Abstract
Objectives
Genotypic HIV drug-resistance testing is typically 60%–65% predictive of response to combination antiretroviral therapy (ART) and is valuable for guiding treatment changes. Genotyping is unavailable in many resource-limited settings (RLSs). We aimed to develop models that can predict response to ART without a genotype and evaluated their potential as a treatment support tool in RLSs.
Methods
Random forest models were trained to predict the probability of response to ART (≤400 copies HIV RNA/mL) using the following data from 14 891 treatment change episodes (TCEs) after virological failure, from well-resourced countries: viral load and CD4 count prior to treatment change, treatment history, drugs in the new regimen, time to follow-up and follow-up viral load. Models were assessed by cross-validation during development, with an independent set of 800 cases from well-resourced countries, plus 231 cases from Southern Africa, 206 from India and 375 from Romania. The area under the receiver operating characteristic curve (AUC) was the main outcome measure.
Results
The models achieved an AUC of 0.74–0.81 during cross-validation and 0.76–0.77 with the 800 test TCEs. They achieved AUCs of 0.58–0.65 (Southern Africa), 0.63 (India) and 0.70 (Romania). Models were more accurate for data from the well-resourced countries than for cases from Southern Africa and India (P < 0.001), but not Romania. The models identified alternative, available drug regimens predicted to result in virological response for 94% of virological failures in Southern Africa, 99% of those in India and 93% of those in Romania.
Conclusions
We developed computational models that predict virological response to ART without a genotype with comparable accuracy to genotyping with rule-based interpretation. These models have the potential to help optimize antiretroviral therapy for patients in RLSs where genotyping is not generally available.
Keywords: antiretroviral therapy, computer models, HIV drug resistance, predictions, treatment outcomes, resource-limited settings.
Introduction
The roll-out of combination antiretroviral therapy (ART) in resource-limited settings (RLSs) is delivering major benefits in terms of both morbidity and mortality.1 Nevertheless, there are a number of issues to be addressed if these benefits are to be maximized over the long term. Many patients on ART are likely to experience treatment failure at some point, requiring a treatment change to re-establish virological suppression. This should be done promptly to minimize the risk of disease progression, the development of drug-resistant variants and, consequently, the spread of drug-resistant virus in the population.2,3
To enable the rapid scale-up of ART in RLSs, WHO recommends a public health approach based on standardized, affordable drug regimens, limited laboratory monitoring and decentralized service delivery.4 This includes the use of clinical criteria and CD4 cell counts to diagnose ART failure and guide treatment changes, rather than viral load monitoring, which is the norm in well-resourced settings.4 However, this strategy has been shown to be associated with deferred treatment switching, accumulation of resistance and increased morbidity and mortality.5–10
In well-resourced settings, following treatment failure a genotypic resistance test is routinely performed to identify resistance-associated mutations.11 This information is typically interpreted using one of many rule-based interpretation systems available via the Internet and/or expert advice.12 These systems indicate whether the virus is likely to be resistant or susceptible to each individual drug, but do not provide any indication of the relative antiviral effects of combinations of drugs. With 25 or more drugs licensed for use in combination and >100 mutations involved in drug resistance, the selection of the optimum new therapy can be demanding. The HIV Resistance Response Database Initiative (RDI) was established in 2002 to address this and has developed computational models that use genotype, viral load, CD4 count and treatment history variables to predict response to drug combinations with ∼80% accuracy.13–15 This compares favourably with the 50%–70% predictive accuracy found by the RDI for genotypic sensitivity scores derived from genotyping with rule-based interpretation for the same cases and with independent studies of the use of genotyping to predict reponse.15–17
The models have been used to power a free experimental web-based HIV treatment response prediction system (HIV-TRePS) assessed by experienced HIV physicians in two clinical pilot studies as a useful aid to clinical practice.18 An alternative modelling system trained with a European dataset has also been evaluated and shown to be comparable to estimates of short-term response provided by HIV physicians.19
Such a system could be a useful tool to support optimal regimen sequencing in RLSs, particularly as additional drugs enter clinical practice. However, genotypic resistance tests are not generally available in RLSs so we decided to explore the possibility of modelling response without a genotype. In the absence of substantial data from RLSs, prototype random forest (RF) models were developed with cases from well-resourced settings. The accuracy of the ‘no-genotype’ models was only slightly diminished, at 78%–82%, compared with those using a genotype.20,21 Similarly, a group using European data confirmed that models developed without a genotype can achieve levels of accuracy that are encouraging.22 However, previous studies have shown that models are most accurate for patients from ‘familiar’ settings, i.e. settings from where the training data were collected.16 Since these models were trained with data from well-resourced settings, a concern was that their performance might be diminished when the models were applied to cases in RLSs.
In this study we set out to develop models trained with data from a wider range of sources than before, to maximize generalizability and to evaluate these models, not only during cross-validation and with test data from the same settings, but with cases from clinical practice in different RLSs in Eastern Europe, sub-Saharan Africa and India. The aim was to develop models that can predict virological response to ART without a genotype and evaluate their potential as a generalizable treatment support tool that could minimize treatment failure in RLSs.
Methods
Clinical data
At the time of the study the RDI database held anonymized data from ∼84 000 patients from >30 countries worldwide. The package of data collected when antiretroviral therapy is changed, for whatever reason, is termed a treatment change episode (TCE).16 TCEs were extracted that had all the following data available (Figure 1): on-treatment baseline plasma viral load (sample taken ≤8 weeks prior to treatment change); on treatment baseline CD4 cell count (≤12 weeks prior to treatment change); baseline regimen (the drugs the patient was taking prior to the change); antiretroviral treatment history; drugs in the new drug regimen; a follow-up plasma viral load determination taken between 4 and 48 weeks following introduction of the new regimen and the time to that follow-up viral load (in order that the models can be trained to predict responses at different times).
Figure 1.
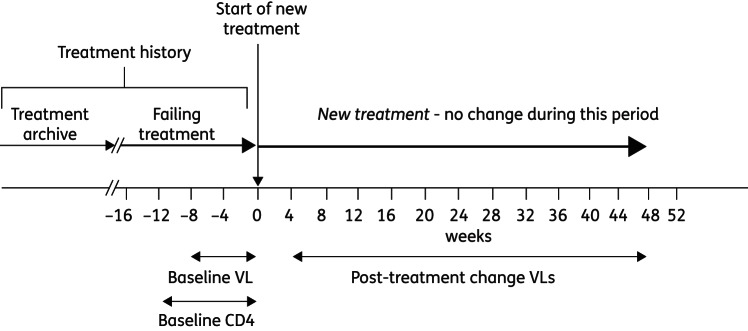
Anatomy of a treatment change episode (TCE). VL, viral load.
These TCEs were censored using the following rules established in previous studies:15 no more than three TCEs from the same change of therapy were permitted (using multiple follow-up viral loads to generate additional TCEs), with viral load determinations ≥4 weeks apart; TCEs involving drugs no longer in current use were excluded; TCEs involving drugs not adequately represented in the database (tipranavir, raltegravir and maraviroc) were excluded; TCEs that include an unboosted protease inhibitor (PI) other than nelfinavir, or ritonavir as the only PI, in the failing or new regimen positions were excluded (but permitted as treatment history variables); TCEs with an undetectable viral load (<400 copies) at baseline were excluded as the models were designed to predict responses to treatment changes following virological failure; TCEs with viral load values of the form ‘<X’, where X is >400 or 2.6 log copies, were excluded as the absolute values were not known.15
Computational model development
The qualifying TCEs were used to train a committee of 10 RF models to predict the probability of the follow-up viral load being ≤400 copies/mL, using methodology described in detail elsewhere.14,15 The input variables used were: baseline viral load (log10 copies HIV RNA/mL); baseline CD4 count (cells/mL); treatment history [five variables determined by previous research to have a significant impact on the accuracy of models, comprising zidovudine, lamivudine/emtricitabine, any non-nucleoside reverse transcriptase inhibitors (NNRTIs), any PIs, enfuvirtide]; antiretroviral drugs in the new regimen [18 variables, comprising zidovudine, didanosine, stavudine, abacavir, lamivudine/emtricitabine, tenofovir DF, efavirenz, nevirapine, etravirine, indinavir, nelfinavir, saquinavir, (fos)amprenavir, lopinavir, atazanavir, darunavir, ritonavir (as a PI booster) and enfuvirtide]; and time from treatment change to the follow-up viral load (number of days).
The output variable was the follow-up viral load coded as a binary variable: ≤2.6 log or 400 copies/mL = 1 (response) and >2.6 log or 400 copies/mL = 0 (failure). The models were trained to produce an estimate of the probability of the follow-up viral load being ≤400 copies/mL.
The performance of the models as predictors of virological response was evaluated by using the models' estimates of the probability of response and the responses observed in the clinics (response versus failure) to plot receiver operating characteristic (ROC) curves and assessing the area under the ROC curve (AUC). In addition, the optimum operating point (OOP) for the models was derived and used to obtain the overall accuracy, sensitivity and specificity of the system.
Data partition
The majority of qualifying TCEs were from North America, Europe, Australia and Japan. The few complete TCEs available to RDI at the time from RLSs that included viral loads (not routinely monitored in RLSs) were set aside to act as test sets. The 15 947 remaining TCEs that met all the criteria described above were partitioned at random into a training set of 14 891 TCEs and a test set of 800 TCEs from 800 patients (∼5% of the available dataset), such that no patient could have TCEs in both sets (additional TCEs from patients in the test set were discarded).
Internal cross-validation
The committee of 10 RF models was developed using a 10× cross-validation scheme whereby 10% of the TCEs were selected at random and the remainder used to train numerous models, and their performance gauged by cross-validation with the 10% that had been left out. Model development continued until further models failed to yield improved accuracy. This process was repeated 10 times until all the TCEs had appeared in a validation set once. With each partition the best-performing RF model was selected as a member of the final committee of models.
External validation
The RF models were validated externally by providing them with the baseline data from the 800 test TCEs, obtaining the predictions of virological response from the models and comparing these with the responses on file. In addition to evaluating the committee of 10 RF models individually, the committee average performance (CAP) was evaluated using the mean of the predictions of the 10 models for each of the test TCEs.
The RF models were then tested using the qualifying TCEs from the following clinics and cohorts in RLSs. In sub-Saharan Africa, data were provided by the Gugulethu Clinic, Desmond Tutu HIV Centre, Cape Town, South Africa (114 TCEs); Ndlovu Medical Centre, Elandsdoorn, South Africa (39 TCEs); and the PASER-M cohort, in the following six sub-Saharan African countries: Kenya, Nigeria, South Africa, Uganda, Zambia and Zimbabwe (78 TCEs).23 Since these individual datasets were small, they were combined to give a Southern African test set of 231 TCEs. In Romania data came from the National Institute of Infectious Diseases “Prof. Dr. Matei Balş” and the “Dr. Victor Babes” Hospital for Infectious and Tropical Diseases, both in Bucharest (375 TCEs). In India, the data originated from an HIV cohort study in the district of Anantapur (206 TCEs).24
Finally, a subset of 55 of the original 800 test TCEs that most resembled the TCEs from RLSs in terms of their treatment history and drugs in the new regimen were used to test the models as a control for the content of the TCEs versus the ‘unfamiliarity’ of the settings from which the data were obtained.
The differences in performance of the models with the different test sets were tested for statistical significance using DeLong's test.25
In silico analysis to identify effective alternatives to failed regimens
In order to assess the potential of the models to help avoid treatment failure as a generalizable tool in different settings, they were used to identify antiretroviral regimens that were predicted to be effective for the treatment failures that occurred in the clinic in each of the datasets. The baseline data for these cases were used by the models to make predictions of response for alternative three-drug regimens comprising drugs that were in use in these settings at the time, as summarized in Table 1. For all these tests the OOP (the cut-off above which the models' prediction is classified as a prediction of response) that was derived during model development was used, as a test of how generalizable the system is with cases from unfamiliar settings.
Table 1.
Drug availability (those in use at the time of the TCEs)
| Ndlovu | Gugulethu | PASER-M | Romania | India | |
|---|---|---|---|---|---|
| N(t)RTIs | |||||
| abacavir | X | • | • | • | • |
| didanosine | • | • | • | • | • |
| emtricitabine | X | X | • | • | • |
| lamivudine | • | • | • | • | • |
| stavudine | • | • | • | • | • |
| tenofovir | • | X | • | • | • |
| zidovudine | • | • | • | • | • |
| NNRTIs | |||||
| efavirenz | • | • | • | • | • |
| etravirine | X | X | X | • | X |
| nevirapine | • | • | • | • | • |
| PIs | |||||
| amprenavir | X | X | X | • | X |
| atazanavir | X | X | X | • | X |
| darunavir | X | X | X | • | X |
| indinavir | X | X | X | • | X |
| lopinavir | • | • | • | • | • |
| nelfinavir | X | X | X | • | • |
| ritonavir | • | • | • | • | • |
| saquinavir | X | X | X | • | X |
| tipranavir | X | X | X | • | X |
| Fusion inhibitors | |||||
| enfuvirtide | X | X | X | • | X |
| Integrase inhibitors | |||||
| raltegravir | X | X | X | X | X |
Symbols: •, drugs that were in use; X, drugs that were not in use.
Results
Characteristics of the datasets
The baseline, treatment and response characteristics of the datasets are summarized in Table 2. The training and main RDI test sets were highly comparable, as would be expected as they were partitioned at random from the pool of qualifying TCEs. A total of 2013 of the training cases gave rise to a single TCE, with 1615 having two TCEs (using two follow-up viral loads at different timepoints) and 3216 having three TCEs. The mean number of linked TCEs per case was 2.18.
Table 2.
Characteristics of the TCEs in the training and test sets
| Training set | RDI Test set | Ndlovu | Gugulethu | PASER-M | Southern Africa | Romania | India | RDI subset | |
|---|---|---|---|---|---|---|---|---|---|
| TCEs | 14 891 | 800 | 39 | 114 | 78 | 164 | 375 | 206 | 55 |
| Patients | 4878 | 800 | 38 | 104 | 78 | 153 | 234 | 165 | 55 |
| Male | 3625 | 602 | 12 | 28 | 44 | 48 | 111 | 87 | 44 |
| Female | 778 | 137 | 26 | 76 | 34 | 105 | 109 | 78 | 10 |
| Gender unknown | 475 | 61 | 0 | 0 | 0 | 0 | 14 | 0 | 1 |
| Median age (years) | 40 | 42 | 32 | 32 | 39 | 34 | 20 | 28 | 39 |
| Baseline data | |||||||||
| median (IQR) baseline VL (log10 copies/mL) | 3.77 (2.67–4.71) | 3.79 (2.72–4.70) | 4.31 (3.88–4.65) | 3.95 (3.46–4.55) | 4.64 (3.97–5.16) | 4.04 (3.57–4.59) | 4.07 (2.6–5.07) | 4.75 (4.16–5.27) | 4.17 (3.27–4.68) |
| median (IQR) baseline CD4 (cells/mm3) | 260 (135–417) | 260 (130–403) | 214 (132–367) | 239 (138–332) | 84 (29–180) | 228 (130–334) | 285 (152–480) | 274 (147–478) | 262 (142–372) |
| Treatment history | |||||||||
| number of previous drugs (median) | 5 | 5 | 4 | 4 | 4 | 4 | 4 | 3 | 3 |
| N(t)RTI experience (%) | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| NNRTI experience (%) | 68 | 67 | 100 | 99 | 100 | 99 | 47 | 100 | 89 |
| PI experience (%) | 83 | 81 | 0 | 4 | 6 | 4 | 76 | 6 | 0 |
| Failures (>2.6 log10 copies/mL follow-up VL) | 6501 | 309 | 14 | 41 | 8 | 57 | 176 | 74 | 14 |
| percent | 44 | 39 | 36 | 36 | 10 | 35 | 47 | 36 | 25 |
| Responses | 8390 | 491 | 25 | 73 | 70 | 107 | 199 | 132 | 41 |
| percent | 56 | 61 | 64 | 64 | 90 | 65 | 53 | 64 | 75 |
| New regimens | |||||||||
| 2 N(t)RTIs + PI (%) | 28 | 31 | 92 | 88 | 91 | 87 | 46 | 55 | 75 |
| 2 N(t)RTIs + NNRTI (%) | 17 | 18 | 8 | 12 | 1 | 10 | 5 | 0 | 4 |
| 3 N(t)RTIs (%) | 8 | 8 | 0 | 0 | 8 | 3 | 1 | 0 | 1 |
| 3 N(t)RTIs + PI (%) | 10 | 10 | 0 | 0 | 0 | 0 | 3 | 45 | 13 |
| other (%) | 37 | 33 | 0 | 0 | 0 | 0 | 45 | 0 | 7 |
VL, viral load.
Males outnumbered females in the RDI datasets by around 5 : 1, whereas there were more women than men in the African data and a gender balance in the Romanian and Indian data. The patients from RLSs were younger than those from the RDI datasets (median 39–42 years). This was particularly true of Romania (median 20 years) and India (median 28 years). The median age of the southern African patients was 34 years.
The data from RLSs had somewhat higher baseline viral loads, with median values ranging from 3.95 (Gugulethu) to 4.79 log10 copies/mL (India) compared with 3.77 log10 copies/mL for the training data. This is consistent with patients in RLSs switching after a greater degree of virological failure, often based on clinical symptoms or a decrease in CD4 count. Interestingly, however, there was less of a difference in baseline CD4 cell counts, with the exception of PASER-M, where the median was 84 cells/mm3 at baseline compared with 260 for the training data and 182–285 for the other RLS datasets. The PASER dataset includes patients from 13 sites in six countries that were switched to second line after prolonged failure with no routine virological monitoring.7 This may explain the low CD4 counts and is likely to represent the reality in many RLSs in Africa.
Almost all (99–100%) of the cases from sub-Saharan Africa and India had received nucleoside/nucleotide reverse transcriptase inhibitors [N(t)RTIs] and NNRTIs in their history, with 0%–6% having experience of PIs. Accordingly, 88%–92% of the Southern African cases and 54% of the Indian cases had been switched onto two N(t)RTIs + PI. The remaining Indian cases had been switched onto three N(t)RTIs and a PI, all but two of which included tenofovir. The Romanian cases included heavily treatment-experienced patients, with 76% having PI experience. Nevertheless, 70% were switched onto a PI-based triple therapy. The data used to train the models included a very diverse range of new regimens comprising 94 distinct types (defined in terms of the number of drugs from different classes) involving between one and eight drugs. The most frequent were: two N(t)RTIs + PI (28%), two N(t)RTIs + NNRTI (17%), three N(t)RTIs + 1 PI (10%), three N(t)RTIs (8%), three N(t)RTIs + NNRTI (6%) and two N(t)RTIs + PI + NNRTI (4%).
The RDI subset selected as resembling the cases from RLSs had a median baseline viral load of 4.17 copies/mL, which was more comparable to that of the Southern African TCEs than were the main training and test sets. Seventy-six percent were switched onto two N(t)RTIs + PI, 4% switched to two N(t)RTIs + NNRTI, 13% to three N(t)RTIs + PI and 7% to other combinations.
Results of the modelling
Cross-validation
The performance characteristics from the ROC curves of the 10 individual models during cross-validation and independent testing are summarized in Table 3. The models achieved an AUC during cross-validation ranging from 0.74 to 0.81, with a mean of 0.77. The overall accuracy ranged from 68% to 76% (mean 72%), sensitivity from 67% to 77% (mean 72%) and specificity from 63% to 75% (mean 71%).
Table 3.
Performance of the models during cross-validation and testing with the 800 RDI test TCEs
| Model | Performance in cross-validation |
Performance with the 800 RDI test set |
||||||
|---|---|---|---|---|---|---|---|---|
| AUC | overall accuracy (%) | sensitivity (%) | specificity (%) | AUC | overall accuracy (%) | sensitivity (%) | specificity (%) | |
| 1 | 0.81 | 74 | 77 | 70 | 0.77 | 71 | 76 | 63 |
| 2 | 0.78 | 72 | 73 | 70 | 0.77 | 71 | 71 | 71 |
| 3 | 0.76 | 71 | 67 | 74 | 0.77 | 72 | 72 | 71 |
| 4 | 0.80 | 76 | 77 | 75 | 0.76 | 73 | 75 | 70 |
| 5 | 0.79 | 74 | 76 | 71 | 0.76 | 70 | 74 | 63 |
| 6 | 0.77 | 71 | 72 | 70 | 0.76 | 70 | 73 | 64 |
| 7 | 0.78 | 71 | 71 | 72 | 0.77 | 71 | 73 | 67 |
| 8 | 0.76 | 70 | 70 | 71 | 0.77 | 72 | 73 | 70 |
| 9 | 0.76 | 71 | 70 | 72 | 0.76 | 70 | 70 | 70 |
| 10 | 0.74 | 68 | 72 | 63 | 0.76 | 70 | 74 | 64 |
| Mean | 0.77 | 72 | 72 | 71 | ||||
| 95% CI | (0.76, 0.79) | (70, 73) | (70, 75) | (69, 73) | ||||
| Committee average performance | 0.77 | 71 | 71 | 70 | ||||
| 95% CI | (0.73, 0.80) | (68, 74) | (67, 75) | (64, 75) | ||||
Testing with the independent set of 800 TCEs
The 10 models achieved an AUC ranging from 0.76 to 0.77, with a committee average of 0.77. The overall accuracy ranged from 70% to 73% (committee average 71%), sensitivity ranged from 70% to 76% (committee average 71%) and specificity from 63% to 71% (committee average 70%). The ROC curve for the CAP is presented in Figure 2.
Figure 2.
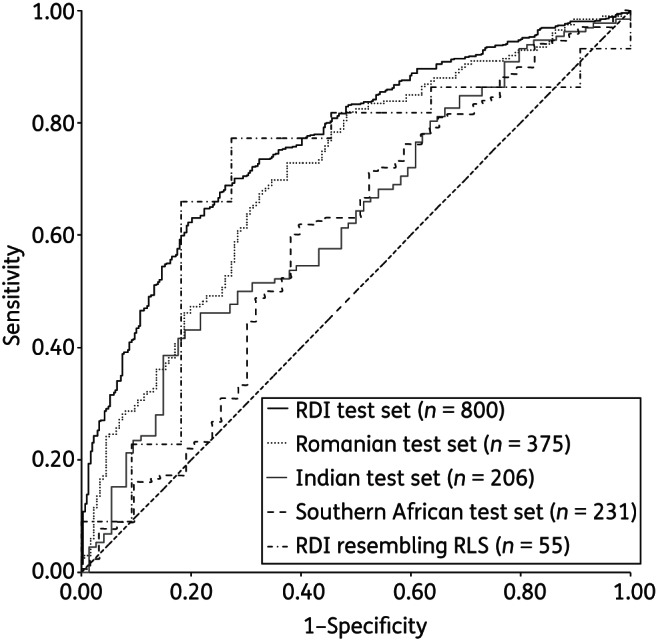
ROC curves for the RF models' predictions for each of the test datasets.
Testing the models with data from RLSs
The accuracy of prediction of the RF committee was tested with each of the test sets from RLSs and the 55 RDI TCEs that resembled the cases from RLSs, with the results presented in Table 4 and the ROC curves in Figure 2. For the data from southern Africa, the AUC ranged from 0.58 for PASER-M to 0.65 for Gugulethu. For the combined southern African data the AUC was 0.60. This compares with 0.77 for the RDI 800 test set and 0.70 for the subset that resembled the RLS cases. Overall accuracy ranged from 60% (PASER-M) to 64% (Ndlovu), compared with 71% and 75% for the RDI 800 set and the 55 TCE subset. Sensitivity ranged from 60% to 68% and specificity from 57% to 63%. This compares with sensitivity of 71% for the RDI 800 and 75% for the subset and specificity of 70% and 73% respectively.
Table 4.
Combined RF model performance with independent test sets from RLSs
| Dataset | AUC | Overall accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
| Ndlovu (n = 39) | 0.61 | 64% | 68% | 57% |
| 95% CI | (0.40, 0.73) | (47%, 79%) | (46%, 85%) | (29%, 82%) |
| Gugulethu (n = 114) | 0.65 | 62% | 62% | 63% |
| 95% CI | (0.55, 0.76) | (57%, 75%) | (50%, 73%) | (47%, 78%) |
| PASER-M (n = 78) | 0.58 | 60% | 60% | 63% |
| 95% CI | (0.38, 0.77) | (49%, 71%) | (48%, 72%) | (24%, 91%) |
| Southern Africa combined (n = 231) | 0.60 | 61% | 60% | 62% |
| 95% CI | (0.52, 0.69) | (53%, 67%) | (52%, 68%) | (49%, 74%) |
| Romania (n = 375) | 0.71 | 67% | 67% | 68% |
| 95% CI | (0.66, 0.76) | (62%, 72%) | (60%, 74%) | (60%, 74%) |
| India (n = 206) | 0.63 | 57% | 55% | 61% |
| 95% CI | (0.55, 0.71) | (50%, 64%) | (47%, 63%) | (49%, 72%) |
| RDI RLS subset (n = 55) | 0.70 | 75% | 75% | 73% |
| 95% CI | (0.51, 0.88) | (60%, 85%) | (60%, 87%) | (39%, 94%) |
For the Romanian cases, the performance was better overall, with an AUC of 0.71, overall accuracy of 67%, sensitivity of 67% and specificity of 68%. For India, the AUC was 0.63, overall accuracy 57%, sensitivity 55% and specificity 61%.
The models' predictions were statistically significantly more accurate for the 800 RDI test set than for the Southern African cases and the Indian cases (P < 0.001), but not the Romanian cases.
In silico analysis
The proportion of cases from the different regions who experienced virological failure following the introduction of a new regimen varied from 10% in PASER-M to 47% in Romania, reflecting the different stages of treatment of the patients (Table 5).
Table 5.
Results of in silico analysis in which the models were used to identify potentially effective regimens for cases of actual treatment failure
| Ndlovu (n = 39) | Gugulethu (n = 114) | PASER-M (n = 78) | Southern Africa (n = 231) | Romania (n = 375) | India (n = 206) | RDI test set (n = 800) | |
|---|---|---|---|---|---|---|---|
| No. (%) of all cases for which the models were able to identify a regimen that was predicted to be effective | 38 (97) | 111 (97) | 68 (87) | 217 (94) | 362 (97) | 205 (99.5) | 767 (96) |
| No. (%) of cases that failed in the clinic | 14 (36) | 41 (36) | 8 (10) | 63 (27) | 176 (47) | 74 (36) | 309 (39) |
| No. (%) of actual failures for which alternative regimens were found that were predicted to be effective | 14 (100) | 39 (95) | 6 (75) | 59 (94) | 164 (93) | 73 (99) | 288 (93) |
| No. (%) for which alternative regimens were found with higher predictions of response | 14 (100) | 41 (100) | 8 (100) | 63 (100) | 176 (100) | 74 (100) | 307 (99) |
The models were able to identify a regimen that was predicted to be effective, comprising only those drugs represented in the data provided to RDI by each centre, in between 87% (PASER-M) and 99.5% (India) of all cases. For those cases where the new regimen prescribed in the clinic failed, the models identified alternative three-drug regimens that were predicted to elicit virological responses in between 75% (PASER) and 100% (Ndlovu) of failures. Alternative regimens with higher predicted probability of response than the failing regimens were identified for all cases from all RLSs.
Discussion
The computational models reported here predicted virological response to a change in antiretroviral therapy following virological failure, without the results of genotypic resistance testing, with a level of accuracy that was comparable to genotyping with rule-based interpretation, which is encouraging. Models that do not require a genotype to make accurate predictions of response have potential utility in RLSs, where resistance testing is often not available. This was the first attempt to test such models using clinical cases from RLSs, and the results confirmed this potential.
The results were fairly consistent for cases from several clinics in sub-Saharan Africa and India and somewhat better for Romania, suggesting that this accuracy may be generalizable to different RLSs in different regions. While Romania is not as resource-constrained as most of sub-Saharan Africa or India, genotyping is not generally available and Romania and other countries of Eastern Europe could, therefore, also benefit from such an approach.
The models were able to identify alternative three-drug regimens comprising locally available drugs that were predicted to produce a virological response for a substantial proportion of the failures observed. This proportion ranged from 75% in the PASER cohort, where the number of drugs available was fairly restricted, to 93% in Romania, 99% in India, where drug availability was greater, and 100% in Ndlovu, where drug availability was highly restricted. The models were able to identify regimens with a higher predicted probability of success than the regimen that failed in all cases from RLSs. This suggests that, had physicians been able to use the system to assist their treatment decisions (by choosing regimens with the highest estimated probability of success according to the system) the number of virological failures could have been reduced. If so, the models could have considerable utility, even in settings where treatment options are highly restricted, and this utility will increase as treatment options expand.
It should be noted that one of the input variables for these models was the plasma viral load, which previous studies by our group and others have shown to be critically important for the predictive accuracy of the models.20,22 Although the viral load of patients on ART is not monitored routinely in most RLSs, it is common practice to evaluate the viral load to confirm virological failure before switching to second-line ART in cases who meet criteria for clinical or immunological failure.4 Moreover, challenges related to the implementation of viral load monitoring in RLSs are increasingly surmountable, as recent technological advances have led to lower test costs and simpler equipment requiring less infrastructure, maintenance and technical expertise.26
The models were more accurate for patients from the familiar settings providing training data than for cases from unfamiliar RLSs, other than Romania, which is consistent with results of previous studies.16 Analyses have so far failed to identify any significant differences between datasets that can explain this. Interestingly, the accuracy of the models was comparable for those cases from familiar settings selected as being highly comparable to those from RLSs and for the RLS cases themselves. HIV-1 subtype diversity is unlikely to be the primary explanation as this phenomenon has been observed with familiar and unfamiliar datasets from the same region as the training data and, in general, there are no substantial differences in ART responses between different clades of HIV.27 These latest results provide further evidence that this phenomenon is not only reproducible, but likely to be due to differences that are either very subtle or related to factors not included in the training data. The development of more accurate models in the future may benefit from the collection of sufficient data from RLSs to develop region- or country-specific models, or at least sufficient for representation in the training data.
During cross-validation and testing with a large independent test dataset from the same settings as the training data, the models were only ∼5% less accurate than is typical for models that use a genotype in their predictions. This accuracy was comparable to that achieved by the EuResist group with models developed and tested using European data only.22 The study extends those findings by testing models with data from RLSs for the first time and by using the models to identify potentially more effective alternatives to the regimens used in the clinic.
The study has some limitations. Firstly, it was retrospective and, as such, no firm claims can be made for the clinical benefit that use of the system as a treatment support tool could provide. The RDI's relative shortage of complete TCEs that include plasma viral loads from RLSs meant that the test sets were relatively small and also prevented the training of models using data from RLSs only, specifically for use in these settings. Nevertheless, the results were positive for clinics and cohorts in many different countries across several disparate regions of the world, which is encouraging in terms of generalizability. Another possible shortcoming inherent in such studies is that the cases used are, by definition, those with complete data around a change of therapy and therefore may not be truly representative of the general patient population. For example, they may be more adherent, which would tend to lead the models to overestimate the probability of response for a population containing a higher proportion of non-adherent patients. Conversely, since we have little information in our database on adherence, the inclusion of some non-adherent patients' data in the training set is unavoidable and likely to lead to underestimation of the probability of response for an adherent individual.
Conclusions
This study is the first to demonstrate that large datasets can be used to develop computational models that predict virological response to antiretroviral therapy for patients in RLSs without a genotype, with accuracy comparable to that of genotyping with rule-based interpretation. The models were able to identify potentially effective alternative regimens for the great majority of cases of treatment failure in RLSs using only those drugs available at each location at the time.
Full validation of this approach would require a prospective, controlled clinical trial and the current results indicate that the accuracy of the models might be significantly improved if data from RLSs were included in the training of future models. Nevertheless, the results suggest this approach has the potential to reduce virological failure and improve patient outcomes in RLSs. It can provide clinicians with a practical tool to support optimized treatment decision making in the absence of resistance tests and where expertise may be lacking in the context of a public health approach to ART rollout and management.
RDI data and study group
The RDI wishes to thank all the following individuals and institutions for providing the data used in training and testing its models.
Cohorts
Frank De Wolf and Joep Lange (ATHENA, The Netherlands); Julio Montaner and Richard Harrigan (BC Center for Excellence in HIV & AIDS, Canada); Tobias Rinke de Wit, Raph Hamers and Kim Sigaloff (PASER-M cohort, The Netherlands); Brian Agan, Vincent Marconi and Scott Wegner (US Department of Defence); Wataru Sugiura (National Institute of Health, Japan); Maurizio Zazzi (MASTER, Italy); Adrian Streinu-Cercel (National Institute of Infectious Diseases “Prof. Dr. Matei Balş”, Bucharest, Romania).
Clinics
Jose Gatell and Elisa Lazzari (University Hospital, Barcelona, Spain); Brian Gazzard, Mark Nelson, Anton Pozniak and Sundhiya Mandalia (Chelsea and Westminster Hospital, London, UK); Lidia Ruiz and Bonaventura Clotet (Fundacion IrsiCaixa, Badelona, Spain); Schlomo Staszewski (Hospital of the Johann Wolfgang Goethe-University, Frankfurt, Germany); Carlo Torti (University of Brescia); Cliff Lane and Julie Metcalf (National Institutes of Health Clinic, Rockville, USA); Maria-Jesus Perez-Elias (Instituto Ramón y Cajal de Investigación Sanitaria, Madrid, Spain); Andrew Carr, Richard Norris and Karl Hesse (Immunology B Ambulatory Care Service, St Vincent's Hospital, Sydney, NSW, Australia); Dr Emanuel Vlahakis (Taylor's Square Private Clinic, Darlinghurst, NSW, Australia); Hugo Tempelman and Roos Barth (Ndlovu Care Group, Elandsdoorn, South Africa); Carl Morrow and Robin Wood (Desmond Tutu HIV Centre, Cape Town, South Africa); Luminita Ene (“Dr. Victor Babes” Hospital for Infectious and Tropical Diseases, Bucharest, Romania).
Clinical trials
Sean Emery and David Cooper (CREST); Carlo Torti (GenPherex); John Baxter (GART, MDR); Laura Monno and Carlo Torti (PhenGen); Jose Gatell and Bonventura Clotet (HAVANA); Gaston Picchio and Marie-Pierre deBethune (DUET 1 & 2 and POWER 3); Maria-Jesus Perez-Elias (RealVirfen).
Funding
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E. This research was supported by the National Institute of Allergy and Infectious Diseases.
Transparency declarations
None to declare.
Disclaimer
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Acknowledgements
References
- 1.Jahn FS, Floyd S, Crampin AC, et al. Population-level effect of HIV on adult mortality and early evidence of reversal after introduction of antiretroviral therapy in Malawi. Lancet. 2008;371:1603–11. doi: 10.1016/S0140-6736(08)60693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen MS, Chen YQ, McCauley M, et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365:493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta RK, Hill A, Sawyer AW, et al. Virological monitoring and resistance to first-line highly active antiretroviral therapy in adults infected with HIV-1 treated under WHO guidelines: a systematic review and meta-analysis. Lancet Infect Dis. 2009;9:409–17. doi: 10.1016/S1473-3099(09)70136-7. [DOI] [PubMed] [Google Scholar]
- 4.WHO. Antiretroviral Therapy for HIV Infection in Adolescents and Adults: Recommendations for a Public Health Approach–2010 Revision. Geneva: WHO; 2010. [PubMed] [Google Scholar]
- 5.DART Trial Team. Routine versus clinically driven laboratory monitoring of HIV antiretroviral therapy in Africa (DART): a randomised non-inferiority trial. Lancet. 2010;375:123–31. doi: 10.1016/S0140-6736(09)62067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosseinipour MC, van Oosterhout JJ, Weigel R, et al. The public health approach to identify antiretroviral therapy failure: high-level nucleoside reverse transcriptase inhibitor resistance among Malawians failing first-line antiretroviral therapy. AIDS. 2009;23:1127–34. doi: 10.1097/QAD.0b013e32832ac34e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sigaloff KCE, Hamers RL, Wallis CL, et al. Unnecessary antiretroviral treatment switches and accumulation of HIV resistance mutations; two arguments for viral load monitoring in Africa. J Acquir Immune Defic Syndr. 2011;58:23–31. doi: 10.1097/QAI.0b013e318227fc34. [DOI] [PubMed] [Google Scholar]
- 8.Zhou J, Li P, Kumarasamy N, Boyd M, et al. Deferred modification of antiretroviral regimen following documented treatment failure in Asia: results from the TREAT Asia HIV Observational Database (TAHOD) HIV Med. 2010;11:31–9. doi: 10.1111/j.1468-1293.2009.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keiser O, Chi BH, Gsponer T, et al. Outcomes of antiretroviral treatment in programmes with and without routine viral load monitoring in southern Africa. AIDS. 2011;25:1761–9. doi: 10.1097/QAD.0b013e328349822f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barth RE, Aitken SC, Tempelman H, et al. Accumulation of drug resistance and loss of therapeutic options precede commonly used criteria for treatment failure in HIV-1 subtype-C-infected patients. Antivir Ther. 2012;17:377–86. doi: 10.3851/IMP2010. [DOI] [PubMed] [Google Scholar]
- 11.Thompson MA, Aberg JA, Cahn P, et al. Antiretroviral treatment of adult HIV infection: 2010 recommendations of the International AIDS Society-USA Panel. JAMA. 2010;304:321–33. doi: 10.1001/jama.2010.1004. [DOI] [PubMed] [Google Scholar]
- 12.Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis. 2011;42:1608–18. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larder BA, DeGruttola V, Hammer S, et al. The international HIV resistance response database initiative: a new global collaborative approach to relating viral genotype treatment to clinical outcome. Antivir Ther. 2002;7:S111. [Google Scholar]
- 14.Wang D, Larder B, Revell A, et al. A comparison of three computational modelling methods for the prediction of virological response to combination HIV therapy. Artif Intell Med. 2009;47:63–74. doi: 10.1016/j.artmed.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Revell AD, Wang D, Boyd MA, et al. The development of an expert system to predict virological response to HIV therapy as part of an online treatment support tool. AIDS. 2011;25:1855–63. doi: 10.1097/QAD.0b013e328349a9c2. [DOI] [PubMed] [Google Scholar]
- 16.Larder BA, Wang D, Revell A, et al. The development of artificial neural networks to predict virological response to combination HIV therapy. Antivir Ther. 2007;12:15–24. [PubMed] [Google Scholar]
- 17.Frentz D, Boucher C, Assel M, et al. Comparison of HIV-1 genotypic resistance test interpretation systems in predicting virological outcomes over time. PLoS One. 2010;5:e11505. doi: 10.1371/journal.pone.0011505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larder BA, Revell A, Mican JM, et al. Clinical evaluation of the potential utility of computational modeling as an HIV treatment selection tool by physicians with considerable HIV experience. AIDS Patient Care STDS. 2011;25:29–36. doi: 10.1089/apc.2010.0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zazzi M, Kaiser R, Sönnerborg A, et al. Prediction of response to antiretroviral therapy by human experts and by the EuResist data-driven expert system (the EVE study) HIV Med. 2010;12:211–8. doi: 10.1111/j.1468-1293.2010.00871.x. [DOI] [PubMed] [Google Scholar]
- 20.Revell AD, Wang D, Harrigan R, et al. Modelling response to HIV therapy without a genotype: an argument for viral load monitoring in resource-limited settings. J Antimicrob Chemother. 2010;65:605–7. doi: 10.1093/jac/dkq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Revell AD, Wang D, Harrigan R, et al. Computational models developed without a genotype for resource-poor countries predict response to HIV treatment with 82% accuracy. Antivir Ther. 2009;14(Suppl 1):A38. [Google Scholar]
- 22.Prosperi MCF, Rosen-Zvi M, Altman A, et al. Antiretroviral therapy optimisation without genotype resistance testing: a perspective on treatment history based models. PLoS One. 2010;5:e13753. doi: 10.1371/journal.pone.0013753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamers RL, Oyomopito R, Kityo C, et al. Cohort profile: the PharmAccess African (PASER-M) and the TREAT Asia (TASER-M) monitoring studies to evaluate resistance—HIV drug resistance in sub-Saharan Africa and the Asia-Pacific. Int J Epidemiol. 2012;41:43–54. doi: 10.1093/ije/dyq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alvarez-Uria G, Midde M, Pakam R, et al. Gender differences, routes of transmission, socio-demographic characteristics and prevalence of HIV related infections of adults and children in an HIV cohort from a rural district of India. Infect Dis Rep. 2012:e19. doi: 10.4081/idr.2012.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45. [PubMed] [Google Scholar]
- 26.Stevens WS, Scott LE, Crowe SM. Quantifying HIV for monitoring antiretroviral therapy in resource-poor settings. J Infect Dis. 2010;201(Suppl 1):S16–26. doi: 10.1086/650392. [DOI] [PubMed] [Google Scholar]
- 27.Kantor R, Katzenstein DA, Efron B, et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med. 2005;2:e112. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]


