Summary
Epithelial organ morphogenesis involves reciprocal interactions between epithelial and mesenchymal cell types to balance progenitor cell retention and expansion with cell differentiation for evolution of tissue architecture. Underlying submandibular salivary gland branching morphogenesis is the regulated proliferation and differentiation of perhaps several progenitor cell populations, which have not been characterized throughout development, and yet are critical for understanding organ development, regeneration, and disease. Here we applied a serial multiplexed fluorescent immunohistochemistry technology to map the progressive refinement of the epithelial and mesenchymal cell populations throughout development from embryonic day 14 through postnatal day 20. Using computational single cell analysis methods, we simultaneously mapped the evolving temporal and spatial location of epithelial cells expressing subsets of differentiation and progenitor markers throughout salivary gland development. We mapped epithelial cell differentiation markers, including aquaporin 5, PSP, SABPA, and mucin 10 (acinar cells); cytokeratin 7 (ductal cells); and smooth muscle α-actin (myoepithelial cells) and epithelial progenitor cell markers, cytokeratin 5 and c-kit. We used pairwise correlation and visual mapping of the cells in multiplexed images to quantify the number of single- and double-positive cells expressing these differentiation and progenitor markers at each developmental stage. We identified smooth muscle α-actin as a putative early myoepithelial progenitor marker that is expressed in cytokeratin 5-negative cells. Additionally, our results reveal dynamic expansion and redistributions of c-kit- and K5-positive progenitor cell populations throughout development and in postnatal glands. The data suggest that there are temporally and spatially discreet progenitor populations that contribute to salivary gland development and homeostasis.
Key words: Salivary gland, Development, Progenitor cells, Multiplex analysis
Introduction
How complex three dimensional structures such as tissues and organs are formed from precursor cells is one of the most fundamental questions in developmental biology. Organogenesis requires two distinct but overlapping processes: morphogenesis, the growth and physical rearrangement of cells to form complex three dimensional structures, and cytodifferentiation, the process by which these cells assume specialized functions. Branching morphogenesis is a conserved developmental mechanism required for the formation of many vertebrate and invertebrate organs, including most exocrine glands such as salivary and mammary glands (Lu et al., 2006; Lu and Werb, 2008). During this process, a primary epithelial bud or tube undergoes coordinated cellular rearrangements to generate branched structures that greatly increase the epithelial surface area for secretion or absorption. Concurrent with branching morphogenesis, the cells undergo cytodifferentiation to produce the multiple epithelial subtypes required for adult organ function. The submandibular salivary gland (SMG) is a classical model system to study morphogenesis and differentiation that undergoes branching morphogenesis during embryonic and post-natal development (Patel et al., 2006; Tucker, 2007). All of the mature epithelial cell subtypes are derived from the epithelial progenitors in the primary rudiment; however, the spatio-temporal progression of cell differentiation has not been mapped through all developmental stages.
The salivary gland epithelium undergoes differentiation to produce multiple sub-types of epithelial cells. These cells can be generally classified as secretory acinar cells that produce the saliva, ductal cells that both modify and transport the saliva, or myoepithelial cells that are thought to both provide the contractile force to induce saliva secretion out of the acini and to maintain tissue architecture (Ogawa, 2003; Mitani et al., 2011), similar to the role of myoepithelial cells in the mammary gland (Hu et al., 2008; Moumen et al., 2011). Two progenitor cell markers have been identified in the mouse SMG that are expressed by cells that can give rise to all of the epithelial lineages of the gland in certain contexts. Cytokeratin 5 (K5) is a basal epithelial cell progenitor marker, and a K5-expressing progenitor cell population that is responsive to parasympathetic innervation was reported to produce both acinar and ductal SMG epithelial cells (Knox et al., 2010). C-kit is a hematopoietic stem cell marker and a progenitor marker in several solid tissues including salivary glands (Kent et al., 2008; Lim et al., 2009), and a c-kit-positive cell population isolated from SMG was reported to differentiate into acinar cells in vitro and to functionally restore saliva secretion in vivo by repopulating the acinar and ductal populations (Lombaert et al., 2008). In the SMG, the developmental origin of the myoepithelial cell population, which surrounds the acinar secretory cells, is less clear. The spatio-temporal developmental distribution of cells expressing these progenitor cell markers and the relationship between these markers has not been reported. Additionally, the distribution of the early differentiation markers of acinar epithelial cells throughout development has not been reported.
In this study, we profiled the spatio-temporal expression patterns of the K5 and c-kit epithelial progenitor markers together with epithelial differentiation markers throughout SMG development. To accomplish this, we utilized a quantitative serial multiplexed immunohistochemistry technology, referred to as multiplexed immunofluorescence microscopy (MxIF). We used image analysis algorithms to identify single cells and quantify protein expression of 20 proteins within individual cells in the same tissue sections throughout a developmental time-course. Using these methods, together with Pearson's correlation analysis coupled to a visual display of the image data, we performed pairwise comparisons of multiple markers in the same tissue sections to quantify the spatio-temporal distribution of cells positive for multiple progenitor and differentiation markers over time. Our results highlight the progressive association of the epithelial and mesenchymal cell populations throughout development that is maintained into adulthood, and identify a likely myoepithelial progenitor population in the developing gland. Our results indicate that the progenitor populations surveyed have differential contributions to SMG development, and that likely cooperate to maintain gland homeostasis.
Materials and Methods
Tissue microarray (TMA) preparation
Submandibular salivary glands (or salivary glands) were excised from timed-pregnant CD-1 mice (Charles River Laboratories) at embryonic days 12 (E12) through E18 and from postnatal day 1 (P1), P5, and P20 following protocols approved by the University at Albany IACUC committee, as previously described (Daley et al., 2009), with day of vaginal plug discovery defined as E = 0. Glands were immediately fixed in 10% neutral buffered formalin (Sigma HT5011), dehydrated, and embedded in paraffin wax using a tissue processor (Shandon Citadel 2000) following standard methods at the University at Albany Histology Core Facility. Cores from paraffin blocks were used to construct a developmental tissue microarray (TMA) using at least three sections of salivary glands from embryonic days E12, 13, 14, 15, 16, 17, 18 and post-natal days P1, 5 and 20. To construct the 104 spot array, 1.5 mm diameter tissue plugs were removed from paraffin blocks and placed into a donor paraffin block in a random arrangement by a commercial vendor (Pantomics, Inc, Richmond, CA). Each developmental stage was represented by an average of 7 tissue plugs (range: 3–11). 5 µm sections of each tissue array were cut from the TMAs and were placed onto Superfrost Plus Slides (Electron Microscopy Sciences 71869-10) by Pantomics.
Antibody validation
Since antibody specificity is required for MxIF, antibody specificity was verified through a series of experiments, including Western analysis and immunohistochemistry in submandibular salivary gland tissues of an appropriate stage. To predict the timing of protein expression, RNA expression was examined using the Salivary Gland Molecular Anatomy Map http://sgmap.nidcr.nih.gov/sgmap/sgexp.html. When peptides representing the epitope were available, peptide preabsorbed antibodies were exposed to salivary gland formalin-fixed, paraffin-embedded (FFPE) sections to verify disappearance of the staining pattern (data not shown). All staining patterns on FFPE sections were also verified in whole mount salivary gland tissues fixed in 4% paraformaldehyde and 5% sucrose in 1× PBS, subjected to immunocytochemistry, and imaged using laser scanning confocal microscopy (510 Meta, Zeiss or SP5, Leica) (Larsen et al., 2003; Daley et al., 2009; Daley et al., 2011; Daley et al., 2012; Sequeira et al., 2012). The antibodies used in this study are listed in Table 1, and the order in which the immunohistochemistry steps were performed is listed in supplementary material Table S1. The antibody to Mucin 10 was generated against the peptide C-QFPVRKYLEDPRY by Everest Biotech for this study. The antibody recognizing SABPA was raised against a mouse SABPA cDNA GST-fusion protein (Dr Lily Mirels, personal communication).
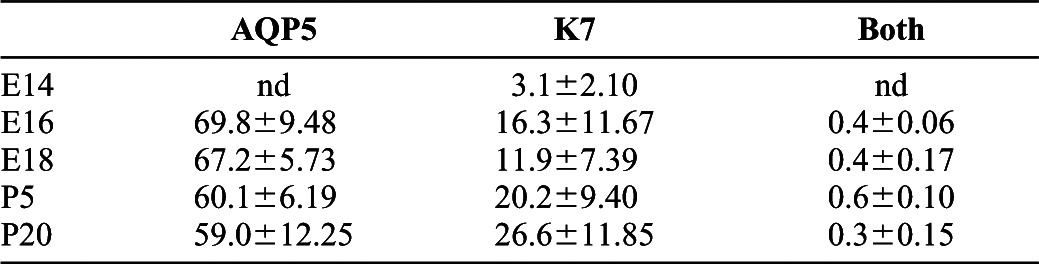
Table 2. Percentages of acinar and ductal marker-expressing cells throughout SMG development.
Single cell analysis data from the MxIF of the SMG developmental TMAs and overlays of statistical data onto the immunostains were used to calculate the percentages of total epithelial cells segmented expressing the proacinar/acinar and ductal cell markers AQP5 and K7, respectively, as represented in </emph>Fig. 3. Averages of three counts for three positions for each developmental stage are shown with standard deviations as total percent of cells positive for each marker alone and total percent positive for both makers. nd denotes conditions where immunoreactivity was not detected. Note the near complete lack of co-expression of these lineage markers throughout development.
Antibody conjugation and validation
Except for antibodies used in the first round by standard indirect immunochemistry using fluorophore-conjugated secondary antibodies (Jackson Immunoresearch) and one antibody that was detected using a zenon-based detection (Life Technologies), all antibodies that passed the specificity tests were directly conjugated to a cyanine dye (Cy3 or Cy5) at available lysines using N-hydroxysuccinimide chemistry using antibody labeling kits as per the manufacturer’s instruction (GE Healthcare). Two dye to antibody ratios were tested for each antibody to achieve optimal sensitivity. To verify specificity of the direct conjugates, appropriate tissue sections were exposed to direct conjugates in phosphate-buffered saline solution (PBS) for 45 min at room temperature in a humidified chamber and the staining pattern compared to that produced with indirect immunohistochemistry. The optimal antibody dilution was determined in independent experiments. Stained slides were then exposed to a chemical inactivation agent to confirm the elimination of fluorescent signal, which is necessary for MxIF.
Serial multiplexed immunofluorescence microscopy (MxIF)
In preparation for MxIF, slides were heated at 60°C for 90 min. Paraffin was removed using Histochoice Clearing agent (Amresco) (2×, 10 min), 100% EtOH (2×, 10 min), 95% EtOH (2×, 10 min), 70% EtOH (2× 10 min), 0.3% Triton X-100 in PBS (10 min), and PBS (3×, 10 min). Antigen retrieval was performed in a pressure cooker in a proprietary antigen retrieval solution developed at the GE Global Research Center (US patent #8,067,241). Sections were blocked in a solution containing 3% BSA (Sigma) and 10% donkey serum (Jackson ImmunoResearch) in PBS for 2 hours at RT. Antibodies were diluted in a 3% BSA solution in PBS, incubated on sections at room temperature for 45 min in a humid chamber, washed, and incubated in 0.5 µM 4′,6-diamidino-2-phenylindole (DAPI) to stain nuclei (Life Technologies). Antigens were detected by serial applications of Cy3 and Cy5 conjugated antibody pairs followed by dye inactivation prior to the next round of antibody application. Dye inactivation was performed by a 10 min incubation at RT with dye inactivation solution (US patent #7,741,045) followed by a 10 min wash in PBS. Bleaching was confirmed by capturing images of the same region at the same exposure time required to capture the original image. Details of antibody suppliers are provided in Table 1 and the sequence of antibody application is detailed in supplementary material Table S1.
Table 1. Antibody information.
Antibody targets and abbreviations used, suppliers, catalog numbers, and formats for direct conjugates.
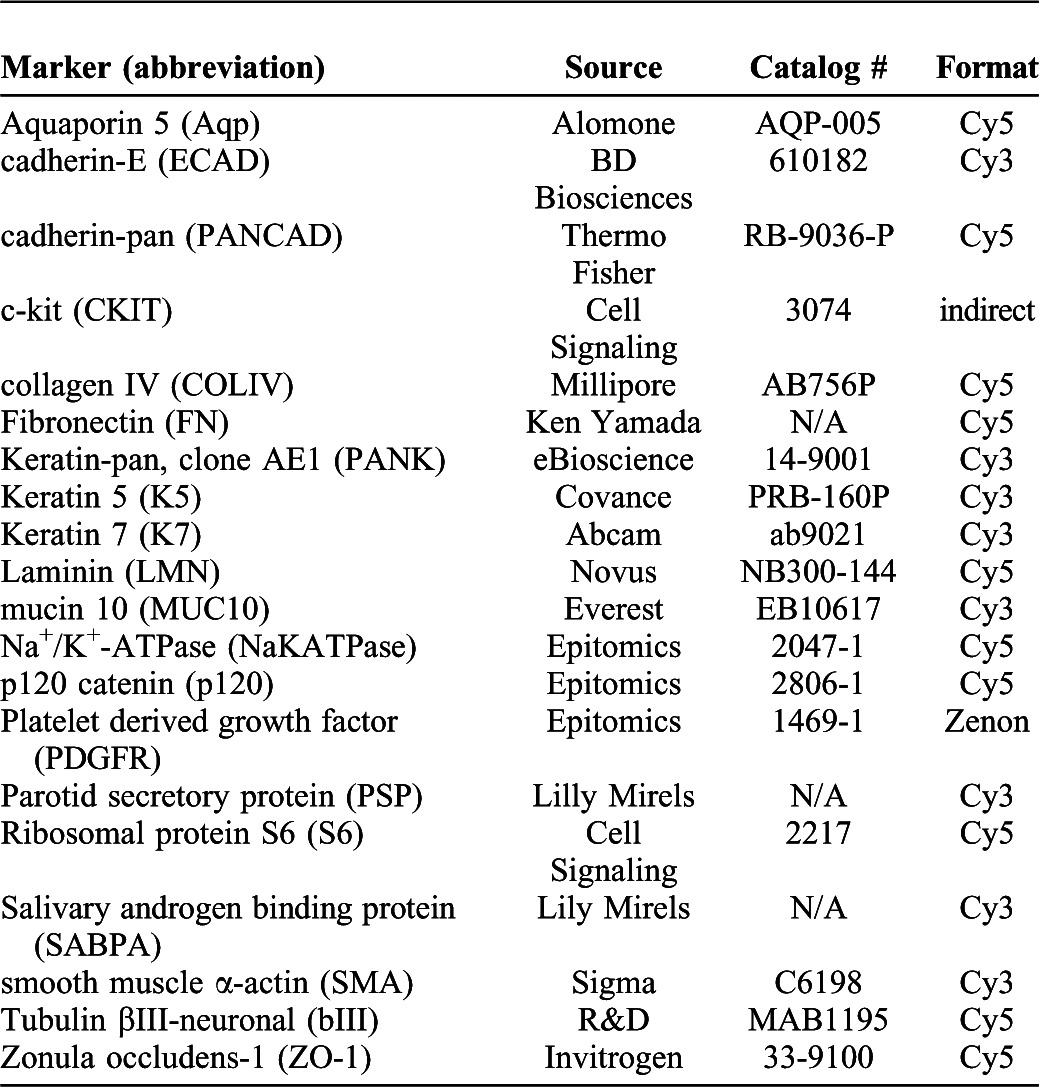
Image capture was performed, as described, using an automated Olympus IX-81 microscope that was outfitted with software developed at GE Global Research to drive acquisition utilizing a piezo-driven automated stage (Prior Scientific) and a Peltier-cooled CCD camera (Q Imaging RET-4000DC-F-M-12). Multiple locations were identified (one representative location per spot on the TMA), and this (x,y) coordinate TMA map was saved. In the first round, the DAPI-stained image was used to detect cells but images were captured using all channels. This first set of images was used for background subtraction for each channel except for the DAPI channel. For subsequent rounds, the TMA map was recalled and the slide adjusted to correspond with the starting position of the first spot. One optimized image exposure time was applied to all locations on the TMA for detection of any given antigen so that staining intensities could be directly compared at all stages of development. Due to the large dynamic range of some expression patterns throughout development, it was not possible to obtain a single exposure time that was optimal for all locations; therefore, some low intensity staining was lost at specific locations and some regions were slightly overexposed. A Brenner gradient-based autofocusing routine (Yazdanfar et al., 2008) was performed prior to image capture at each location. Images were acquired using an Olympus U-PLAN S-APO 20X, 0.75 N.A. objective and saved in TIFF format. Collection of images at 20× with a high N.A. objective allows both representation of the tissue structure with larger numbers of cells and quantitative analysis of the expression patterns in individual cells.
Single cell segmentation and quantification
Images captured at the University at Albany were transferred to a GE fully automatic and high-throughput image analysis system for quantitative image processing, as described. Cell segmentation was performed by the following steps: 1) alignment of all of the image sets via the bleaching-resistant DAPI channel (Can et al., 2008), 2) removal of autofluorescence using the first round of images, and 3) reconstruction of the epithelial tissue architecture at the sub-cellular level. The epithelial region was segmented using the staining pattern produced by both pan-cytokeratin and E-cadherin antibodies. Epithelial plasma membranes were detected using a combination of the staining patterns represented by membrane markers (Na+/K+-ATPase and pan-cadherin), while cytoplasm was detected using ribosomal protein S6 and nuclei by DAPI. Using a variation of a watershed algorithm, we segmented individual cells, assigning a unique ID to each epithelial cell. A wavelet-based nuclei detection algorithm was applied to segment the nuclei (Padfield et al., 2008) and a variation of the probabilistic method described by Can et al. was applied to segment the membrane and cytoplasm (Can et al., 2009). All pixels were digitally compartmentalized within epithelial cells as: nuclei, membrane, or cytoplasm using detection algorithms previously described (Can et al., 2008). A similar image analysis routine was applied to a study on c-Met distribution in colon cancer patients (Ginty et al., 2008).
We quantified both morphological (cell level) and protein-specific features (sub-cellular level) in the epithelial cells. Structures that did not meet criteria for designation of a cell were not included in statistical results; however, parameters were optimized to avoid loss of cells. We computed protein features from the autofluorescence-removed images to assign a unique sub-cellular compartment (nuclei, membrane or cytoplasm) to each detected pixel, as described (Ginty et al., 2008). We then validated the cell segmentation algorithm by visually inspecting the segmented image results with the staining patterns used for the segmentation by overlaying the segmentation masks onto biomarker images using visualization software.
Subcellular compartment data was also used to computationally generate a virtual representation of the tissue structure analogous to a standard hematoxylin and eosin (H&E)-stained tissue section, where the nuclei are shown in purple to simulate hematoxylin staining and the non-nuclear tissues are shown in pink to simulate eosin staining.
Analysis of image staining patterns and creation of multiple channel overlays of up to 10 biomarker images per spot of the TMA was performed using visualization software. The color assigned to each biomarker, the contribution of the biomaker to the final color image, and the transparency of the stain were changed to produce color blended images that highlighted staining patterns in the tissue spot. Image overlays were exported from the visualization software as png files, cropped, and processed using the levels command in Adobe Photoshop (Adobe) to produce the final figures; some color overlays were prepared using Adobe Photoshop.
Statistical analysis and quantification
Low quality images were eliminated from analysis using an image quality algorithm to identify images lacking a stain or out of focus images. Statistical analysis was performed on the remaining images using the median pixel intensity at each location on a per cell basis. An additional processing step was performed to eliminate staining within structures that did not meet all criteria to be considered a cell. Pearson's correlation analysis was performed for all cells per spot and the correlation coefficients were then averaged across all the spots for that stage. Correlation results for each dataset were incorporated into an image viewing software tool, and visualized as an overlay onto the relevant marker immunostain pattern. Thresholds for each marker were set by visual comparison of the overlay with the corresponding immunostain for each image, minimizing the likelihood of false positives. The number of positive cells or double-positive cells exceeding the chosen thresholds was quantified.
Website
A website was created where users can view a subset of the dataset described in this manuscript that includes all time-points analyzed: http://sgdatlas.rit.albany.edu. Users can either compare staining patterns produced from antibodies across multiple stages or can compare multiple antibodies within a single stage. One representative image for each marker from each developmental stage is accessible through the web tool.
Results
Multiplex analysis of submandibular salivary gland morphogenesis
To examine the spatio-temporal distribution of differentiating epithelial cells and their progenitors during SMG development, we constructed a tissue microarray (TMA) encompassing the mouse SMG initial bud stage at embryonic day 12 (E12) through the juvenile stage just prior to sexual dimorphism, postnatal day 20 (P20), collecting tissues at 24 hour increments. Serial MxIF enables multiple rounds of immunohistochemistry to be performed sequentially on a single tissue section to examine the distribution of large numbers of antigens in the exact same cells. Directly conjugated antibodies to detect tissue compartments, cell differentiation state, and progenitor cell markers were applied sequentially to the SMG developmental TMA with all markers and corresponding antibody probes listed in Table 1 in the order listed in supplementary material Table S1. Following image processing, developmental time-points representing major stages of embryonic and post-natal SMG development were selected for further analysis: E14/branching morphogenesis/pseudoglandular stage, E16/onset of cytodifferentiation/canalicular stage, E18/expansion of terminal tubule proacinar cells/terminal bud stage, P5/immature, and P20/juvenile prior to overt sexual dimorphism, as previously defined (Melnick and Jaskoll, 2000; Tucker, 2007). Expression profiles for 20 individual immunohistochemical markers are shown for these developmental stages in supplementary material Fig. S1. Protein expression patterns for these antigens at all of the developmental time-points can be viewed at the interactive website http://sgdatlas.rit.albany.edu. Overlays of a subset of markers depicting structural evolution of the glands are shown at two different zoom levels in Fig. 1A, using antibodies to E-cadherin, laminin, and βIII tubulin to detect the epithelium, basement membrane, and neuronal cells, respectively. Platelet derived growth factor receptor (PDGFR) was used to identify salivary gland mesenchymal cells (Yamamoto et al., 2008). Profiles for the basement membrane protein, collagen IV, and the basement membrane and mesenchymal extracellular matrix protein, fibronectin, mirrored the expression profiles for laminin and PDGFR, respectively (supplementary material Fig. S1). Original full size overlays with boxes indicating the regions used for the first zoom level in Fig. 1 are shown in supplementary material Fig. S2. Significantly, the interactions between the stromal fibroblasts and neurons became progressively tighter and more extensive during mesenchyme condensation and epithelial innervation, and these epithelial mesenchyme interactions were maintained throughout development and gland maturation.
Fig. 1. MxIF analysis of mouse submandibular salivary gland morphogenesis.
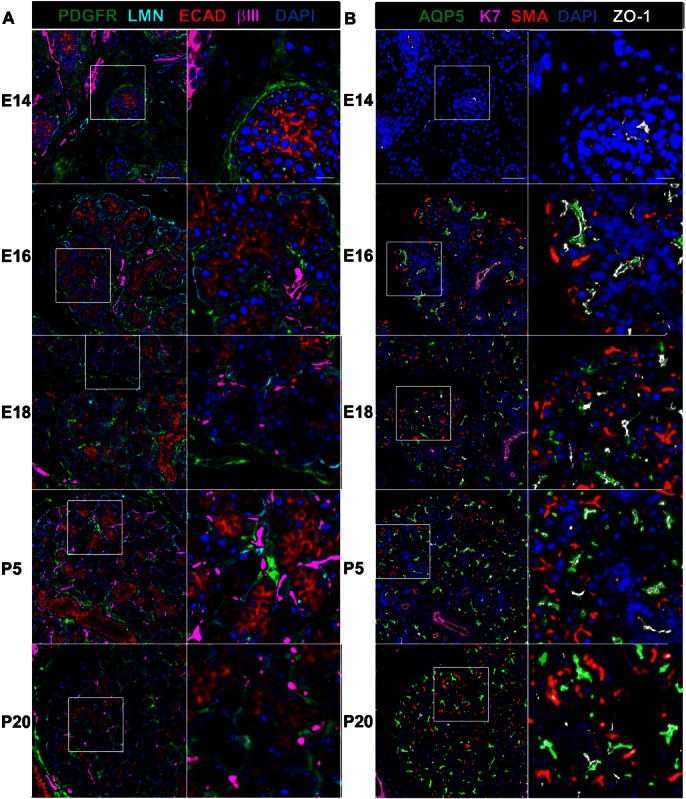
MxIF of a developmental TMA including embryonic stages (E14, E16, E18) and postnatal stages (P5 and P20) was performed using sequential application of directly conjugated antibodies to detect multiple markers of tissue structures and cell types on the same tissue sections. (A) Tissue compartments. The epithelium, mesenchyme, neurons, and basement membranes was detected using antibodies directed towards E-cadherin (ECAD, red), platelet-derived growth factor (PDGFR, green), βIII tubulin (bIII, magenta), and laminin (LMN, cyan), respectively. (B) Epithelial differentiation. Maturation of the proacinar, ductal, and myoepithelial cell types was detected using antibodies to aquaporin 5 (AQP5, green), cytokeratin 7 (K7, magenta), and smooth muscle α-actin (SMA, red); maturation of the cell–cell adhesions was monitored using an antibody to zonula occludens-1 (ZO-1, white). DAPI was used to stain the nuclei in both A and B. Scale bars: 50 µm zoom level one and 10 µm zoom level two.
From one TMA subjected to MxIF, image overlays of the ductal marker cytokeratin 7 (K7) (Walker et al., 2008), the proacinar and acinar cell marker aquaporin 5 (Aqp5) (Larsen et al., 2011), and the myoepithelial marker smooth muscle α-actin (SMA) (Ogawa, 2003; Mitani et al., 2011), shown at two different levels of zoom, demonstrate the evolution of the epithelial compartment from a simple primary bud connected to a single solid stalk of cells to a more complex interconnected structure containing extensive arrays of acini connected by a network of hollow ducts (Fig. 1B). Original full size overlays with boxes indicating the regions used for the first zoom level in Fig. 1 are shown in supplementary material Fig. S2. The tight junction protein zonula occludens-1 (ZO-1), which is also expressed in the endothelial cells of blood vessels in the stroma, demonstrates the progressive formation of apical surfaces in the epithelium from proximal to distal regions in the presumptive ducts followed by the terminal end buds and developing acini, as reported previously (Hieda et al., 1996; Hashizume et al., 2004; Hashizume and Hieda, 2006; Walker et al., 2008). Variation in the intensity of the general epithelial markers changes over time with E-cadherin and pan-cytokeratin generally showing greater reactivity towards cells having a ductal morphology. K7 was detected primarily at the apical membrane of larger ductal structures, but was not expressed at high enough levels to be detected in many of the presumptive ducts and smaller ductal structures that demonstrated higher levels of pan-cytokeratin. The cadherin-associated protein, p120 catenin, also showed greater immunoreactivity in mature ducts than in mature acinar cells, in addition to its expression in the stromal vasculature and neurons (supplementary material Fig. S1).
Interestingly, epithelial SMA protein expression was detected in the outer cells of the epithelial buds beginning at E16 as the morphology of the outer cells transitioned from columnar to a more cuboidal shape. By E18, these outer cells that consistently maintain contact with the basement membrane had assumed the compact, extended shape characteristic of myoepithelial cells. This suggests that SMA is a marker for both committed submandibular salivary gland myoepithelial progenitor cells and mature myoepithelial cells, consistent with its expression by proliferating unipotent myoepithelial progenitor cells that have been described in developing mammary glands (Van Keymeulen et al., 2011) and by isolated multipotent mammary progenitor cells in vitro (Zhao et al., 2012). Concomitant detection of these epithelial subtypes provides a platform to address additional molecular markers in the context of the intact tissue.
Quantitative single cell detection
To systematically quantify the representation of each epithelial cell type in developing salivary glands, it was necessary to develop algorithms that could identify the epithelial cells and quantify the number of cells in each subpopulation as a percentage of the total. MxIF provides the opportunity to perform quantitative single cell analysis from image data, since individual epithelial cells can be identified using segmentation algorithms and multiple marker expression patterns, and levels can be compared in the same tissue sections. The epithelial cell markers pan-cytokeratin and E-cadherin were used together to define the epithelial compartment and computationally generate an epithelial mask (Fig. 2A). To perform quantitative single cell analysis, an algorithm was developed to recognize a cell as a structure that has a nucleus (identified by DAPI staining), a plasma membrane (identified using both Na+/K+-ATPase and pan-cadherin), and cytoplasm (recognized by the cytoplasmic ribosomal protein S6). Additional measured morphological properties were used to improve the recognition mechanisms by excluding partial cells. This provided systematic segmentation of the individual epithelial cells for further analysis. A subset of this data was used to computationally generate a pseudocolored hematoxylin and eosin (H&E)-like image to verify the tissue architecture in standard histological fashion (Fig. 2B). Full size images were used for all subsequent quantitative analyses to maximize sample sizes.
Fig. 2. Single cell segmentation of the epithelium and comparison with simulated hematoxylin and eosin (H&E) stained images.
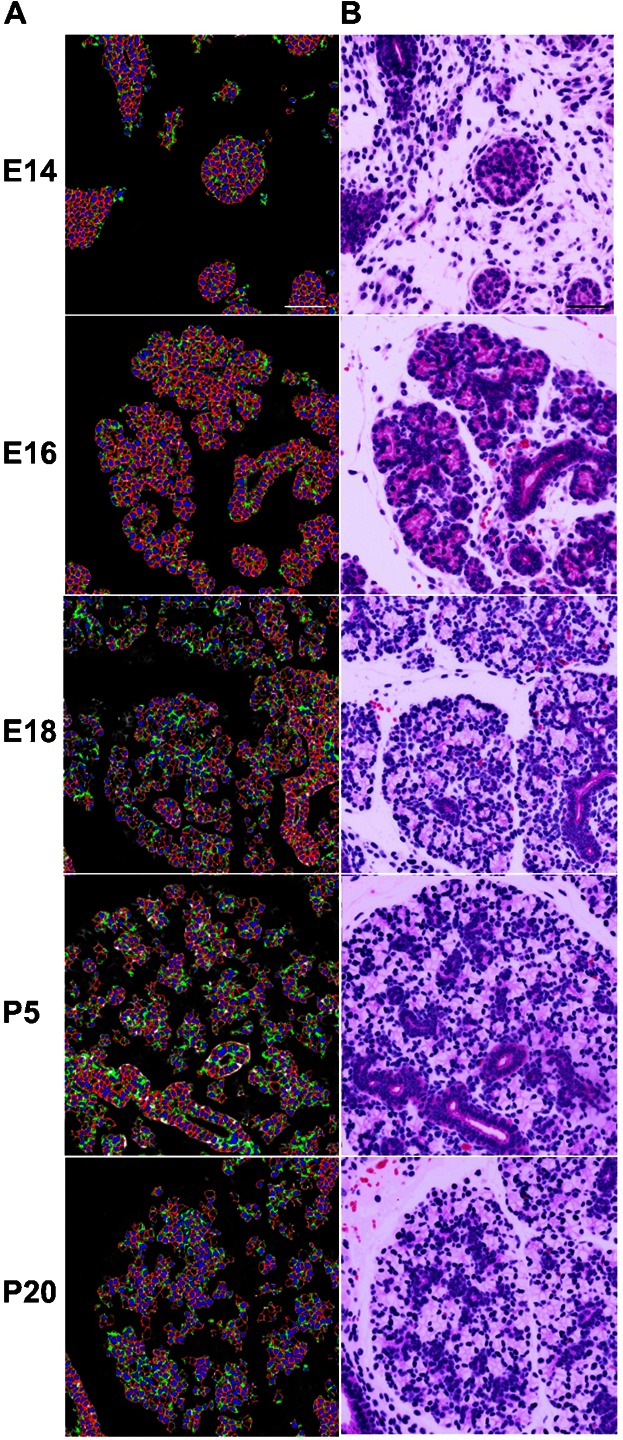
(A) Using a developmental TMA, MxIF was used to identify markers of cell type and cell subcompartments: epithelium (E-cadherin and pan-keratins), plasma membrane (Na+/K+-ATPase and pan-cadherin), cytoplasm (S6), and nuclei (DAPI) (supplementary material Fig. S1). An epithelial mask was computed using the E-cadherin and pan-cytokeratin stains and used to identify cells with the algorithm that uses the cell membrane, cytoplasm, and nuclei stains to identify individual cells. Computationally segmented epithelial cell membranes are displayed in (red), cytoplasm (green), and nuclei (blue), and are displayed on top of an image of E-cadherin to identify the epithelial cells. Each cell was assigned a unique identifier (data not shown) for quantification. (B) The cell segmentation in A was used to computationally generate corresponding simulated histological H&E images, which are displayed for comparison to illustrate the tissue morphology. Scale bars: 50 µm.
Quantitative single cell analysis of acinar vs ductal cell differentiation
The acinar cell lineage is of interest as the cell type that produces saliva and loses function with salivary hypofunction. The water channel protein aquaporin 5 (Aqp5) is an early marker of proacinar cells in developing SMG, whose expression is retained in mature acinar cells and at much lower levels in acinar-proximal intercalated duct cells but is absent in other mature duct cells (Larsen et al., 2011). In the developmental TMAs, Aqp5 protein was first detected at E15 (data not shown), after the onset of the ductal differentiation marker K7, with a primarily membranous localization that became more concentrated at the apical surface by E16 (Fig. 1; supplementary material Fig. S1). Using the cell segmentation algorithms and the pixel values for acinar and ductal markers, we computationally classified the epithelium as presumptive/mature ductal and proacinar/acinar cell populations using staining patterns for K7 and Aqp5 (Fig. 3). Using Pearson's correlation coefficient calculations to compare pairs of markers, we quantified the number of cells expressing ductal and proacinar markers. With ductal cell differentiation markers preceding acinar cell differentiation markers, only ductal and non-differentiated epithelial cells are detected at E14, with K7-positive cells comprising only 3% of the population. Note that early SMG epithelial cells are largely negative for acinar, ductal, and myoepithelial cell differentiation markers, consistent with their developmental plasticity (Wei et al., 2007). As development proceeds, the percentage of cells expressing acinar and ductal lineage markers expands rapidly and is sustained throughout development with virtually no overlap of ductal and acinar marker expression (Fig. 3; Table 2).
Fig. 3. Quantitative spatio-temporal analysis of proacinar and ductal cell populations during submandibular development.
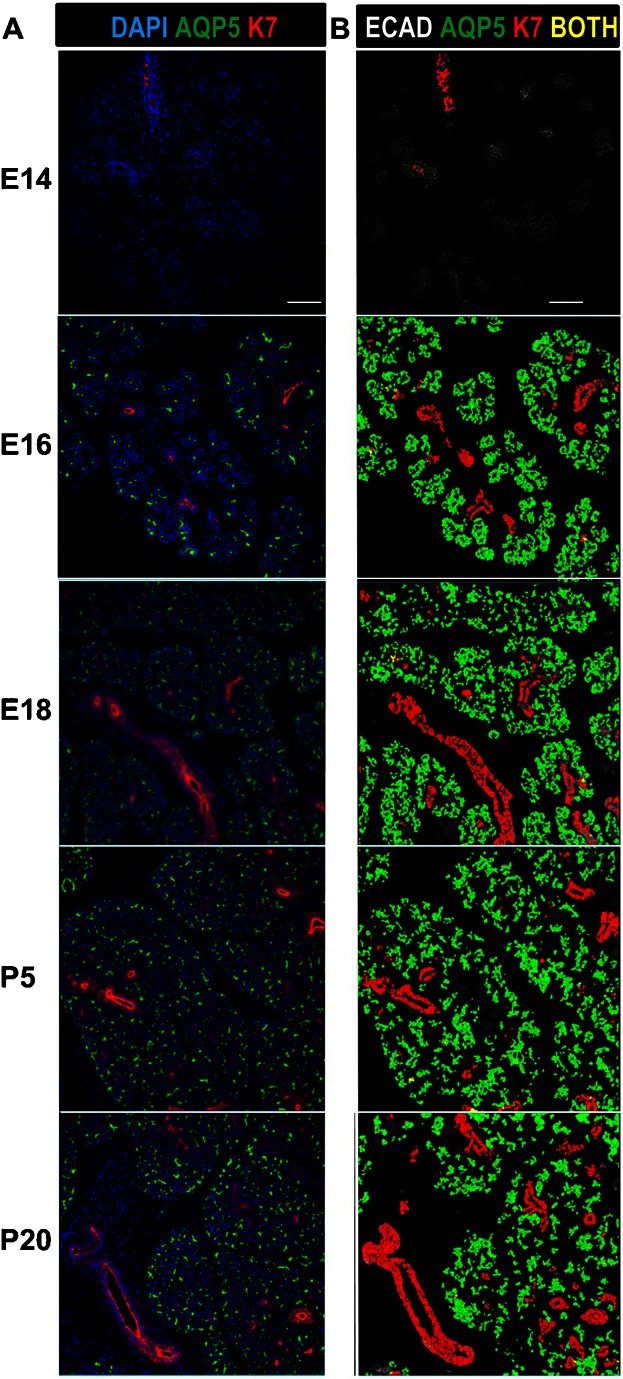
(A) The proacinar and ductal SMG cell populations were identified using antibodies to detect AQP5 (green) or K7 (red), respectively, with DAPI staining of total nuclei (blue). (B) Statistical outputs from A were overlaid on E-cadherin-stained images (white) to produce overlays of the computationally identified proacinar (green) and ductal (red) epithelial cell populations. Cells that segregate both as acinar and ductal are labeled in yellow. Note that almost all cells are either proacinar or ductal, with very little overlap detected between these lineage markers. Developmental stages are as indicated. Numerical data for the quantitative cell analysis is shown in Table 2. Scale bars: 100 µm.
Quantitative single cell analysis of secretory acinar cell differentiation
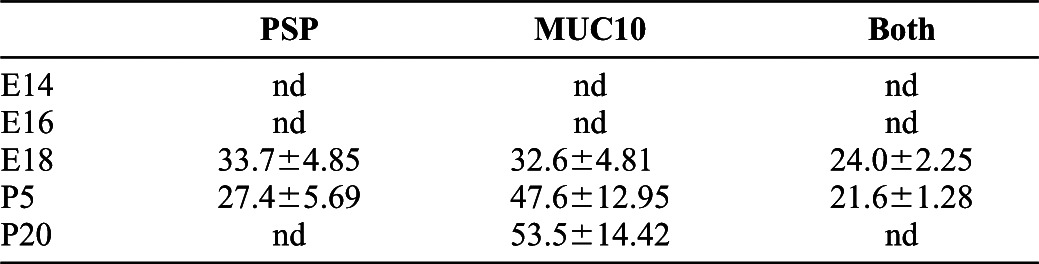
Secretory acinar differentiation can be tracked using antibodies to detect SMG secretory proteins. Parotid secretory protein (PSP)/BPIFA2E is expressed transiently in a secretory progenitor population in developing SMG, but is not expressed in mature acinar cells (Ball et al., 2003). Mucin 10 (MUC10) is expressed in developing and mature mucous acinar SMG cells (Denny et al., 1996; Melnick et al., 2001), and SABPA is a mature SMG serous acinar secretory protein (Wickliffe et al., 2002). Our MxIF images revealed that PSP was first detected at E17/E18 in Aqp5-positive proacinar cells (Fig. 4 and data not shown), and expression was subsequently lost as the acini mature with no expression detectable by P20. The mature acinar protein, MUC 10, was detectable by E17 (data not shown) and prenatal expression was largely overlapping with the transiently expressed PSP (Fig. 4; Table 3), indicating that the mature secretory acinar cells develop from cells that co-express the transiently expressed secretory protein PSP. Additionally, SABPA and MUC10 were co-expressed in developing and mature acinar cells (supplementary material Fig. S3), highlighting the mixed seromucous cell phenotype of rodent SMG epithelial cells (Denny et al., 1997; Okumura et al., 2012), differing from human submandibular glands where serous and mucinous acini are discrete cell populations.
Fig. 4. Quantitative spatio-temporal analysis of secretory acinar cell differentiation during submandibular development.
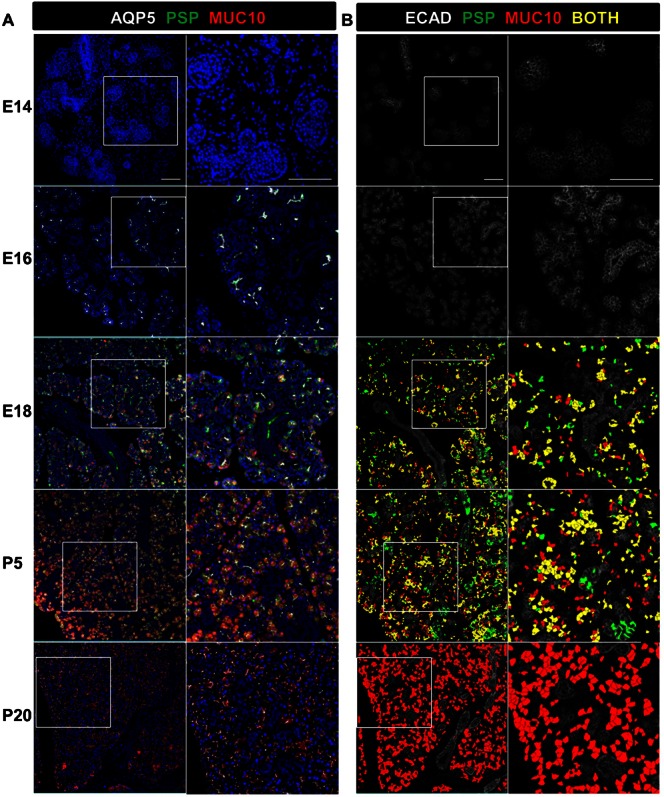
(A) Maturation of SMG epithelial secretory cell differentiation is shown by MxIF of the secretory proteins PSP (green) and MUC10 (red) together with DAPI staining of total nuclei (blue) and AQP5 expression in the acinar lineage (white) in a low and high magnification view. Inset areas are shown with white dashed lines. (B) Statistical outputs from A were overlaid on E-cadherin-stained images (white) to produce overlays of the computationally identified secretory cells expressing PSP (green) and MUC10 (red). Cells that co-express the transient perinatal protein PSP and the mature secretory protein MUC10 are shown in yellow. Note the late onset of secretory protein expression and extensive co-expression of these secretory products followed by extinction of PSP expression as the glands approach maturity at P20. Developmental stages are as indicated. Numerical data for the quantitative cell analysis is shown in Table 3. Scale bars: 100 µm zoom level one and 50 µm zoom level two.
Table 3. Percentages of secretory acinar marker-expressing cells throughout SMG development.
Single cell analysis data from the MxIF of the SMG developmental TMAs and overlays of statistical data onto the immunostains were used to calculate the percentages of total epithelial cells expressing the transient perinatal secretory protein, PSP, and the mature SMG secretory protein, MUC10, respectively, as shown in </emph>Fig. 4. Averages of three counts from three positions for each developmental stage are shown with standard deviations for total percent of cells positive for each marker alone and total percent positive for both makers. nd indicates not detected. Extensive co-expression of these secretory markers was revealed during the perinatal stages of development.
Quantitative single cell analysis of progenitor cell distributions
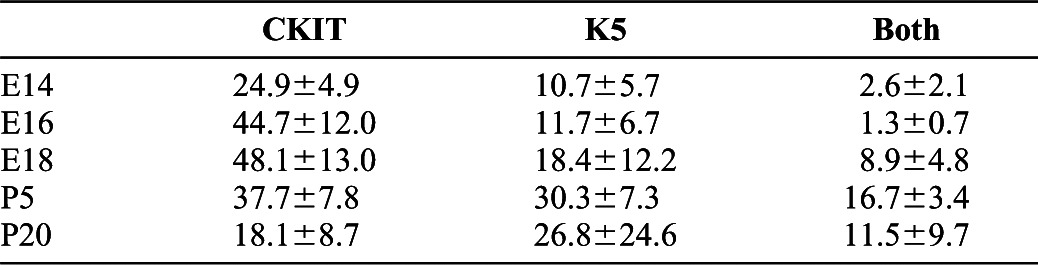
The SMG progenitor cell populations have not been precisely defined during development, although K5-positive and c-kit-positive cells have been reported to produce both acinar and ductal populations under specific circumstances. Serial MxIF was used to define the relationships between the K5- and c-kit-positive progenitor cell populations (Fig. 5), using quantitative single cell analysis to determine the percentages of single and double positive progenitor marker expressing cells in the epithelium at each developmental stage (Table 4). Original full size overlays used for quantitative analyses with boxes indicating the regions used for the first zoom level in Fig. 5 are shown in supplementary material Fig. S4. Shown at two different zoom levels in Fig. 5, in early development (E13–E16), anti-K5 and anti-c-kit antibodies labeled largely distinct cell populations, with K5-positive cells primarily localized to the presumptive ducts and c-kit-positive cells primarily localized to the end buds where they partially co-localized with Aqp5 at the onset of Aqp5 expression (supplementary material Fig. S1 and data not shown). In contrast, partial co-localization of K5 and c-kit was observed as c-kit began partitioning into the ducts later in development (E18). Both markers were primarily ductal and partially overlapping in the mature glands except for the K5-positive, SMA-positive myoepithelial cells that are c-kit negative, although difficulties with concise segmentation of the elongate and stellar K5 basal cells may result in undersegmentation of this population and overestimation of the percent overlap of K5 and c-kit at later developmental stages. These data highlight the dynamic expression patterns of these two SMG progenitor cell markers throughout gland development. Whereas K5 was primarily localized in the developing and mature ducts, c-kit was found to be largely restricted to the end buds and proacinar cells during early development, with a striking and progressive partitioning into the ducts during late prenatal and postnatal development.
Fig. 5. Dynamic expression of epithelial progenitor cell markers during submandibular gland development.
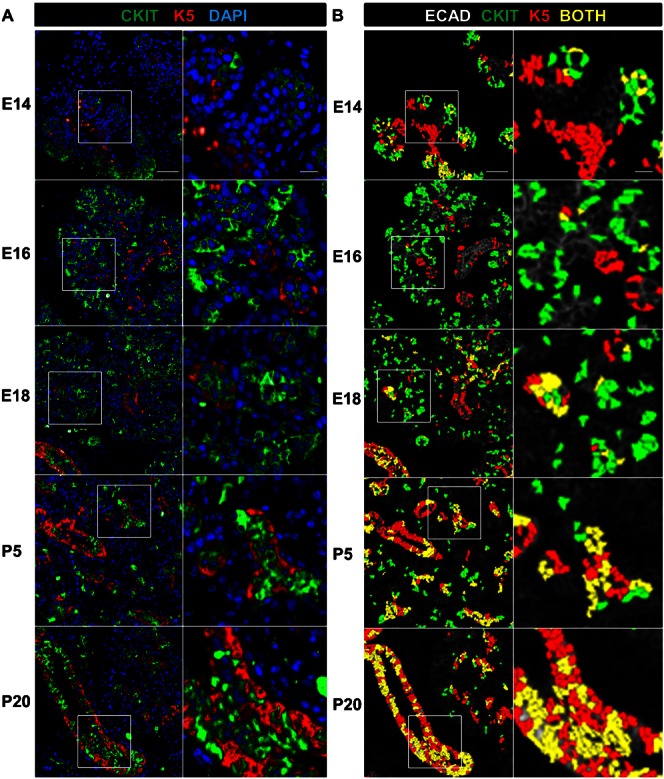
(A) The spatio-temporal distribution of the functional epithelial progenitor markers c-kit (CKIT, green) and keratin 5 (K5, red) are shown throughout SMG salivary gland development, with total nuclei stained with DAPI (blue) in a low and high magnification view. Inset areas are shown with white lines. (B) Statistical outputs from A were overlaid on E-cadherin-stained images (white) to produce overlays of the computationally identified CKIT-(green) and K5-(red) expressing epithelial progenitor cell populations. Cells that segregate as expressing both as c-kit and K5 are shown in yellow. Note the early expression of c-kit primarily in the end buds followed by progressive partitioning to the ducts, whereas the K5 expressing cells are largely ductal throughout development. Developmental stages are as indicated. Numerical data for the quantitative cell analysis is shown in Table 4. Scale bars: 50 µm zoom level one and 10 µm zoom level two.
Table 4. Percentages of progenitor marker-expressing cells throughout SMG development.
Single cell analysis data from the MxIF of the SMG developmental TMAs and overlays of statistical data onto the immunostains were used to calculate the percentages of total epithelial cells segmented expressing the progenitor cell markers c-kit (CKIT) and keratin 5 (K5), as shown in </emph>Fig. 5. Averages of three counts for three positions for each developmental stage are shown with standard deviations for total single- and double-positive cells. Note the largely exclusive expression of these progenitor markers early in development and the partial overlap later in development as c-kit becomes progressively restricted to the ducts.
Discussion
In this study we performed a quantitative single cell analysis of progenitor and differentiation markers within the context of submandibular salivary glands over a developmental time course. This is the first study to quantitatively delineate the spatio-temporal distribution of multiple cell differentiation and progenitor cell markers together in the same cells during SMG development. We tracked the spatio-temporal expression of multiple previously reported epithelial cell differentiation markers, including aquaporin 5, PSP, mucin 10, and SABPA (acinar), K7 (ductal), and smooth muscle α-actin (myoepithelial). Since c-kit- and K5-expressing cells have been shown to be capable of producing or reconstituting the epithelial compartment in submandibular salivary glands and are also of interest as potential diagnostic and/or therapeutic targets in cancers (Chu and Weiss, 2002), we examined the spatio-temporal distribution of cells expressing these markers. K5 expression was generally restricted to the ductal cells throughout embryonic and postnatal development, consistent with previous studies suggesting that progenitor cells reside in the ducts in mature submandibular glands (Denny et al., 1997; Man et al., 2001; Lombaert et al., 2008). In contrast, c-kit was not restricted to ducts, but rather was expressed by a distinct population throughout development and a percentage of these cells were found to reside in ductal cells. We also identified SMA-positive, K5-negative cells that are basally restricted in proacinar salivary gland end buds that may be proliferating myoepithelial progenitors that are functionally distinct from progenitor cells for the secretory acinar and ductal cells.
Interestingly, our data indicate that the spectrum of cell types that are c-kit positive changes developmentally and suggests that multiple progenitor cell populations may contribute to SMG development and tissue homeostasis. Additional studies in salivary and other glands have also revealed that multipotent embryonic progenitor cells seem to temporally transition to more restricted progenitor cell types to regulate normal gland homeostasis; however, the multi-lineage potential of such cells can be revealed during tissue regeneration or following transplantation. In studies by Kishi et al., the colony-forming potential of cell populations derived from neonatal rat SMG had greater multi-lineage differentiation ability than did adult SMG cell populations (Kishi et al., 2006). Interestingly, lineage analyses in the mammary and sweat glands have demonstrated that early embryonic multipotent progenitor populations transition to multiple distinct restricted progenitor populations later in development to control normal gland homeostasis, but that these cells can revert to a more embryonic or multi-potent state upon injury or in transplantation scenarios (Van Keymeulen et al., 2011; Lu et al., 2012). Further, cancer cells frequently upregulate progenitor markers, including K5 and c-kit. Thus, our data supports the model that the developmental plasticity of a cell expressing a given progenitor marker is a function of its maturation stage and its tissue context.
Although myoepithelial cells have long been known to encircle the acinar epithelial cells in the submandibular gland (Ogawa, 2003), a myoepithelial progenitor population has not previously been described in this organ. We found that SMA is expressed early in an apparent myoepithelial progenitor population in the developing SMG epithelium adjacent to the basement membrane, prior to both K5 protein expression and terminal myoepithelial morphological differentiation. In the mammary gland, myoepithelial cell differentiation has been investigated in detail. Prior to birth, K5-positive basal cells of the ramified ducts are SMA-negative, while SMA-expressing myoepithelial cells develop postnatally and expand during alveolar development during pregnancy and lactation. During puberty, the cap cells of the terminal buds that are proliferating myoepithelial progenitors are SMA positive, but K5 is expressed only very weakly in these cells relative to the rest of the basal epithelia (Moumen et al., 2011). Myoepithelial progenitor cells derived from bipotent K5-positive, SMA-negative mammary progenitors in vitro also display an SMA-positive, K5-negative phenotype (Zhao et al., 2012), and bipotent K5-positive cells that give rise to myoepithelial and luminal cells have been described in the SMG and prostate (Knox et al., 2010), as well as in the mammary glands (Van Keymeulen et al., 2011). Thus, the SMA-positive, K5-negative cells identified in our study that are basally restricted in proacinar salivary gland end buds may be proliferating myoepithelial progenitors that are functionally distinct from progenitor cells for the secretory acinar and ductal cells, similar to the terminal bud cap cells in mammary glands. Although we did not perform time-lapse imaging or lineage tracking in this study, the outer columnar cell population of the immature buds that directly contacts the basement membrane appears to transition into the myoepithelial cell type. Since myoepithelial cells can contribute to both acinar and ductal cell repopulation in rat SMG regeneration models and may again respond to signaling from parasympathetic innervation (Denny et al., 1997; Proctor and Carpenter, 2007; Cotroneo et al., 2008), these cells are of interest in regenerative medicine approaches.
Other progenitor cell markers have been described that are likely to be important for salivary gland development and homeostasis and may be useful in regenerative therapies (Bullard et al., 2008; Okumura et al., 2012; Rugel-Stahl et al., 2012; Arany et al., 2011; Banh et al., 2011; Lombaert et al., 2011; Nanduri et al., 2011; Purwanti et al., 2011; Tran et al., 2011; Palmon et al., 2012). Examination of the relationship of other progenitor markers to c-kit and K5 and to each other awaits further study. Ultimately, functional studies will be required to determine the potency of specific progenitor cell populations under specific growth conditions and the relationship of the progenitor markers to cell lineage, although the context of such lineage analyses can have profound effects on the potential of the progenitor populations being studied. Since marker-expressing cell populations are heterogeneous during development and in pathologies, such as cancer (Potts et al., 2012), and since tissue location is a critical determinant of cell behavior (Pizzo et al., 2005; Johnson et al., 2007), the single cell analysis methods described here will be useful for quantitative characterization of cell subpopulations both during development and disease states. As organ development and homeostasis are orchestrated by dynamic interactions between the progenitor cells, basement membrane, mesenchymal fibroblasts, vasculature, and neurons, our studies further highlight the importance of context for characterizing progenitor cell populations. Extension of these studies will allow for characterization of the progenitor cell niches and the molecular mechanisms that regulate progenitor cell function during development and disease, as a prerequisite for development of regenerative medicine approaches.
Supplementary Material
Acknowledgments
The authors thank Dr David Tieman, Dr Deepa Chitre, Swami Manickam, Christopher Hammond, Dr Zhengu Pang, Dr Colin McCulloch, Dr Meagan Rothney, Dr Kashan Shaikh, and Iza Ferreira for excellent technical assistance and suggestions. The authors would also like to thank Dr Jo Ellen Welch and Donald Matthews at the University at Albany Center for Cancer Genomics for assistance with histology and for use of tissue processing equipment. The authors thank Drs Lily Mirels and Kenneth M. Yamada for generous gifts of antibodies and for valuable conversations. The anti-MUC10 antibody was provided through a grant from Everest Biotech. The authors thank Drs James Castracane and Nathaniel Cady for use of the Leica SP5 confocal microscope. This work was funded by the NIH/NIDCR RC1DE020402 to M.L. and the General Electric Global Research Center, by NIH/NIDCR RO1DE019244 to M.L., and NIH C06 RR015464 to University at Albany, SUNY.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Arany S., Catalán M. A., Roztocil E., Ovitt C. E. (2011). Ascl3 knockout and cell ablation models reveal complexity of salivary gland maintenance and regeneration. Dev. Biol. 353, 186–193 10.1016/j.ydbio.2011.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball W. D., Mirels L., Hand A. R. (2003). Psp and Smgb: a model for developmental and functional regulation in the rat major salivary glands. Biochem. Soc. Trans. 31, 777–780 10.1042/BST0310777 [DOI] [PubMed] [Google Scholar]
- Banh A., Xiao N., Cao H., Chen C. H., Kuo P., Krakow T., Bavan B., Khong B., Yao M., Ha C.et al. (2011). A novel aldehyde dehydrogenase-3 activator leads to adult salivary stem cell enrichment in vivo. Clin. Cancer Res. 17, 7265–7272 10.1158/1078-0432.CCR-11-0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard T., Koek L., Roztocil E., Kingsley P. D., Mirels L., Ovitt C. E. (2008). Ascl3 expression marks a progenitor population of both acinar and ductal cells in mouse salivary glands. Dev. Biol. 320, 72–78 10.1016/j.ydbio.2008.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Can A., Bello M., Tao X., Seel M., Gerdes M. (2008). TMA-Q: a tissue quality assurance tool for sequentially multiplexed Tmas. In Abstracts of the 3rd Intercontinental Congress of Pathology. My 18-21, 2008. Virchows Arch. 452 Suppl. 1, S11 10.1007/s00428-008-0613-x [DOI] [PubMed] [Google Scholar]
- Can A., Bello M., Cline H. E., Tao X., Mendonca P., Gerdes M. (2009). A unified segmentation method for detecting subcellular compartments in immunofluroescently labeled tissue images. International Workshop on Microscopic Image Analysis with Applications in Biology. Sept. 3,4, 2009. [Google Scholar]
- Chu P. G., Weiss L. M. (2002). Expression of cytokeratin 5/6 in epithelial neoplasms: an immunohistochemical study of 509 cases. Mod. Pathol. 15, 6–10 10.1038/modpathol.3880483 [DOI] [PubMed] [Google Scholar]
- Cotroneo E., Proctor G. B., Paterson K. L., Carpenter G. H. (2008). Early markers of regeneration following ductal ligation in rat submandibular gland. Cell Tissue Res. 332, 227–235 10.1007/s00441-008-0588-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley W. P., Gulfo K. M., Sequeira S. J., Larsen M. (2009). Identification of a mechanochemical checkpoint and negative feedback loop regulating branching morphogenesis. Dev. Biol. 336, 169–182 10.1016/j.ydbio.2009.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley W. P., Kohn J. M., Larsen M. (2011). A focal adhesion protein-based mechanochemical checkpoint regulates cleft progression during branching morphogenesis. Dev. Dyn. 240, 2069–2083 10.1002/dvdy.22714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley W. P., Gervais E. M., Centanni S. W., Gulfo K. M., Nelson D. A., Larsen M. (2012). ROCK1-directed basement membrane positioning coordinates epithelial tissue polarity. Development 139, 411–422 10.1242/dev.075366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny P. C., Mirels L., Denny P. A. (1996). Mouse submandibular gland salivary apomucin contains repeated N-glycosylation sites. Glycobiology 6, 43–50 10.1093/glycob/6.1.43 [DOI] [PubMed] [Google Scholar]
- Denny P. C., Ball W. D., Redman R. S. (1997). Salivary glands: a paradigm for diversity of gland development. Crit. Rev. Oral Biol. Med. 8, 51–75 10.1177/10454411970080010301 [DOI] [PubMed] [Google Scholar]
- Ginty F., Adak S., Can A., Gerdes M., Larsen M., Cline H., Filkins R., Pang Z., Li Q., Montalto M. C. (2008). The relative distribution of membranous and cytoplasmic met is a prognostic indicator in stage I and II colon cancer. Clin. Cancer Res. 14, 3814–3822 10.1158/1078-0432.CCR-08-0180 [DOI] [PubMed] [Google Scholar]
- Hashizume A., Hieda Y. (2006). Hedgehog peptide promotes cell polarization and lumen formation in developing mouse submandibular gland. Biochem. Biophys. Res. Commun. 339, 996–1000 10.1016/j.bbrc.2005.11.106 [DOI] [PubMed] [Google Scholar]
- Hashizume A., Ueno T., Furuse M., Tsukita S., Nakanishi Y., Hieda Y. (2004). Expression patterns of claudin family of tight junction membrane proteins in developing mouse submandibular gland. Dev. Dyn. 231, 425–431 10.1002/dvdy.20142 [DOI] [PubMed] [Google Scholar]
- Hieda Y., Iwai K., Morita T., Nakanishi Y. (1996). Mouse embryonic submandibular gland epithelium loses its tissue integrity during early branching morphogenesis. Dev. Dyn. 207, 395–403 [DOI] [PubMed] [Google Scholar]
- Hu M., Yao J., Carroll D. K., Weremowicz S., Chen H., Carrasco D., Richardson A., Violette S., Nikolskaya T., Nikolsky Y.et al. (2008). Regulation of in situ to invasive breast carcinoma transition. Cancer Cell 13, 394–406 10.1016/j.ccr.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. R., Leight J. L., Weaver V. M. (2007). Demystifying the effects of a three-dimensional microenvironment in tissue morphogenesis. Methods Cell Biol. 83, 547–583 10.1016/S0091-679X(07)83023-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent D., Copley M., Benz C., Dykstra B., Bowie M., Eaves C. (2008). Regulation of hematopoietic stem cells by the steel factor/KIT signaling pathway. Clin. Cancer Res. 14, 1926–1930 10.1158/1078-0432.CCR-07-5134 [DOI] [PubMed] [Google Scholar]
- Kishi T., Takao T., Fujita K., Taniguchi H. (2006). Clonal proliferation of multipotent stem/progenitor cells in the neonatal and adult salivary glands. Biochem. Biophys. Res. Commun. 340, 544–552 10.1016/j.bbrc.2005.12.031 [DOI] [PubMed] [Google Scholar]
- Knox S. M., Lombaert I. M., Reed X., Vitale–Cross L., Gutkind J. S., Hoffman M. P. (2010). Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis. Science 329, 1645–1647 10.1126/science.1192046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M., Hoffman M. P., Sakai T., Neibaur J. C., Mitchell J. M., Yamada K. M. (2003). Role of PI 3-kinase and PIP3 in submandibular gland branching morphogenesis. Dev. Biol. 255, 178–191 10.1016/S0012-1606(02)00047-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen H. S., Aure M. H., Peters S. B., Larsen M., Messelt E. B., Galtung H. K. (2011). Localization of AQP5 during development of the mouse submandibular salivary gland. J. Mol. Histol. 42, 71–81 10.1007/s10735-010-9308-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E., Vaillant F., Wu D., Forrest N. C., Pal B., Hart A. H., Asselin–Labat M. L., Gyorki D. E., Ward T., Partanen A.kConFab (2009). Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 15, 907–913 10.1038/nm.2000 [DOI] [PubMed] [Google Scholar]
- Lombaert I. M., Brunsting J. F., Wierenga P. K., Faber H., Stokman M. A., Kok T., Visser W. H., Kampinga H. H., de Haan G., Coppes R. P. (2008). Rescue of salivary gland function after stem cell transplantation in irradiated glands. PLoS ONE 3, e2063 10.1371/journal.pone.0002063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaert I. M., Knox S. M., Hoffman M. P. (2011). Salivary gland progenitor cell biology provides a rationale for therapeutic salivary gland regeneration. Oral Dis. 17, 445–449 10.1111/j.1601-0825.2010.01783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Werb Z. (2008). Patterning mechanisms of branched organs. Science 322, 1506–1509 10.1126/science.1162783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Sternlicht M. D., Werb Z. (2006). Comparative mechanisms of branching morphogenesis in diverse systems. J. Mammary Gland Biol. Neoplasia 11, 213–228 10.1007/s10911-006-9027-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C. P., Polak L., Rocha A. S., Pasolli H. A., Chen S. C., Sharma N., Blanpain C., Fuchs E. (2012). Identification of stem cell populations in sweat glands and ducts reveals roles in homeostasis and wound repair. Cell 150, 136–150 10.1016/j.cell.2012.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man Y. G., Ball W. D., Marchetti L., Hand A. R. (2001). Contributions of intercalated duct cells to the normal parenchyma of submandibular glands of adult rats. Anat. Rec. 263, 202–214 10.1002/ar.1098 [DOI] [PubMed] [Google Scholar]
- Melnick M., Jaskoll T. (2000). Mouse submandibular gland morphogenesis: a paradigm for embryonic signal processing. Crit. Rev. Oral Biol. Med. 11, 199–215 10.1177/10454411000110020401 [DOI] [PubMed] [Google Scholar]
- Melnick M., Chen H., Zhou Y., Jaskoll T. (2001). An alternatively spliced Muc10 glycoprotein ligand for putative L-selectin binding during mouse embryonic submandibular gland morphogenesis. Arch. Oral Biol. 46, 745–757 10.1016/S0003-9969(01)00027-9 [DOI] [PubMed] [Google Scholar]
- Mitani Y., Li J., Weber R. S., Lippman S. L., Flores E. R., Caulin C., El–Naggar A. K. (2011). Expression and regulation of the ΔN and TAp63 isoforms in salivary gland tumorigenesis clinical and experimental findings. Am. J. Pathol. 179, 391–399 10.1016/j.ajpath.2011.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moumen M., Chiche A., Cagnet S., Petit V., Raymond K., Faraldo M. M., Deugnier M. A., Glukhova M. A. (2011). The mammary myoepithelial cell. Int. J. Dev. Biol. 55, 763–771 10.1387/ijdb.113385mm [DOI] [PubMed] [Google Scholar]
- Nanduri L. S., Maimets M., Pringle S. A., van der Zwaag M., van Os R. P., Coppes R. P. (2011). Regeneration of irradiated salivary glands with stem cell marker expressing cells. Radiother. Oncol. 99, 367–372 10.1016/j.radonc.2011.05.085 [DOI] [PubMed] [Google Scholar]
- Ogawa Y. (2003). Immunocytochemistry of myoepithelial cells in the salivary glands. Prog. Histochem. Cytochem. 38, 343–426 10.1016/S0079-6336(03)80001-3 [DOI] [PubMed] [Google Scholar]
- Okumura K., Shinohara M., Endo F. (2012). Capability of tissue stem cells to organize into salivary rudiments. Stem Cells Int. 2012, 502136 10.1155/2012/502136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padfield D. R., Rittscher J., Roysam B. (2008). Spatio-temporal cell segmentation and tracking for automated screening. In 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 14–17 May, 2008, Paris, France , pp. 376–379 10.1109/ISBI.2008.4541011 [DOI] [Google Scholar]
- Palmon A., David R., Neumann Y., Stiubea–Cohen R., Krief G., Aframian D. J. (2012). High-efficiency immunomagnetic isolation of solid tissue-originated integrin-expressing adult stem cells. Methods 56, 305–309 10.1016/j.ymeth.2011.10.002 [DOI] [PubMed] [Google Scholar]
- Patel V. N., Rebustini I. T., Hoffman M. P. (2006). Salivary gland branching morphogenesis. Differentiation 74, 349–364 10.1111/j.1432-0436.2006.00088.x [DOI] [PubMed] [Google Scholar]
- Pizzo A. M., Kokini K., Vaughn L. C., Waisner B. Z., Voytik–Harbin S. L. (2005). Extracellular matrix (ECM) microstructural composition regulates local cell-ECM biomechanics and fundamental fibroblast behavior: a multidimensional perspective. J. Appl. Physiol. 98, 1909–1921 10.1152/japplphysiol.01137.2004 [DOI] [PubMed] [Google Scholar]
- Potts S. J., Krueger J. S., Landis N. D., Eberhard D. A., Young G. D., Schmechel S. C., Lange H. (2012). Evaluating tumor heterogeneity in immunohistochemistry-stained breast cancer tissue. Lab. Invest. 92, 1342–1357 10.1038/labinvest.2012.91 [DOI] [PubMed] [Google Scholar]
- Proctor G. B., Carpenter G. H. (2007). Regulation of salivary gland function by autonomic nerves. Auton. Neurosci. 133, 3–18 10.1016/j.autneu.2006.10.006 [DOI] [PubMed] [Google Scholar]
- Purwanti N., Tsuji D., Azlina A., Karabasil M. R., Javkhlan P., Hasegawa T., Yao C., Akamatsu T., Itoh K., Hosoi K. (2011). Induction of Sca-1 in the duct cells of the mouse submandibular gland by obstruction of the main excretory duct. J. Oral Pathol. Med. 40, 651–658 10.1111/j.1600-0714.2011.01011.x [DOI] [PubMed] [Google Scholar]
- Rugel–Stahl A., Elliott M. E., Ovitt C. E. (2012). Ascl3 marks adult progenitor cells of the mouse salivary gland. Stem Cell Res. 8, 379–387 10.1016/j.scr.2012.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira S. J., Soscia D. A., Oztan B., Mosier A. P., Jean–Gilles R., Gadre A., Cady N. C., Yener B., Castracane J., Larsen M. (2012). The regulation of focal adhesion complex formation and salivary gland epithelial cell organization by nanofibrous PLGA scaffolds. Biomaterials 33, 3175–3186 10.1016/j.biomaterials.2012.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran S. D., Sumita Y., Khalili S. (2011). Bone marrow-derived cells: A potential approach for the treatment of xerostomia. Int. J. Biochem. Cell Biol. 43, 5–9 10.1016/j.biocel.2010.10.010 [DOI] [PubMed] [Google Scholar]
- Tucker A. S. (2007). Salivary gland development. Semin. Cell Dev. Biol. 18, 237–244 10.1016/j.semcdb.2007.01.006 [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A., Rocha A. S., Ousset M., Beck B., Bouvencourt G., Rock J., Sharma N., Dekoninck S., Blanpain C. (2011). Distinct stem cells contribute to mammary gland development and maintenance. Nature 479, 189–193 10.1038/nature10573 [DOI] [PubMed] [Google Scholar]
- Walker J. L., Menko A. S., Khalil S., Rebustini I., Hoffman M. P., Kreidberg J. A., Kukuruzinska M. A. (2008). Diverse roles of E-cadherin in the morphogenesis of the submandibular gland: insights into the formation of acinar and ductal structures. Dev. Dyn. 237, 3128–3141 10.1002/dvdy.21717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Larsen M., Hoffman M. P., Yamada K. M. (2007). Self-organization and branching morphogenesis of primary salivary epithelial cells. Tissue Eng. 13, 721–735 10.1089/ten.2006.0123 [DOI] [PubMed] [Google Scholar]
- Wickliffe J. K., Lee V. H., Smith E., Tandler B., Phillips C. J. (2002). Gene expression, cell localization, and evolution of rodent submandibular gland androgen-binding protein. Eur. J. Morphol. 40, 257–260. [DOI] [PubMed] [Google Scholar]
- Yamamoto S., Fukumoto E., Yoshizaki K., Iwamoto T., Yamada A., Tanaka K., Suzuki H., Aizawa S., Arakaki M., Yuasa K.et al. (2008). Platelet-derived growth factor receptor regulates salivary gland morphogenesis via fibroblast growth factor expression. J. Biol. Chem. 283, 23139–23149 10.1074/jbc.M710308200 [DOI] [PubMed] [Google Scholar]
- Yazdanfar S., Kenny K. B., Tasimi K., Corwin A. D., Dixon E. L., Filkins R. J. (2008). Simple and robust image-based autofocusing for digital microscopy. Opt. Express 16, 8670–8677 10.1364/OE.16.008670 [DOI] [PubMed] [Google Scholar]
- Zhao X., Malhotra G. K., Band H., Band V. (2012). Derivation of myoepithelial progenitor cells from bipotent mammary stem/progenitor cells. PLoS ONE 7, e35338 10.1371/journal.pone.0035338 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


