. Hum Mol Genet. 2007;16:2359–2365. doi: 10.1093/hmg/ddm193. [DOI] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annual review of cell and developmental biology. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO reports. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Mizuno Y, Fujita Y, Takatama M, Nakazato Y, Okamoto K. Optineurin in neurodegenerative diseases. Neuropathology. 2011;31:569–574. doi: 10.1111/j.1440-1789.2011.01199.x. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjorkoy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. The Journal of cell biology. 2010;188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. The Journal of biological chemistry. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Sahlender DA, Roberts RC, Arden SD, Spudich G, Taylor MJ, Luzio JP, Kendrick-Jones J, Buss F. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. The Journal of cell biology. 2005;169:285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarfarazi M, Rezaie T. Optineurin in primary open angle glaucoma. Ophthalmol Clin North Am. 2003;16:529–541. doi: 10.1016/s0896-1549(03)00061-0. [DOI] [PubMed] [Google Scholar]
- Schwab C, Yu S, McGeer EG, McGeer PL. Optineurin in Huntington’s disease intranuclear inclusions. Neurosci Lett. 2012;506:149–154. doi: 10.1016/j.neulet.2011.10.070. [DOI] [PubMed] [Google Scholar]
- Sugihara K, Maruyama H, Kamada M, Morino H, Kawakami H. Screening for OPTN mutations in amyotrophic lateral sclerosis in a mainly Caucasian population. Neurobiol Aging. 2011;32:1923, e1929–1910. doi: 10.1016/j.neurobiolaging.2011.03.024. [DOI] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- Tumer Z, Bertelsen B, Gredal O, Magyari M, Nielsen KC, Lucamp, Gronskov K, Brondum-Nielsen K. Novel heterozygous nonsense mutation of the OPTN gene segregating in a Danish family with ALS. Neurobiol Aging. 2012;33:208, e201–205. doi: 10.1016/j.neurobiolaging.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nature reviews. Molecular cell biology. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- van Blitterswijk M, van Vught PW, van Es MA, Schelhaas HJ, van der Kooi AJ, de Visser M, Veldink JH, van den Berg LH. Novel optineurin mutations in sporadic amyotrophic lateral sclerosis patients. Neurobiol Aging. 2012;33:1016, e1011–1017. doi: 10.1016/j.neurobiolaging.2011.05.019. [DOI] [PubMed] [Google Scholar]
- Wagner S, Carpentier I, Rogov V, Kreike M, Ikeda F, Lohr F, Wu CJ, Ashwell JD, Dotsch V, Dikic I, et al. Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene. 2008;27:3739–3745. doi: 10.1038/sj.onc.1211042. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Tanaka M. p62/SQSTM1 in autophagic clearance of a non-ubiquitylated substrate. Journal of cell science. 2011;124:2692–2701. doi: 10.1242/jcs.081232. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio) University of Oregon Press; Eugene: 2000. [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witan H, Kern A, Koziollek-Drechsler I, Wade R, Behl C, Clement AM. Heterodimer formation of wild-type and amyotrophic lateral sclerosis-causing mutant Cu/Zn-superoxide dismutase induces toxicity independent of protein aggregation. Hum Mol Genet. 2008;17:1373–1385. doi: 10.1093/hmg/ddn025. [DOI] [PubMed] [Google Scholar]
- Xi Y, Noble S, Ekker M. Modeling neurodegeneration in zebrafish. Curr Neurol Neurosci Rep. 2011;11:274–282. doi: 10.1007/s11910-011-0182-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerbury JJ, Gower D, Vanags L, Roberts K, Lee JA, Ecroyd H. The small heat shock proteins alphaB-crystallin and Hsp27 suppress SOD1 aggregation in vitro. Cell stress & chaperones. 2012 doi: 10.1007/s12192-012-0371-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Wu CJ, Zhao Y, Ashwell JD. Optineurin negatively regulates TNFalpha-induced NF-kappaB activation by competing with NEMO for ubiquitinated RIP. Curr Biol. 2007;17:1438–1443. doi: 10.1016/j.cub.2007.07.041. [DOI] [PubMed] [Google Scholar]