Summary
Recently, gene editing with transcription activator-like effector nucleases (TALENs) has been used in the life sciences. TALENs can be easily customized to recognize a specific DNA sequence and efficiently introduce double-strand breaks at the targeted genomic locus. Subsequent non-homologous end-joining repair leads to targeted gene disruption by base insertion, deletion, or both. Here, to readily evaluate the efficacy of TALENs in Xenopus laevis embryos, we performed the targeted gene disruption of tyrosinase (tyr) and pax6 genes that are involved in pigmentation and eye formation, respectively. We constructed TALENs targeting tyr and pax6 and injected their mRNAs into fertilized eggs at the one-cell stage. Expectedly, introduction of tyr TALEN mRNA resulted in drastic loss of pigmentation with high efficiency. Similarly, for pax6, TALENs led to deformed eyes in the injected embryos. We confirmed mutations of the target alleles by restriction enzyme digestion and sequence analyses of genomic PCR products. Surprisingly, not only biallelic but also paralogous, gene disruption was observed. Our results demonstrate that targeted gene disruption by TALENs provides a method comparable to antisense morpholinos in analyzing gene function in Xenopus F0 embryos, but also applies beyond embryogenesis to any life stage.
Key words: Eye, Pax6, Pigmentation, TALEN, Tyrosinase, Xenopus laevis
Introduction
Amphibians are attractive experimental models for elucidating the mechanisms underlying animal development. However, the functional analysis of genes is limiting in amphibians as compared with those of other model animals, because there has been no way to efficiently introduce targeted gene disruption in eggs and germ cells. Therefore, innovation of targeted and heritable mutagenesis techniques has been long wished in the research area.
To generate gene knockout organisms, gene editing using transcription activator-like effector nucleases (TALENs) technique has been introduced to various model animals including worm, cricket, zebrafish, and rat (Wood et al., 2011; Huang et al., 2011; Tesson et al., 2011; Sander et al., 2011; Watanabe et al., 2012). Recently, it has been reported that targeted gene disruption by TALENs succeeded with high efficiency in Xenopus tropicalis and Xenopus laevis (Lei et al., 2012; Ishibashi et al., 2012). TALENs consist of two functional domains: a customized repeat array of TAL DNA-binding domains that recognize a specific DNA sequence and nuclease domain of FokI, a class IIS restriction enzyme (Miller et al., 2011). A pair of the customized TALENs binds to its target sequence, allowing FokI nuclease domains to dimerize and introduce double-strand breaks (DSBs). When DSBs are efficiently repaired by error-prone non-homologous end-joining (NHEJ) in cells, insertion, deletion, and insertion and/or deletion frequently occur at the TALEN-mediated DSB site. Subsequently, NHEJ repair leads to target gene disruption by frame shift. Therefore, the TALEN technique readily modifies target loci and disrupts gene functions. However, this technique requires optimization for each organism. For example, to increase the efficiency of TALEN in vivo, multiple TALEN scaffolds have recently been improved and examined with some modifications: truncation of N- and C-termini, hetero- or homodimers of FokI nuclease domains, and backbone vectors (Miller et al., 2011; Mussolino et al., 2011; Cade et al., 2012).
In this study, utilizing recent information, we generated modified high-efficiency TALENs based on the Golden Gate Method (Cermak et al., 2011; Sakuma et al., 2013). Here, we examine the efficiency of gene disruption by our modified TALENs in X. laevis early embryogenesis, using two endogenous genes, tyrosinase and pax6, involved in pigmentation and eye formation.
Materials and Methods
Generation of TALENs
Pairs of TALENs against tyr and pax6 were generated using the Golden Gate TALEN kit and TAL effector Kit (Addgene) (Cermak et al., 2011) with some modifications (Sakuma et al., 2013). TALEN target sites were determined by TAL effector Nucleotide Targeter Ver. 2.0 (Doyle et al., 2012). After construction of TALE arrays against each gene, they were subcloned into a destination vector based on pcDNA3.1 containing the truncated N- and C-terminal domains of TALE (+153/+47) and wild-type FokI nuclease domain derived from pTAL3 (Cermak et al., 2011; Mussolino et al., 2011; Sakuma et al., 2013). We screened for a more efficient TALENs set by the Single Stranded Annealing (SSA) assay (Ochiai et al., 2010). TALEN mRNAs were synthesized by mMessage mMachine T7 Ultra Kit (Life Technologies) after linearization by XmaI (NEB).
Manipulation of Xenopus laevis eggs
Fertilized eggs were obtained from breeding pairs of X. laevis injected with human chorionic gonadotropin (Sive et al., 2000). Eggs were dejellyed with 2% cysteine treatment and then were moved into 3% Ficoll in 0.3× MMR. Approximately 550 pg of each right and left or 1100 pg of only right TALENs mRNA were injected into eggs at the one-cell stage with 4.6 nl volume using Nano-ject II (Drummond). Injected embryos were cultured in 0.1× MMR containing gentamycin from the blastula to the swimming tadpole stages at 18°C. The animals were handled in accordance with the guidelines of Hiroshima University for the use and care of experimental animals.
Mutation analysis
Genomic DNA samples of individual embryos were extracted with DNeasy kit (Qiagen). Genomic regions containing the TALEN target site of each gene were amplified with specific primer sets (described in supplementary material Table S1). PCR was carried out with KOD FX Neo or LA Taq (ToYoBo and TaKaRa, respectively). To examine the efficiency of target mutations in the injected embryos, we performed restriction enzyme digestion of PCR products using HinfI (TaKaRa) or Hpy188I (NEB). To confirm the presence of TALEN-mediated mutations, PCR products were subcloned into pCR2.1/TOPO (Life Technologies), and then positive clones were selected by colony PCR. Colony PCR products were sequenced using BigDye Terminator Ver. 3.1 (Life Technologies).
Results and Discussion
Tyrosinase gene disruption by TALENs
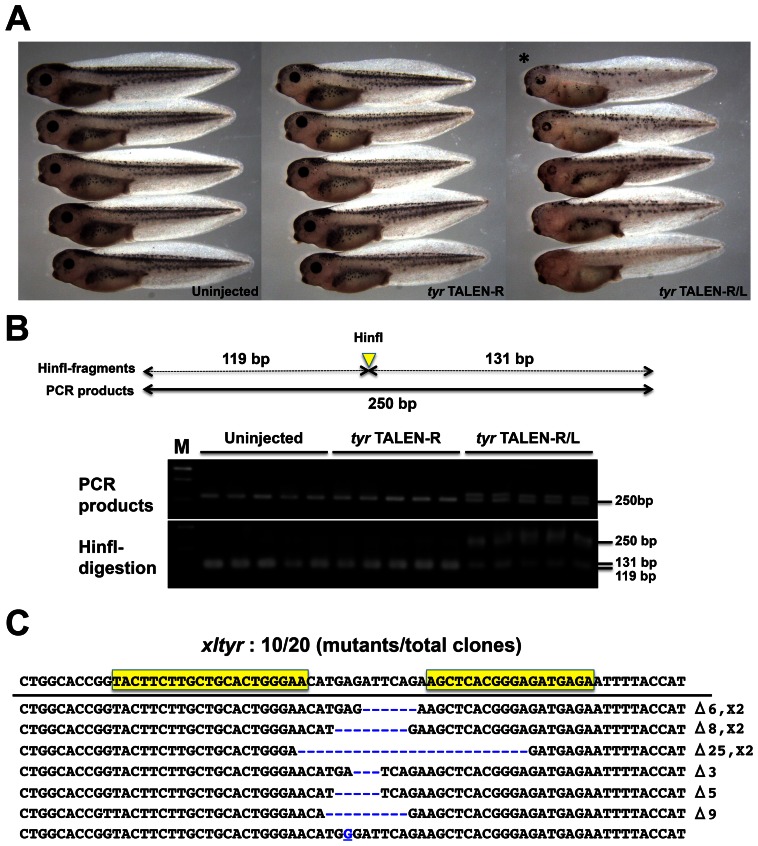
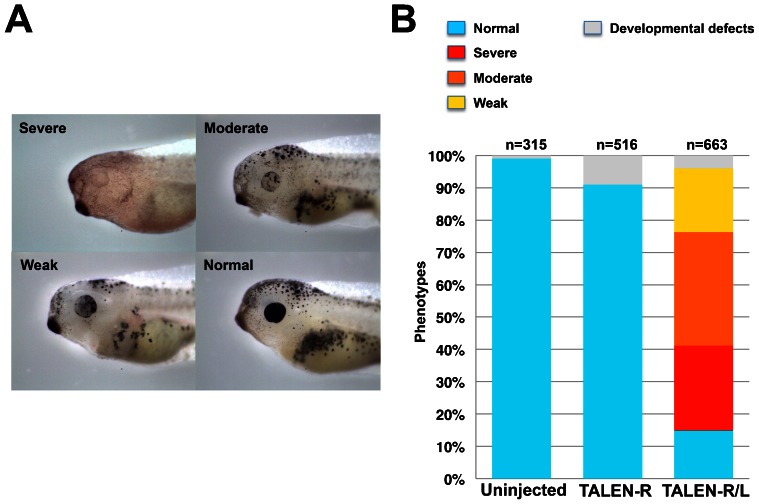
To examine whether our TALENs effectively induce targeted gene disruption in X. laevis, we generated two TALEN sets against tyrosinase (oculocutaneous albinism IA; tyr) involved in melanin synthesis and is specifically expressed in retinal pigment epithelium (RPE) and melanophores (Kumasaka et al., 2003). If this gene is effectively disrupted by TALENs, we can visually identify tyr phenotypes as albino. X. laevis has an allotetraploid genome, but only one tyr cDNA sequence is deposited in the public databases, NCBI Refseq and Xenbase (http://www.xenbase.org/common). Two pairs of TALENs against the first exon of the tyr locus were generated, and we selected a set with higher activity by the SSA assay (Ochiai et al., 2010). Next, we injected 550 pg of each right and left TALEN pair (TALEN-R/L) mRNA into the fertilized eggs at the one-cell stage. As a negative control, we also injected 1100 pg of the right TALEN mRNA (TALEN-R) only. As expected, many albino phenotypes were observed in the tyr TALEN-R/L-injected embryos, while there was no change of pigmentation in both the TALEN-R-injected and uninjected control embryos (Fig. 1A). In particular, pigmentation in the RPE was effectively lost by tyr TALENs. Similar to previous data in X. tropicalis, tyr TALENs effectively induced albinism in the RPE of F0 embryos (Ishibashi et al., 2012). We confirmed the targeted gene disruption in the phenotypes by genomic PCR with restriction enzyme digestion. The genomic region containing tyr TALEN site was amplified by PCR with a specific primer set (Fig. 1B, upper; supplementary material Table S1). PCR products of TALEN-R/L-injected embryos appear as a doublet compared with those of TALEN-R-injected and uninjected embryos, possibly due to heteroduplex formation of mutant and normal alleles (Fig. 1B, middle). The spacer region between TALEN target sequences contains the HinfI recognition site. Therefore, if this site is disrupted, the restriction enzyme cannot recognize and cut it. Most PCR product of TALEN-R/L-injected embryos became resistant to HinfI; in contrast, those of TALEN-R-injected and uninjected samples were almost completely digested (Fig. 1B, lower). Moreover, a PCR sample from one of the phenotypes was subcloned and subjected to DNA sequencing analysis. The rate of tyr mutation was 50% (10 mutants/20 total clones; Fig. 1C). The same mutation rate was also obtained from genomic PCR clones of the pooled phenotypes (supplementary material Fig. S1). In addition, 30% of total mutations were in-frame mutations and may not affect enzymatic activity of tyr. Phenotypes could be divided into three groups: severe phenotypes showed loss of pigmentation in RPE; moderate phenotypes showed mosaic but more than half loss of pigmentation in RPE; weak phenotypes showed slight loss of pigmentation in RPE (Fig. 2A,B). More than 70% of the TALEN-R/L-injected embryos show some pigment disruption. Patchy loss of pigmentation was observed, suggesting that mosaic but biallelic tyr disruption by frameshift in the pigmentless RPE and melanophores.
Fig. 1. Disruption of tyrosinase (tyr) in TALEN mRNA-injected embryos.
(A) Phenotypes of tyr TALEN-injected embryos: Uninjected control embryos; TALEN-R, embryos injected with 1100 pg right tyr TALEN mRNA; TALEN-R/L, embryos injected with 550 pg of each right and left tyr TALEN mRNA. Injected embryos were reared to the hatching stage. (B) Detection of tyr mutations by genomic PCR and digestion with a restriction enzyme. Five embryos each were collected from the three experimental groups (Uninjected, TALEN-R, and TALEN-R/L). Upper image: a schematic drawing of tyr genomic PCR product. Genomic DNA of each embryo was prepared and then was subjected to PCR with a specific primer set for amplification containing the tyr TALEN target site. HinfI site is located on spacer sequence of the TALEN site. Middle image: a gel electrophoresis image of tyr genomic PCR products. Lower image: HinfI-digestion of PCR products for detecting tyr mutations. PCR products were purified and then were digested by HinfI. HinfI fragments are divided into 131 and 119 bp. M means 100 bp ladder marker. (C) Sequences and frequencies of tyr mutations. PCR product from the TALEN-R/L-injected embryo (asterisk in panel A) was subcloned and was subjected to DNA sequencing analysis. Mutant sequences were aligned to that of wild type. Sequence of wild-type allele is shown at the top. Yellow boxes indicate tyr TALEN target site. Gaps generated by deletion are shown as blue dashes. A substitution is shown as an underlined blue character. Types and frequencies of each mutation (deletion and insertion) are shown at the right of the panel.
Fig. 2. Phenotypes of tyr TALEN-injected embryos.
Embryos were divided into four groups according to their phenotypes. (A) Representative phenotypes: Severe, near complete loss of pigmentation in retina pigmented epithelium (RPE); Moderate, more than half loss of pigmentation in RPE; Weak, less than half loss of pigmentation in RPE; Normal, no alteration of pigmentation. (B) Percentage of phenotypes in the three experimental groups, Uninjected, TALEN-R, and TALEN-R/L. Total numbers of individuals in this experiment are shown at the top of each graph.
Our results are slightly different from that of the previous report in X. tropicalis (Ishibashi et al., 2012); TALEN mediated tyr disruption in X. tropicalis seemed to be more efficient than in X. laevis. This disparity may reflect differences of species or TALEN platforms. Another key issue for applying TALENs in Xenopus is toxicity. In zebrafish, heterodimeric TALENs show less toxic than homodimeric TALENs (Cade et al., 2012). However, in some cases, introduction of a high dose of TALEN mRNA with heterodimer type FokI, (500∼800 pg) exhibits high toxicity and rate of developmental defects in X. tropicalis (Lei et al., 2012). On the other hand, our results and Ishibashi et al. demonstrate that even if a high dose of TALEN mRNA is injected, its toxicity and induction of developmental defects are low in X. laevis and X. tropicalis (Ishibashi et al., 2012). Actually, most of the TALEN-injected embryos developed into tadpoles and adults with albinism (supplementary material Fig. S2). Therefore, there is room for further optimization of TALEN platforms to increase the efficiency and decrease toxicity in Xenopus.
pax6 gene disruption by TALENs
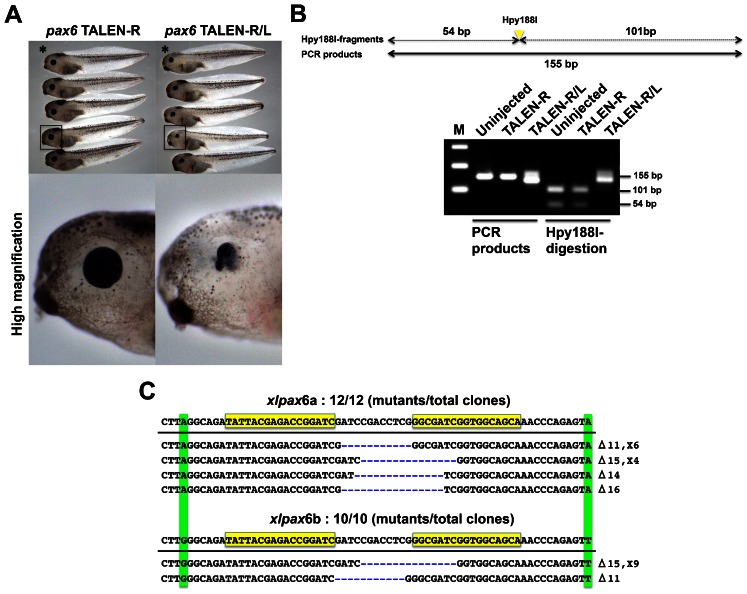
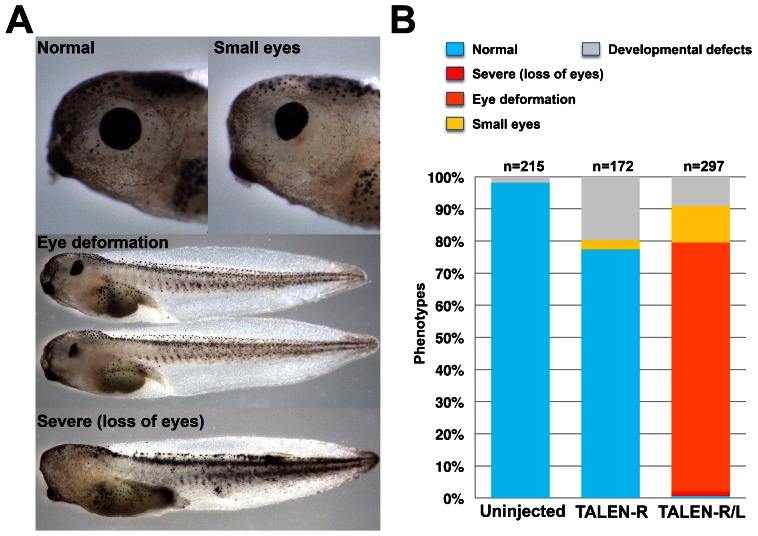
Next, we generated TALEN sets against pax6 involved in eye lens formation (Rungger-Brändle et al., 2010). Due to its allotetraploid genome, there are two paralogs, pax6a and pax6b, in X. laevis in the public databases. We generated a pair of TALENs that recognize the conserved sequences of the putative third exon between pax6a and pax6b. We validated the activity of this TALEN set by SSA assay and injected their mRNA into the fertilized eggs. Expectedly, many phenotypes showed disturbance of eye development in the TALEN-R/L mRNA-injected embryos (Fig. 3A). The phenotypes closely resemble with those of pax6 morphants (Rungger-Brändle et al., 2010), suggesting that injection of TALENs recapitulates the knockdown of pax6 by antisense morpholinos. Also, pax6 protein was greatly reduced in the TALEN-R/L-injected embryos as compared with control siblings (supplementary material Fig. S3). We also confirmed the targeted gene disruption by genomic PCR with restriction enzyme digestion (Fig. 3B). Genomic region containing pax6 TALEN site was amplified with a specific primer set (Fig. 3B, upper). PCR products of the TALEN-R/L-injected embryos tend to be slightly lower molecular weight than those of the TALEN-R-injected and uninjected embryos, possibly due to deletions mediated by TALENs (see other experimental examples; supplementary material Fig. S4). PCR products from TALEN-R and uninjected control samples were completely digested by Hpy188I. In contrast, TALEN-R/L PCR products could not be digested, indicating that the spacer sequence containing Hpy188I site is mutated. To confirm the mutations by pax6 TALENs, we carried out DNA sequencing analysis. Surprisingly, sequences of all clones exhibited deletion mutations in a TALEN-R/L-injected embryo (22 mutants/22 total clones; Fig. 3C). The similar result was obtained about the high mutation rate from the pooled phenotypes (supplementary material Fig. S5). Interestingly, both pax6a and pax6b mutation was confirmed, suggesting that the TALENs almost perfectly introduced bi-allelic and paralogous mutation in X. laevis. The TALEN target site is located at the sequence encoding linker region in paired domain of PAX6, which is a crucial domain for DNA binding (Xu et al., 1999). Therefore, even if deletions and insertions occur in multiples of 3 bp, in-frame mutations of pax6 by TALENs could perturb eye formation. Phenotypes could be divided into three groups: severe phenotypes showed loss of eyes; moderate phenotypes showed deformation of eyes; weak phenotypes showed small eyes (Fig. 4A). More than 85% of the TALEN-R/L-injected embryos showed some perturbation of eye formation, whereas most of TALEN-R-injected and uninjected embryos presented normal phenotypes (Fig. 4B). pax6 TALENs also exhibited low toxicity in injected embryos. Rate of developmental defects is within 20% in the TALEN-R- and TALEN-R/L-injected embryos.
Fig. 3. Disruption of pax6 in the TALEN mRNA-injected embryos.
(A) Phenotypes of pax6 TALEN-injected embryos: TALEN-R, embryos injected with 1100 pg right pax6 TALEN mRNA; TALEN-R/L, embryos injected with 550 pg of each right and left pax6 TALEN mRNA. Injected embryos were reared to the hatching stage. Upper image: low magnification of each experimental group. Lower image: high magnification of representative phenotypes from TALEN-R- and TALEN-R/L-injected embryos noted by squares in the upper image. (B) Detection of pax6 mutations by genomic PCR and digestion with a restriction enzyme. One embryo was collected from each of the three experimental groups (asterisks in panel A): Uninjected, TALEN-R, and TALEN-R/L. Upper image: a schematic drawing of pax6 genomic PCR product. Genomic DNA of each embryo was prepared and then was subjected to PCR with a specific primer set for amplification containing the pax6 TALEN target site. Hpy188I site is located on spacer sequence of the TALEN site. Left image: a gel electrophoresis image of pax6 genomic PCR products. Right image: Hpy188I-digestion of PCR products for detecting pax6 mutations. PCR products were purified and then digested by Hpy188I. Hpy188I fragments are divided into 101 and 54 bp. M means 100 bp ladder marker. Uninjected indicates uninjected control embryo. (C) Sequences and frequencies of pax6 mutation by TALENs. PCR product from the TALEN-R/L-injected embryo (asterisk in panel A) was subcloned and was subjected to DNA sequencing analysis. Mutant sequences were aligned to those of wild type. Sequences of pax6a and pax6b wild-type alleles are shown at the top. Yellow boxes indicate pax6 TALEN target site. Gaps generated by deletion are shown as blue dashes. Types and frequencies of each mutation (deletion and insertion) are shown at the right of the panel. Paralogous substitutions between pax6a and pax6b are highlighted in green.
Fig. 4. Phenotypes of pax6 TALEN-injected embryos.
Embryos were divided into four groups according to their phenotypes. (A) Representative phenotypes: Severe, near complete loss of eyes; Eye deformation, embryos with deformed eyes; Small eyes, embryos with apparently small eyes as compared with the Uninjected and TALEN-R-injected embryos; Normal, no alteration of eye formation and size. (B) Percentage of phenotypes in the three experimental groups, Uninjected, TALEN-R, and TALEN-R/L. Total numbers of individuals in this experiment are shown at the top of each graph.
In this research, we demonstrate that our engineered TALENs effectively disrupt target genes with low toxicity in X. laevis embryos. In addition, our results also suggest that TALENs can introduce not only bi-allelic but also paralogous genome editing in X. laevis, where most genes are duplicated due to polyploidization. Thus, our TALEN architecture is suitable for X. laevis research. TALEN specificity seems to be high; however, there are problems regarding off-target effects when uncharacterized genes are analyzed. Then rescue experiments regarding genes of interest using mRNA injection and transgenic techniques will be needed for precise analysis of gene function in Xenopus. We expect that targeted gene disruption by TALENs will provide an alternative method to analyze functions of genes comparable to antisense morpholino in X. laevis and X. tropicalis research.
Supplementary Material
Acknowledgments
The authors are grateful to Dr Daniel R. Buchholz for the critical reading of our manuscript. This research was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) to K.T.S., T.S. and T.Y.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Cade L., Reyon D., Hwang W. Y., Tsai S. Q., Patel S., Khayter C., Joung J. K., Sander J. D., Peterson R. T., Yeh J. R. (2012). Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 40, 8001–8010 10.1093/nar/gks518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T., Doyle E. L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J. A., Somia N. V., Bogdanove A. J., Voytas D. F. (2011). Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 10.1093/nar/gkr218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle E. L., Booher N. J., Standage D. S., Voytas D. F., Brendel V. P., Vandyk J. K., Bogdanove A. J. (2012). TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 40, W117–W122 10.1093/nar/gks608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Xiao A., Zhou M., Zhu Z., Lin S., Zhang B. (2011). Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 29, 699–700 10.1038/nbt.1939 [DOI] [PubMed] [Google Scholar]
- Ishibashi S., Cliffe R., Amaya E. (2012). Highly efficient bi-allelic mutation rates using TALENs in Xenopus tropicalis. Biol. Open 1, 1273–1276 10.1242/bio.20123228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumasaka M., Sato S., Yajima I., Yamamoto H. (2003). Isolation and developmental expression of tyrosinase family genes in Xenopus laevis. Pigment Cell Res. 16, 455–462 10.1034/j.1600-0749.2003.00064.x [DOI] [PubMed] [Google Scholar]
- Lei Y., Guo X., Liu Y., Cao Y., Deng Y., Chen X., Cheng C. H., Dawid I. B., Chen Y., Zhao H. (2012). Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs). Proc. Natl. Acad. Sci. USA 109, 17484–17489 10.1073/pnas.1215421109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. C., Tan S., Qiao G., Barlow K. A., Wang J., Xia D. F., Meng X., Paschon D. E., Leung E., Hinkley S. J.et al. (2011). A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148 10.1038/nbt.1755 [DOI] [PubMed] [Google Scholar]
- Mussolino C., Morbitzer R., Lütge F., Dannemann N., Lahaye T., Cathomen T. (2011). A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 39, 9283–9293 10.1093/nar/gkr597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai H., Fujita K., Suzuki K., Nishikawa M., Shibata T., Sakamoto N., Yamamoto T. (2010). Targeted mutagenesis in the sea urchin embryo using zinc-finger nucleases. Genes Cells 15, 875–885 10.1111/j.1365-2443.2010.01425.x [DOI] [PubMed] [Google Scholar]
- Rungger–Brändle E., Ripperger J. A., Steiner K., Conti A., Stieger A., Soltanieh S., Rungger D. (2010). Retinal patterning by Pax6-dependent cell adhesion molecules. Dev. Neurobiol. 70, 764–780 10.1002/dneu.20816 [DOI] [PubMed] [Google Scholar]
- Sakuma T., Hosoi S., Woltjen K., Suzuki K. I., Kashiwagi K., Wada H., Ochiai H., Miyamoto T., Kawai N., Sasakura Y.et al. (2013). Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells [Epub ahead of print] 10.1111/gtc.12037 [DOI] [PubMed] [Google Scholar]
- Sander J. D., Cade L., Khayter C., Reyon D., Peterson R. T., Joung J. K., Yeh J. R. (2011). Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 29, 697–698 10.1038/nbt.1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H. L., Grainger R. M., Harland R. M. (2000). Early Development Of Xenopus Laevis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Tesson L., Usal C., Ménoret S., Leung E., Niles B. J., Remy S., Santiago Y., Vincent A. I., Meng X., Zhang L.et al. (2011). Knockout rats generated by embryo microinjection of TALENs. Nat. Biotechnol. 29, 695–696 10.1038/nbt.1940 [DOI] [PubMed] [Google Scholar]
- Watanabe T., Ochiai H., Sakuma T., Horch H. W., Hamaguchi N., Nakamura T., Bando T., Ohuchi H., Yamamoto T., Noji S.et al. (2012). Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases. Nat. Commun. 3, 1017 10.1038/ncomms2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A. J., Lo T. W., Zeitler B., Pickle C. S., Ralston E. J., Lee A. H., Amora R., Miller J. C., Leung E., Meng X.et al. (2011). Targeted genome editing across species using ZFNs and TALENs. Science 333, 307 10.1126/science.1207773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. E., Rould M. A., Xu W., Epstein J. A., Maas R. L., Pabo C. O. (1999). Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev. 13, 1263–1275 10.1101/gad.13.10.1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.