Summary
The Sonic hedgehog (Shh) signaling pathway is well known in patterning of the neural tube during embryonic development, but its emerging role in differentiated neurons is less understood. Here we report that Shh enhances autophagy in cultured hippocampal neurons. Microarray analysis reveals the upregulation of multiple autophagy-related genes in neurons in response to Shh application. Through analysis of the autophagy-marker LC3 by immunoblot analysis and immunocytochemistry, we confirm activation of the autophagy pathway in Shh-exposed neurons. Using electron microscopy, we find autophagosomes and associated structures with a wide range of morphologies in synaptic terminals of Shh-exposed neurons. Moreover, we show that Shh-triggered autophagy depends on class III Phosphatidylinositol 3-kinase complexes (PtdIns3K). These results identify a link between Shh and autophagy pathways and, importantly, provide a lead for further understanding the physiology of Shh signaling activity in neurons.
Key words: Sonic hedgehog, Autophagy, Hippocampal neurons, Synapses
Introduction
The Sonic hedgehog (Shh) signaling pathway is an evolutionarily conserved system that contributes to multiple phases of neural development. At early embryonic phases, Shh signaling plays an essential role in pattern formation of the neural tube (Ingham and McMahon, 2001; Varjosale and Taipale, 2008). At later embryonic stages, Shh is involved in maintenance of interneuron identity in the ventral telencephalon (Xu et al., 2010). Shh signaling has also been appreciated as a growth factor for postnatal cerebellar granule cell precursors (Dahmane and Ruiz i Altaba, 1999; Wallace, 1999; Wechsler-Reya and Scott, 1999) and adult neural progenitors located in several specific stem cell niches (Lai et al., 2003; Palma et al., 2005; Breunig et al., 2008; Han et al., 2008).
While many studies have focused on functions of the Shh signaling pathway on neural progenitors, its function in differentiated neurons is less well understood. However, recent studies have revealed Shh's roles in neurons that no longer have stem-cell-like properties. For example, Shh synergizes with NGF to provide trophic support to cholinergic neurons in the basal forebrain (Reilly et al., 2002), and protects mesencephalic dopaminergic neurons against neurotoxins (Tsuboi and Shults, 2002; Dass et al., 2005). Also, Shh is actively involved in adult nigrostriatal circuitry (Gonzalez-Reyes et al., 2012). Shh, which originates from mesencephalic dopaminergic neurons, is necessary for supporting dopaminergic neurons as well as cholinergic and fast spiking GABAergic neurons of the striatum (Gonzalez-Reyes et al., 2012).
Shh and its receptors, Patched (Ptch) and Smoothened (Smo), are also expressed in hippocampal neurons (Traiffort et al., 1999; Sasaki et al., 2010; Petralia et al., 2011a; Petralia et al., 2011b). In a previous study, we found that Shh induces structural and functional changes in presynaptic terminals of cultured hippocampal neurons (Mitchell et al., 2012). However, the molecular mechanisms underlying Shh signaling in hippocampal neurons are unknown. We conducted a pilot study to survey the global gene expression pattern of cultured hippocampal neurons after adding Shh. We noticed an increased expression of a number of genes related to the autophagy pathway, including the well-established autophagy marker LC3 (supplementary material Table S1). This observation was surprising, given no direct evidence to date linking the Shh signaling pathway to autophagy in neurons. For this reason, we focused this study on addressing the question of whether Shh activates the autophagy pathway in hippocampal neurons. Using immunoblotting, immunocytochemistry and electron microscopy, we found that Shh elicits autophagic activity that requires class III Phosphatidylinositol 3-kinase complexes (PtdIns3K).
Results and Discussion
We investigated autophagy activity in cultured hippocampal neurons that had been exposed to ShhN for 48 hr (9–10 DIV+48 hr) (supplementary material Fig. S1A). Bioactivity and efficacy of the ShhN have been verified by Shh-light2 assay (Mitchell et al., 2012). We focused on the 48 hr-ShhN-exposure because this experimental condition increased expression of autophagy-related genes (supplementary material Table S1), and also levels of Ptch (Mitchell et al., 2012) and Gli1 (supplementary material Fig. S1B) – indicators of an activated Shh signaling pathway (Hillman et al., 2011; Dorn et al., 2012).
Using immunoblot analysis, we examined the conversion of nonlipidated LC3-I to lipidated LC3-II, a widely used autophagy indicator (Klionsky et al., 2012; Menzies et al., 2012). LC3-II level was low in untreated control neurons. In ShhN-treated neurons, LC3-II level was increased approximately 2-fold as compared to control (Fig. 1A,B; supplementary material Fig. S1C). Likewise, the Shh agonist SAG (Shh agonist) (Chen et al., 2002) also increased LC3-II level (Fig. 1A,B; supplementary material Fig. S1C). LC3-II induction was blocked by co-application of the Shh inhibitor robotnikinin (Stanton et al., 2009) (Fig. 1C), suggesting that ShhN accounts for the autophagy induction. We also tested the effects of 10-NCP, a small compound shown to effectively induce neuronal autophagy (Tsvetkov et al., 2010). 10-NCP (10 µM) – in the absence of ShhN – increased LC3-II level in neurons (Fig. 1D), in agreement with results of a previous study (Tsvetkov et al., 2010). When 10-NCP was added to ShhN-treated neurons, it elicited a further LC3-II increase (Fig. 1D, lane 1 and 2; supplementary material Fig. S1A,D). A lower dose of 10-NCP (1 µM) produced a smaller but similar trend of increase in the LC3-II level (Fig. 1D, lane 3 and 4). To determine whether elevated LC3-II level reflects an increase in autophagosome synthesis or a reduction in autophagosome degradation, we tested the effect of ShhN when the last step of autophagy-mediated degradation was inhibited by bafilomycin A (BFA) (Yamamoto et al., 1998). On exposure to BFA, LC3-II level elevated further in ShhN-treated neurons (Fig. 1D, lane 5 and 6; supplementary material Fig. S1A,C), indicating that ShhN induced autopohagy.
Fig. 1. ShhN induces autophagy in hippocampal neurons.
(A) Representative immunoblot of neurons incubated with control medium, or ShhN (10%), or a Shh agonist SAG (100 nM) for 48 hr. (B) Quantification of A. LC3-II level is normalized to total actin (mean±s.e.m., **P<0.01, n = 4). Additional example blot is shown in supplementary material Fig. S1C. (C) The Shh inhibitor robotnikinin (50 µM) prevents the ShhN-induced LC3-II increase. (D) Representative blot of neurons treated with 10-NCP or BFA (for the last 12 hr of the 48 hr-ShhN incubation period). 10-NCP (10):10 µM; 10-NCP (1): 1 µM; BFA: 0.4 µM. Additional blots of NCP and BFA are shown in supplementary material Fig. S1C,D. (E) Representative confocal images of hippocampal neurons treated as indicated and labeled with the LC3 antibody (green) and an antibody to a neuronal marker MAP2 (red). Upper and lower panel show different representative views at high- and low-magnifications, respectively. Scale bars: 10 µm. Additional examples are shown in supplementary material Fig. S2. (F) Quantification of neurons containing LC3 puncta, as shown in E (mean±s.e.m., ***P<0.001, n≥60 neurons from 3 experiments). (G) LC3 punctate structures were scattered along the neurites (white arrowheads). Scale bars: 10 µm. Additional examples are shown in supplementary material Fig. S3. (H) Quantification of LC3 puncta per 10 µm neurite. n≥30 neurites from 3 experiments. (I) Neurons immunolabeled with an antibody to LBPA. Scale bar: 10 µm. Low-magnification examples are shown in supplementary material Fig. S4. (J) Quantification of I. n≥45 neurons from 3 experiments.
We then examined LC3-associated autophagic structures by immunocytochemistry. Neurons were identified based on their distinctive morphology and confirmed by expression of the neuronal marker MAP2. Untreated control neurons displayed little LC3 immunoreactivity (Fig. 1E,F). In contrast, addition of ShhN produced a robust increase in the number of LC3-immunopositive puncta (Fig. 1E,F; see supplementary material Fig. S2A–C for additional examples at high- and low-magnification). Robotnikinin blocked the formation of LC3-positive structures (Fig. 1E,F).
In addition to the soma of the neurons, LC3-positive puncta were seen dotted along neurites of the ShhN-treated neurons (Fig. 1G,H; also supplementary material Fig. S3A). These discrete puncta likely correspond to synaptic terminals of cultured hippocampal neurons (supplementary material Fig. S3B) (Dean and Scheiffele, 2009; Fletcher et al., 1994; Petralia et al., 2013). Indeed, and as discussed below, autophagosomes are found in the synaptic terminals.
We examined another autophagic/lysosomal pathway marker, the late endosome-specific lipid lysobisphosphatidic acid (LBPA) (Matsuo et al., 2004; Pérez-Sala et al., 2009). LBPA-immunolabeled structures were rarely seen in control neurons (Fig. 1I). By contrast, application of ShhN substantially increased the number of LBPA-labeled structures, and this increased even further upon 10-NCP induction (Fig. 1I,J; see supplementary material Fig. S4 for low-magnification images).
In order to identify autophagosomes more definitively and to visualize synaptic terminals directly, we next examined these neurons at higher resolution using electron microscopy. We used older neurons (18–19 DIV+48 hr ShhN) as their synapses are more mature. In synaptic terminals of control neurons, it was uncommon to find clearly identifiable autophagic structures, irrespective of pre- or post-synaptic distinction (data not shown). In contrast, synaptic terminals of ShhN-treated neurons contained a wide range of endocytic structures and autophagic structures (Fig. 2) (Boland et al., 2008; Eskelinen et al., 2011; Fader and Colombo, 2009; Liou et al., 1997; Ylä-Anttila et al., 2009). Endocytic structures included late sorting endosomes (Fig. 2A,E,G), as well as multivesicular bodies, which are characterized by the presence of distinctive intralumenal vesicles (mv in Fig. 2A–D, Fig. 2G). Autophagic structures or vacuoles displayed different morphological characteristics, representing autophagosomes at various stages (Boland et al., 2008; Eskelinen et al., 2011; Fader and Colombo, 2009; Liou et al., 1997; Ylä-Anttila et al., 2009). Fig. 2E shows an example of initial autophagic vacuoles (avi) based on the presence of cytoplasmic constituents surrounded by multiple layers of membranes. Fig. 2C,E show examples of degradative autophagic vacuoles (avd), which are filled with degrading or degraded components. Fig. 2G is a striking example of a ShhN-treated presynaptic terminal that houses a continuous sequence of (from left to right) a late sorting endosome (se), a multivesicular body (mv), and an autophagosome with a portion in the early stages of degradation on the left (avi) and a portion in later stages on the right (avd). Furthermore, Fig. 2F shows an example of a presynaptic terminal containing phagophores (ph) – a fully formed one on the left and a partially formed (U-shaped) one on the right.
Fig. 2. Electron micrographs showing various stages in the formation of autophagosomes and associated structures at synapses of ShhN-treated hippocampal neurons.
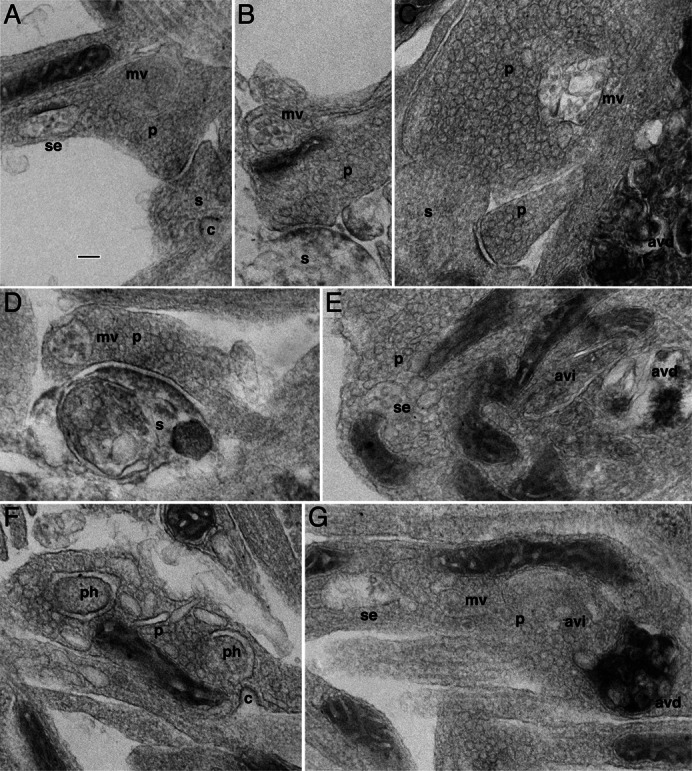
(A) A presynaptic terminal (p) contains a late sorting endosome (se) and a multivesicular body (mv). A postsynaptic spine (s) apposing to the presynaptic terminal has a clathrin-coated pit (c). (B) Another example of a presynaptic terminal containing a multivesicular body. The postsynaptic spine appears to be degenerating. (C) A presynaptic terminal contains a multivesicular body – but this example has additional irregular membranes, possibly representing a combination of a mutivesicular body with an autophagosome to form an amphisome. Notice that a process to the lower right is filled with a late or degradative autophagosome (avd) complex with the characteristic osmiophilic structures. (D) This synapse has a multivesicular body in the presynaptic terminal and an autophagosome complex filling much of the spine. (E) A presynaptic terminal expansion of an axon containing a late sorting endosome (se) on the left, an early stage autophagosome (avi) near the center, and a late stage autophagosome (avd) on the right. (F) Another example of presynaptic terminal expansion but it contains two putative phagophores (ph), and a clathrin-coated pit (c) nearby. (G) A presynaptic terminal expansion contains a continuous sequence (from left to right) of a late sorting endosome, a multivesicular body, an early autophagosome, and a late stage autophagosome filled with osmiophilic contents. Scale bar: 100 nm.
We focused our analysis on the presynaptic terminals of the ShhN-treated neurons (Fig. 3). Many presynaptic compartments were crowded with, or nearly filled by autophagic structures, resulting in the resident synaptic vesicles being pushed against the presynaptic membrane or almost completely obscured (Fig. 3C–F). We surveyed clearly traceable presynaptic compartments (229 for control and ShhN-treated) and counted those containing multivesicular bodies or autophagosomes. ShhN treatment induced a marked increase in the occurrence of these structures in presynaptic terminals (Fig. 3G).
Fig. 3. Electron micrographs showing presynaptic autophagosomes in hippocampal neurons treated with ShhN.

(A–D,F) Examples of early autophagosomes (avi). (D) A synapse with a presynatic terminal (p) and a postsynaptic spine (s). Above this typical synapse, a large process is almost completely occupied by autophagosomes resulting in a few barely visible obscured vesicles. (E) An example of a late autophagosome (avd) based on the characteristic osmiophilic contents. Scale bar: 100 nm. (G) Quantification of presynaptic terminals that contain a multivesicular body or autophagosome. n = 229 presynaptic terminals for control; n = 229 presynaptic terminals for ShhN-treated.
In addition to their presence in the presynaptic terminals (Figs 2, 3), various autophagosomes were also found in other parts of the ShhN-treated neurons (supplementary material Fig. S5). Supplementary material Fig. S5A shows a U-shaped phagophore (ph) in a presynaptic terminal, and a process adjacent to this presynaptic terminal is also filled with multiple autophagosomes. Supplementary material Fig. S5B shows an array of degenerative structures that take up nearly an entire postsynaptic process. Supplementary material Fig. S5C shows a phagophore located within a postsynaptic spine. Supplementary material Fig. S5D shows clusters of late autophagosomes occupying a process of unknown nature. Finally, supplementary material Fig. S5E shows distinctive autophagosomes with whirls of multilayered membranes residing in a cell soma. Taken together, our observations suggest that Shh induces autophagy pathway activation in hippocampal neurons.
We went on to test whether the observed Shh-induced autophagy was mediated through phosphatidylinositol 3-kinase complexes (PtdIns3K) that are known to be essential during autophagy (Chen and Klionsky, 2011). Different classes of PtdIns3Ks exert distinctly different effects: class I Ptdlns3K inhibits autophagy, whereas class III Ptdlns3K stimulates autophagy (Petiot et al., 2000). The Ptdlns3K signaling has been also linked to the Shh signaling pathway (Kenney et al., 2004; Riobó et al., 2006). We first asked whether class I Ptdlns3K was involved in ShhN-induced autophagy. Upon ShhN- or SAG-treatment, while increased LC3-II was once again detected, the level of Akt phosphorylation was not noticeably different between treated neurons and untreated control neurons (Fig. 4A). These results indicate that class I Ptdlns3K is not directly involved or does not play a prominent role in Shh-induced autophagy in the hippocampal neurons.
Fig. 4. Shh-induced autophagy requires class III Ptdlns3K activity.

(A) Hippocampal neurons were treated with ShhN (10%), SAG (100 nM), or ShhN in the presence of BFA (0.4 µM). Cell lysates were analysed by immunoblots with the indicated antibodies. Brain tissue from PTEN (phosphatase and tensin homologue deleted from chromosome 10) deletion mouse (Di Cristofano et al., 1998; Groszer et al., 2001) was used as a positive control for phosphorylated Akt (pAkt). (B) Immunoblots showing LC3-II level from control neurons, neurons treated with ShhN and BFA, or ShhN and class III PtdlnsK inhibitor 3-MA (10 mM). Notice that 3-MA blocks the ShhN-induced LC3-II increase. (C) Example confocal images of hippocampal neurons transfected with FYVE-DsRed and cultured in control medium, or ShhN, or ShhN plus 3-MA. Neurons are marked with a MAP2 antibody (turquoise in lower panel). Scale bar: 10 µm. Additional examples are shown in supplementary material Fig. S6. (D) Quantification of FYVE fluorescence intensity (a.u., arbitrary unit) in the soma of neurons (n≥45 neurons from 3 experiments). ***P<0.001 by Student's t test.
We then examined class III Ptdlns3K by adding 3-methyladenine (3-MA) to ShhN-treated neurons. 3-MA blocks the class III PtdlnsK activity necessary for autophagy induction (Seglen and Gordon, 1982). Addition of 3-MA effectively prevented ShhN-elicited LC3 elevation (Fig. 4B; supplementary material Fig. S1D), indicating that Shh-induced autophagy in hippocampal neurons is possibly mediated through class III Ptdlns3K. We also used a fluorescent reporter, FYVE-DsRed. The FYVE domain is a zinc finger protein domain that binds Ptdlns(3)P specifically (Kutateladze et al., 1999). It has been used as a reporter for levels of intracellular Ptdlns(3)P, and thus is a sensor for activity of class III Ptdlns3K (Zhang et al., 2007). We transiently transfected neurons (9–10 DIV) with FYVE-DsRed and treated these neurons with ShhN (10%) for 48 hr. Confocal microscope images showed that, without addition of ShhN, transfected neurons have diffuse fluorescence signals with occasional punctate structures (Fig. 4C; supplementary material Fig. S6A). By contrast, with addition of ShhN, transfected neurons displayed large punctate FYVE-DsRed structures and quantification showed an approximately 2-fold increase in FYVE score (Fig. 4C,D; additional examples in supplementary material Fig. S6B). Moreover, the increased FYVE was completely blocked by 3-MA (Fig. 4C,D; also supplementary material Fig. S6B). Thus, our data suggest that Shh-triggered autophagy requires class III Ptdlns3K activity.
Our results present evidence that hippocampal neurons can respond to Shh signaling by upregulating autophagy activity. In a previous study while addressing the subcellular location of the Shh's receptor Ptch, we repeatedly – and puzzlingly – noticed that Ptch immunolabeling was often located in close vicinity to autophagosomes that were found in normal hippocampal neurons of adult brain (Petralia et al., 2011a). This intimate physical association of Ptch and autophagosomes is not, in and of itself, sufficient to determine whether Ptch affects autophagosome formation or functions, or whether Ptch is being processed or degraded by autophagosomes. The new data described here imply that the association may be more than merely physical: that a functional association between Ptch – or the Shh signaling pathway – and autophagosomes probably exists. While our findings do not directly address this question, they provide the impetus to design functional experiments in future studies.
Two recent studies reported that Shh signaling indeed regulates autophagy (Li et al., 2012; Jimenez-Sanchez et al., 2012). One discrepancy, however, between these two studies was how the autophagy pathway responded to Shh signaling activity. While the study by Li et al., found enhanced autophagy by Shh that is consistent with our findings, the study by Jimenez-Sanchez et al., reported the opposite – suppressed autophagosome synthesis by Shh signaling. This contradiction could result from differences in experimental conditions and designs such as the extent and duration of Shh pathway activation, the basal autophagy level, the method used in inducing autophagy, and perhaps most physiologically relevant, the type of cells studied.
The biological significance of the interaction between the Shh signaling pathway and autophagy in neurons is not yet clear. In neural stem cells, Shh serves as a mitogen (Lai et al., 2003; Palma et al., 2005; Breunig et al., 2008; Han et al., 2008), although it is unknown if it is mediated through autophagy. In differentiated neurons, Shh-enhanced autophagy could be responsible for some of Shh's functions, including the newly described role in presynaptic differentiation of hippocampal neurons (Mitchell et al., 2012), and in circuit formation and maintenance of other types of neurons in the adult brain (Gonzalez-Reyes et al., 2012; Harwell et al., 2012). The chief function of autophagy is to remove damaged proteins and organelles (Mizushima, 2007). In addition, autophagy participates in normal turnover of various intracellular components (Mizushima, 2007), a critical process that takes place in developing synaptic terminals, and is used by growing neurons. Indeed, autophagy has been reported to promote synapse formation in Drosophila (Shen and Ganetzky, 2009), although whether Shh activity is involved has not been tested. Given that autophagy also serves as a cellular adaptive response to stress, it is not surprising that autophagy has been shown to protect neurons against degeneration in animal models of Alzheimer's disease (Yang et al., 2011) and ischemic stroke (Wang et al., 2012). Finally, one might speculate that Shh-induced autophagy could also control or modify Shh signaling activity, a feedback regulatory mechanism analogous to the mechanism used by the Wnt signaling pathway (Gao et al., 2010). In total, this study demonstrates that the interaction between Shh signaling and autophagy may be involved not only in autophagy regulation, but also in regulation of Shh signaling activity in neurons.
Materials and Methods
Hippocampal neuron culture and reagents
All animal procedures were approved by the NIA Animal Care and Use Committee, and complied with the NIH Guide for Care and Use of Laboratory Animals. Cultures of hippocampal neurons were prepared from embryonic day 18 rat brains, and dissociated neurons were grown in Neurobasal medium/B27 as described (Mattson et al., 1989; Kaech and Banker, 2006; Bushlin et al., 2008).
Shh-N-conditioned medium from HEK 293 cells was prepared as described (Chen et al., 2002). Control medium was prepared from non-transfected HEK 293 cells. Throughout this study, we use ShhN to refer to Shh-N-conditioned medium. ShhN- or control-medium was used at 10%. The efficacy of ShhN was validated using the Shh-light2 assay (Taipale et al., 2000; Mitchell et al., 2012). SAG (Shh agonist) was purchased from Axxora (ALX-270–426) and used at 100 nM (Chen et al., 2002). Robotnikinin was from BioVision and used at 50 µM (Stanton et al., 2009). 10-NCP was from EMD (Calbiochem, 124020). BFA and 3-MA were from Sigma. LC3 antibody was kindly provided by Dr Jay Debnath (UCSF) and also purchased from Cell Signaling. LBPA antibody was from Echelon Biosciences, Inc. (Z-SLBPA). MAP2 antibody was from Sigma. FYVE-DsRed was a kind gift of Dr Junying Yuan (Harvard Medical School).
Immunoblotting, immunocytochemistry, transfection, and immunofluorescence microscopy
Lysates of cultured neurons were prepared as described (Petralia and Yao, 2007). Immunoblot analysis was conducted using 4–12% gradient gels followed by standard blotting procedure. Blots were scanned and quantified by ImageJ.
Immunofluorescence labeling was carried out as described (Bushlin et al., 2008).
Briefly, following fixation with 4% paraformaldehyde, neurons were permeabilized in 0.2% Triton X-100 and incubated with primary antibody (LC3, 1:200; LBPA, 1:100; MAP2, 1:2,500). Neurons were then incubated with appropriate secondary antibody and mounted in Prolong antifade containing DAPI. For FYVE-DsRed experiments, neurons were transfected using the calcium phosphate kit (Clontech), followed by treatment with ShhN. Neurons were fixed and analyzed 2 days later.
Labeled or transfected neurons were examined using a 40× or a 63× objective on a Zeiss LSM710 laser scanning confocal microscope. Confocal acquisition settings were kept the same for those samples when quantification was performed. Brightness, contrast and levels of images were minimally adjusted (in Adobe Photoshop 8.0).
Electron microscopy
Electron microscopy was performed as described (Petralia and Wenthold, 1999; Petralia et al., 2010; Mitchell et al., 2012). Briefly, neurons were fixed in 2% paraformaldehyde 2% glutaraldehyde, postfixed in 1% osmium tetroxide and embedded in epon. The glass coverslip was removed with hydrofluoric acid and thin sections were stained with lead citrate.
From 2 independent cultures, we photographed 215 randomly selected fields from control neurons and 192 from ShhN-treated neurons. Traceable presynaptic profiles from these micrographs were selected for further analyses (229 for control and 229 for ShhN-treated). The number of profiles containing a clearly identifiable multivesicular body or autophagosome was counted (Fig. 3G).
Data analysis
Statistical comparisons were performed by using Student's t test. All results were expressed as mean±s.e.m.
Supplementary Material
Acknowledgments
We thank Dr Carolyn Marie Ott and Dr Jennifer Lippincott-Schwartz for discussions and reagents; Dr Simonetta Camandola for advice on PtdIns3K related experiments; Dr James K. Chen for Shh-N expressing HEK 293 cells; Dr Jay Debnath for LC3 antibody; Dr Steven Finkbeiner for information on 10-NCP; Dr JunyingYuan for FYVE-DsRed; Dr Fred Indig for confocal microscopy. This work was supported by the Intramural Research Programs of the NIA/NIH and NIDCD/NIH.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Boland B., Kumar A., Lee S., Platt F. M., Wegiel J., Yu W. H., Nixon R. A. (2008). Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J. Neurosci. 28, 6926–6937 10.1523/JNEUROSCI.0800-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig J. J., Sarkisian M. R., Arellano J. I., Morozov Y. M., Ayoub A. E., Sojitra S., Wang B., Flavell R. A., Rakic P., Town T. (2008). Primary cilia regulate hippocampal neurogenesis by mediating sonic hedgehog signaling. Proc. Natl. Acad. Sci. USA 105, 13127–13132 10.1073/pnas.0804558105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushlin I., Petralia R. S., Wu F., Harel A., Mughal M. R., Mattson M. P., Yao P. J. (2008). Clathrin assembly protein AP180 and CALM differentially control axogenesis and dendrite outgrowth in embryonic hippocampal neurons. J. Neurosci. 28, 10257–10271 10.1523/JNEUROSCI.2471-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Klionsky D. J. (2011). The regulation of autophagy – unanswered questions. J. Cell Sci. 124, 161–170 10.1242/jcs.064576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. K., Taipale J., Young K. E., Maiti T., Beachy P. A. (2002). Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. USA 99, 14071–14076 10.1073/pnas.182542899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmane N., Ruiz i Altaba A. (1999). Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 126, 3089–3100. [DOI] [PubMed] [Google Scholar]
- Dass B., Iravani M. M., Huang C., Barsoum J., Engber T. M., Galdes A., Jenner P. (2005). Sonic hedgehog delivered by an adeno-associated virus protects dopaminergic neurones against 6-OHDA toxicity in the rat. J. Neural Transm. 112, 763–778 10.1007/s00702-004-0227-7 [DOI] [PubMed] [Google Scholar]
- Dean C., Scheiffele P. (2009). Imaging synaptogenesis by measuring accumulation of synaptic proteins. Cold Spring Harb. Protoc. 11, pdb.prot5315 10.1101/pdb.prot5315 [DOI] [PubMed] [Google Scholar]
- Di Cristofano A., Pesce B., Cordon–Cardo C., Pandolfi P. P. (1998). Pten is essential for embryonic development and tumour suppression. Nat. Genet. 19, 348–355 10.1038/1235 [DOI] [PubMed] [Google Scholar]
- Dorn K. V., Hughes C. E., Rohatgi R. (2012). A Smoothened-Evc2 complex transduces the Hedgehog signal at primary cilia. Dev. Cell 23, 823–835 10.1016/j.devcel.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen E. L., Reggiori F., Baba M., Kovács A. L., Seglen P. O. (2011). Seeing is believing: the impact of electron microscopy on autophagy research. Autophagy 7, 935–956 10.4161/auto.7.9.15760 [DOI] [PubMed] [Google Scholar]
- Fader C. M., Colombo M. I. (2009). Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ. 16, 70–78 10.1038/cdd.2008.168 [DOI] [PubMed] [Google Scholar]
- Fletcher T. L., De Camilli P., Banker G. (1994). Synaptogenesis in hippocampal cultures: evidence indicating that axons and dendrites become competent to form synapses at different stages of neuronal development. J. Neurosci. 14, 6695–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Cao W., Bao L., Zuo W., Xie G., Cai T., Fu W., Zhang J., Wu W., Zhang X.et al. (2010). Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat. Cell Biol. 12, 781–790 10.1038/ncb2082 [DOI] [PubMed] [Google Scholar]
- Gonzalez–Reyes L. E., Verbitsky M., Blesa J., Jackson–Lewis V., Paredes D., Tillack K., Phani S., Kramer E. R., Przedborski S., Kottmann A. H. (2012). Sonic hedgehog maintains cellular and neurochemical homeostasis in the adult nigrostriatal circuit. Neuron 75, 306–319 10.1016/j.neuron.2012.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszer M., Erickson R., Scripture–Adams D. D., Lesche R., Trumpp A., Zack J. A., Kornblum H. I., Liu X., Wu H. (2001). Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science 294, 2186–2189 10.1126/science.1065518 [DOI] [PubMed] [Google Scholar]
- Han Y. G., Spassky N., Romaguera–Ros M., Garcia–Verdugo J. M., Aguilar A., Schneider–Maunoury S., Alvarez–Buylla A. (2008). Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat. Neurosci. 11, 277–284 10.1038/nn2059 [DOI] [PubMed] [Google Scholar]
- Harwell C. C., Parker P. R., Gee S. M., Okada A., McConnell S. K., Kreitzer A. C., Kriegstein A. R. (2012). Sonic hedgehog expression in corticofugal projection neurons directs cortical microcircuit formation. Neuron 73, 1116–1126 10.1016/j.neuron.2012.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman R. T., Feng B. Y., Ni J., Woo W. M., Milenkovic L., Hayden Gephart M. G., Teruel M. N., Oro A. E., Chen J. K., Scott M. P. (2011). Neuropilins are positive regulators of Hedgehog signal transduction. Genes Dev. 25, 2333–2346 10.1101/gad.173054.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P. W., McMahon A. P. (2001). Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15, 3059–3087 10.1101/gad.938601 [DOI] [PubMed] [Google Scholar]
- Jimenez–Sanchez M., Menzies F. M., Chang Y. Y., Simecek N., Neufeld T. P., Rubinsztein D. C. (2012). The Hedgehog signalling pathway regulates autophagy. Nat. Commun. 3, 1200 10.1038/ncomms2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S., Banker G. (2006). Culturing hippocampal neurons. Nat. Protoc. 1, 2406–2415 10.1038/nprot.2006.356 [DOI] [PubMed] [Google Scholar]
- Kenney A. M., Widlund H. R., Rowitch D. H. (2004). Hedgehog and PI-3 kinase signaling converge on Nmyc1 to promote cell cycle progression in cerebellar neuronal precursors. Development 131, 217–228 10.1242/dev.00891 [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., Abdalla F. C., Abeliovich H., Abraham R. T., Acevedo–Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre–Ghiso J. A.et al. (2012). Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 445–544 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze T. G., Ogburn K. D., Watson W. T., de Beer T., Emr S. D., Burd C. G., Overduin M. (1999). Phosphatidylinositol 3-phosphate recognition by the FYVE domain. Mol. Cell 3, 805–811 10.1016/S1097-2765(01)80013-7 [DOI] [PubMed] [Google Scholar]
- Lai K., Kaspar B. K., Gage F. H., Schaffer D. V. (2003). Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat. Neurosci. 6, 21–27 10.1038/nn983 [DOI] [PubMed] [Google Scholar]
- Li H., Li J., Li Y., Singh P., Cao L., Xu L. J., Li D., Wang Y., Xie Z., Gui Y.et al. (2012). Sonic hedgehog promotes autophagy of vascular smooth muscle cells. Am. J. Physiol. Heart Circ. Physiol. 303, H1319–H1331 10.1152/ajpheart.00160.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou W., Geuze H. J., Geelen M. J., Slot J. W. (1997). The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J. Cell Biol. 136, 61–70 10.1083/jcb.136.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo H., Chevallier J., Mayran N., Le Blanc I., Ferguson C., Fauré J., Blanc N. S., Matile S., Dubochet J., Sadoul R.et al. (2004). Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 303, 531–534 10.1126/science.1092425 [DOI] [PubMed] [Google Scholar]
- Mattson M. P., Murrain M., Guthrie P. B., Kater S. B. (1989). Fibroblast growth factor and glutamate: opposing roles in the generation and degeneration of hippocampal neuroarchitecture. J. Neurosci. 9, 3728–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies F. M., Moreau K., Puri C., Renna M., Rubinsztein D. C. (2012). Measurement of autophagic activity in mammalian cells. Curr. Protoc. Cell Biol. 54, 15.16.1–15.16.25 10.1002/0471143030.cb1516s54 [DOI] [PubMed] [Google Scholar]
- Mitchell N., Petralia R. S., Currier D. G., Wang Y. X., Kim A., Mattson M. P., Yao P. J. (2012). Sonic hedgehog regulates presynaptic terminal size, ultrastructure and function in hippocampal neurons. J. Cell Sci. 125, 4207–4213 10.1242/jcs.105080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. (2007). Autophagy: process and function. Genes Dev. 21, 2861–2873 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- Palma V., Lim D. A., Dahmane N., Sánchez P., Brionne T. C., Herzberg C. D., Gitton Y., Carleton A., Alvarez–Buylla A., Ruiz i Altaba A. (2005). Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development 132, 335–344 10.1242/dev.01567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez–Sala D., Boya P., Ramos I., Herrera M., Stamatakis K. (2009). The C-terminal sequence of RhoB directs protein degradation through an endo-lysosomal pathway. PLoS ONE 4, e8117 10.1371/journal.pone.0008117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petiot A., Ogier–Denis E., Blommaart E. F., Meijer A. J., Codogno P. (2000). Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 275, 992–998 10.1074/jbc.275.2.992 [DOI] [PubMed] [Google Scholar]
- Petralia R. S., Wenthold R. J. (1999). Immunocytochemistry of NMDA receptors. Methods Mol. Biol. 128, 73–92 10.1385/1-59259-683-5:73 [DOI] [PubMed] [Google Scholar]
- Petralia R. S., Yao P. J. (2007). AP180 and CALM in the developing hippocampus: expression at the nascent synapse and localization to trafficking organelles. J. Comp. Neurol. 504, 314–327 10.1002/cne.21454 [DOI] [PubMed] [Google Scholar]
- Petralia R. S., Wang Y. X., Hua F., Yi Z., Zhou A., Ge L., Stephenson F. A., Wenthold R. J. (2010). Organization of NMDA receptors at extrasynaptic locations. Neuroscience 167, 68–87 10.1016/j.neuroscience.2010.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia R. S., Schwartz C. M., Wang Y. X., Mattson M. P., Yao P. J. (2011a). Subcellular localization of patched and smoothened, the receptors for Sonic hedgehog signaling, in the hippocampal neuron. J. Comp. Neurol. 519, 3684–3699 10.1002/cne.22681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia R. S., Wang Y. X., Mattson M. P., Yao P. J. (2011b). Sonic hedgehog distribution within mature hippocampal neurons. Commun. Integr. Biol. 4, 775–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia R. S., Wang Y. X., Indig F. E., Bushlin I., Wu F., Mattson M. P., Yao P. J. (2013). Reduction of AP180 and CALM produces defects in synaptic vesicle size and density. Neuromolecular Med. 15, 49–60 10.1007/s12017-012-8194-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly J. O., Karavanova I. D., Williams K. P., Mahanthappa N. K., Allendoerfer K. L. (2002). Cooperative effects of Sonic hedgehog and NGF on basal forebrain cholinergic neurons. Mol. Cell. Neurosci. 19, 88–96 10.1006/mcne.2001.1063 [DOI] [PubMed] [Google Scholar]
- Riobó N. A., Lu K., Ai X., Haines G. M., Emerson C. P., Jr (2006). Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc. Natl. Acad. Sci. USA 103, 4505–4510 10.1073/pnas.0504337103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki N., Kurisu J., Kengaku M. (2010). Sonic hedgehog signaling regulates actin cytoskeleton via Tiam1-Rac1 cascade during spine formation. Mol. Cell. Neurosci. 45, 335–344 10.1016/j.mcn.2010.07.006 [DOI] [PubMed] [Google Scholar]
- Seglen P. O., Gordon P. B. (1982). 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. USA 79, 1889–1892 10.1073/pnas.79.6.1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Ganetzky B. (2009). Autophagy promotes synapse development in Drosophila. J. Cell Biol. 187, 71–79 10.1083/jcb.200907109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton B. Z., Peng L. F., Maloof N., Nakai K., Wang X., Duffner J. L., Taveras K. M., Hyman J. M., Lee S. W., Koehler A. N.et al. (2009). A small molecule that binds Hedgehog and blocks its signaling in human cells. Nat. Chem. Biol. 5, 154–156 10.1038/nchembio.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J., Chen J. K., Cooper M. K., Wang B., Mann R. K., Milenkovic L., Scott M. P., Beachy P. A. (2000). Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature 406, 1005–1009 10.1038/35023008 [DOI] [PubMed] [Google Scholar]
- Traiffort E., Charytoniuk D., Watroba L., Faure H., Sales N., Ruat M. (1999). Discrete localizations of hedgehog signalling components in the developing and adult rat nervous system. Eur. J. Neurosci. 11, 3199–3214 10.1046/j.1460-9568.1999.00777.x [DOI] [PubMed] [Google Scholar]
- Tsuboi K., Shults C. W. (2002). Intrastriatal injection of sonic hedgehog reduces behavioral impairment in a rat model of Parkinson's disease. Exp. Neurol. 173, 95–104 10.1006/exnr.2001.7825 [DOI] [PubMed] [Google Scholar]
- Tsvetkov A. S., Miller J., Arrasate M., Wong J. S., Pleiss M. A., Finkbeiner S. (2010). A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc. Natl. Acad. Sci. USA 107, 16982–16987 10.1073/pnas.1004498107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varjosalo M., Taipale J. (2008). Hedgehog: functions and mechanisms. Genes Dev. 22, 2454–2472 10.1101/gad.1693608 [DOI] [PubMed] [Google Scholar]
- Wallace V. A. (1999). Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr. Biol. 9, 445–448 10.1016/S0960-9822(99)80195-X [DOI] [PubMed] [Google Scholar]
- Wang P., Guan Y. F., Du H., Zhai Q. W., Su D. F., Miao C. Y. (2012). Induction of autophagy contributes to the neuroprotection of nicotinamide phosphoribosyltransferase in cerebral ischemia. Autophagy 8, 77–87 10.4161/auto.8.1.18274 [DOI] [PubMed] [Google Scholar]
- Wechsler–Reya R. J., Scott M. P. (1999). Control of neuronal precursor proliferation in the cerebellum by Sonic hedgehog. Neuron 22, 103–114 10.1016/S0896-6273(00)80682-0 [DOI] [PubMed] [Google Scholar]
- Xu Q., Guo L., Moore H., Waclaw R. R., Campbell K., Anderson S. A. (2010). Sonic hedgehog signaling confers ventral telencephalic progenitors with distinct cortical interneuron fates. Neuron 65, 328–340 10.1016/j.neuron.2010.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., Tashiro Y. (1998). Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 23, 33–42 10.1247/csf.23.33 [DOI] [PubMed] [Google Scholar]
- Yang D. S., Stavrides P., Mohan P. S., Kaushik S., Kumar A., Ohno M., Schmidt S. D., Wesson D., Bandyopadhyay U., Jiang Y.et al. (2011). Reversal of autophagy dysfunction in the TgCRND8 mouse model of Alzheimer's disease ameliorates amyloid pathologies and memory deficits. Brain 134, 258–277 10.1093/brain/awq341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylä–Anttila P., Vihinen H., Jokitalo E., Eskelinen E. L. (2009). 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5, 1180–1185 10.4161/auto.5.8.10274 [DOI] [PubMed] [Google Scholar]
- Zhang L., Yu J., Pan H., Hu P., Hao Y., Cai W., Zhu H., Yu A. D., Xie X., Ma D.et al. (2007). Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc. Natl. Acad. Sci. USA 104, 19023–19028 10.1073/pnas.0709695104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



