Summary
Fibroblast growth factor (Fgf) signaling plays important roles in various developmental processes including brain development. Here, we identified zebrafish fgf22 predominantly expressed in the posterior midbrain and anterior midbrain–hindbrain boundary (MHB) primordia during early embryonic brain development. To examine roles of Fgf22 in midbrain development, we analyzed fgf22 knockdown embryos. The fgf22 morphants were defective in proper formation of the MHB constriction and the midbrain. The knockdown of fgf22 caused decreased cell proliferation in the midbrain, expanded expression of roof plate and tegmental marker genes, and decreased expression of tectal marker genes, indicating that Fgf22 is required for cell proliferation, roof plate formation, and tectum specification in the midbrain. Fgf receptor 2b (Fgfr2b), a potential receptor for Fgf22, was also required, indicating that Fgf22 signaling is mediated through Fgfr2b. The floor plate and the MHB are crucial for the dorsoventral patterning of the midbrain through Hedgehog (Hh) and Fgf signaling, respectively. The fgf3/fgf8 double morphant phenotype was essentially similar to that of fgf22 morphants, whereas the phenotype caused by inhibition of Hh signaling was not. fgf3 and fgf8 were expressed earlier than fgf22 in the MHB primordium and Fgf3/Fgf8 signaling was required for fgf22 expression in the posterior midbrain. Furthermore, fgf22 partially rescued the fgf3/fgf8 double morphant phenotype. The present results indicate Fgf22 to be involved in midbrain development downstream of Fgf3 and Fgf8 in the MHB but not of Hh in the floor plate.
Key words: Fgf, Fgf22, Fgf3, Fgf8, Zebrafish, Development, MHB, Midbrain, Regionalization, Proliferation
Introduction
During early embryonic brain development in vertebrates, the neural plate is regionalized along the anteroposterior (A/P) and dorsoventral (D/V) axis. Patterning along the A/P and D/V axis of the neural tube is finely regulated by signals that emanate from adjacent tissues and/or from the neuroepithelium itself. The best characterized local organizing centers involved in the refinement of A/P and D/V patterns are the roof plate and floor plate, the anterior neural ridge, the zona limitans intrathalamica, and the isthmic organizer, also referred to as the midbrain–hindbrain boundary (MHB) (reviewed by Altmann and Brivanlou, 2001; Briscoe and Ericson, 2001; Liu and Joyner, 2001; Rhinn and Brand, 2001; Simeone, 2002; Wilson et al., 2002). Among them, the roof plate and floor plate are specialized structures that mark the dorsal and ventral midline of the neural tube, respectively, and are involved in D/V patterning. D/V patterning mechanisms have been best studied in the developing spinal cord and depend on the relative amount of a ventralizing factor, Sonic hedgehog (Shh), provided by the floor plate and notochord and dorsalizing factors, Bone morphogenic proteins (Bmps), produced by the roof plate. In mice lacking Shh gene function, the nervous system shows abnormalities in the development of ventral midline structures like the floor plate and notochord and the differentiation of ventral cell types (Chiang et al., 1996). Conversely, the misexpression of Shh transforms cell fate specification, from dorsal to ventral cells, and induces differentiation into ventral neuronal cell types in the dorsal region (Agarwala et al., 2001). On the other hand, the Bmps coordinate dorsal patterning of the neural tube and the generation of different dorsal neuronal cell types in the spinal cord. The disruption of the Bmp antagonist Noggin induces D/V patterning defects in the neural tube and ventral neurons are missing in noggin mutants (Alexandre and Wassef, 2005). These general mechanisms of D/V patterning are common to the spinal cord and midbrain. However, the exact functions of genes involved in D/V patterning of the midbrain and the interactions between these genes are still not well understood. Furthermore, several observations suggest that midbrain D/V patterning requires additional signals.
Fibroblast growth factors (Fgfs) make up a large family comprising 22 members in mammals. Among them, Fgf22 is a member of the Fgf7 subfamily (Itoh and Ornitz, 2004). Fgf signaling is mediated by Fgf receptor (Fgfr) proteins, which belong to a family of tyrosine kinase-containing transmembrane proteins that bind to Fgf molecules. The Fgfr gene family comprises four members, Fgfr1-Fgfr4 (Itoh and Ornitz, 2004). Fgf22 preferentially binds to a product of the Fgfr2b gene (Zhang et al., 2006). Here, we identified zebrafish fgf22 predominantly expressed in the posterior midbrain and anterior MHB primordia during early embryonic brain development. Fgf22 was critical for cell proliferation, the formation of the roof plate, and the specification of the tectum through Fgfr2b in the midbrain. In addition to the floor plate, the isthmic organizer is crucial for the patterning of the midbrain through the production of several secreted molecules (Alexandre and Wassef, 2005). However, less is known about the influence of the isthmic organizer on the midbrain D/V patterning. fgf3 and fgf8 were expressed at earlier stages than fgf22 in the MHB primordium. Fgf3/Fgf8 signaling was required for fgf22 expression in the posterior midbrain. The fgf3/fgf8 double morphant phenotype was essentially similar to that of fgf22 morphants, and partially rescued by fgf22. However, the phenotype caused by inhibition of Hedgehog (Hh) signaling in the floor plate differed from that of fgf22 morphants. The present results indicate that Fgf22 regulated by Fgf3/Fgf8 signaling but not by Hh signaling is involved in the formation of the roof plate and the specification of the tectum through Fgfr2b in the midbarin. The present findings should provide new insights into roles of Fgf signaling in midbrain development.
Materials and Methods
Fish maintenance
Zebrafish (Danio rerio) were maintained, referring to The Zebrafish Book (Westerfield, 1995). Embryos were obtained by natural spawning and cultured at 28.5°C in Zebrafish Ringer's solution. The developmental stages of the embryos were determined by the hours post fertilization (hpf) and by morphological features, as described by Kimmel et al. (Kimmel et al., 1995).
Isolation and characterization of zebrafish Fgf22 cDNA
Zebrafish fgf22 was identified by BLAST (Basic Local Alignment Search Tool, http://blast.ncbi.nlm.nih.gov/Blast.cgi) – searching zebrafish cDNA and genomic DNA sequences with the amino acid sequence of human FGF22. The full-length cDNA was isolated by polymerase chain reaction (PCR) with zebrafish embryonic cDNA as a template. The GenBank accession number for the fgf22 cDNA is AB254028.
The positions of zebrafish fgf22, bsg, hcn2, and polrmt on chromosome 22 were obtained from the Ensembl Zebrafish Genome Browser (http://www.ensembl.org/Danio_rerio). The map positions of human FGF22, BSG, HCN2, and POLRMT on chromosome 19 were obtained from LocusLink (http://www.ncbi.nlm.nih.gov/genome/guide).
Temporal expression profiles were determined by reverse transcription-polymerase chain reaction (RT-PCR) using the following primers (5′ primer/3′ primer): fgf22, 5′-CATCATGCCGACTGCTTGTGCA-3′/5′-TGATGAAGTGTCCGGCTATGTG-3′ (688 bp fragment) and zebrafish elongation factor 1-a (ef1α) (Miyake et al., 2005).
Whole mount in situ hybridization and sectioning
Digoxigenin- or fluorescein-labeled RNA probes were synthesized by in vitro transcription using T7 or SP6 RNA polymerase. A 0.7-kb fgf22 probe was synthesized using the full-length cDNA-containing plasmid. Other probes used were zebrafish wnt1 (Kelly and Moon, 1995), pax2.1 (Krauss et al., 1991), otx2 (Mori et al., 1994), eng2 (Ekker et al., 1992), her5 (Müller et al., 1996), Fgf8 (Reifers et al., 1998), Fgf3 (Phillips et al., 2001), nkx6.2 (Guner and Karlstrom, 2007), pax7a (Seo et al., 1998), lmx1b.2 (Elsen et al., 2008), bmp5 (Holzschuh et al., 2005), meis2a (Waskiewicz et al., 2001), and msxb (Ekker et al., 1997). Whole mount in situ hybridization was performed according to standard protocols and developed with BM Purple (Roche) and Fast Red (Roche).
Fixed embryos were transferred to 20% sucrose in PBS, mounted in OCT compound, and sectioned at 16 µm.
Injection of morpholino oligonucleotides
Morpholino oligonucleotides (MOs) were synthesized by Gene-Tools, LLC (Corvallis, OR). MOs were diluted in Danieau buffer (Nasevicius and Ekker, 2000). The sequences of MOs used are as follows: fgf22 exon 2/intron 2 splice-blocking MO1, 5′-ATGCGATGTACCTACCGATCCGAAAG-3′; fgf22 exon 1/intron 1 splice-blocking fgf22 MO2, 5′-AGCACTGTGTATCTACTCACTGTCA-3′; fgfr2b exon 7/intron 7 splice-blocking MO1, 5′-CCTGCTTTTTTACCTGGTATGACAA-3′; fgfr2b exon 7/intron 7 splice-blocking MO2, 5′-CCACGCTCCTGCTTTTTTACCTGGT-3′; and universal control MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′. fgf3 MO, fgf8 MO, and tp53 MO were reported previously (Maroon et al., 2002; Miyake et al., 2005; Gerety and Wilkinson, 2011; Miyake et al., 2012). fgf22 MO1 (12 ng), fgf22 MO2 (30 ng), fgfr2b MO1 (12 ng), fgfr2b MO2 (8 ng), or universal control MO (12 ng) was injected into zebrafish 2- to 4-cell embryos. fgf3 MO (10 µg/µl) and fgf8 MO (20 µg/µl) were injected at a volume of 0.15–0.25 nl into one- to two-cell embryos. tp53 MO (13.4 µg/µl) was injected at 0. 35–0.4 nl into 2-cell embryos.
To determine the efficacy of MOs, RNA was isolated from wild-type, fgf22 MO1, fgf22 MO2, fgfr2b MO1, or fgfr2b MO2-injected embryos. cDNA was amplified from the RNA by RT-PCR using the above primers and the following primers (5′ primer/3′ primer): fgfr2b, 5′-GAGCTCGGGCATAAACAGCT-3′/5′-CTGGAGGATAATCCGTCTCG-3′ (176 bp fragment) and fgfr2c, 5′-GACGGCAGGTGTGAACACTA-3′/5′- CTGGAGGATAATCCGTCTCG -3′ (182 bp fragment).
RNA injection
The entire coding region of zebrafish fgf22 cDNA was inserted into a vector, pCS2+ (Turner and Weintraub, 1994). Capped fgf22 mRNA was synthesized using a mMESSAGE mMACHINE kit (Ambion) from a linearized pCS2+ containing fgf22 cDNA. The mRNA was diluted to 10 ng/µl with water and injected in a volume of 1 nl into zebrafish 2- to 4-cell embryos.
H3P antibody staining and TUNEL assay
Proliferating and apoptotic cells were detected using a rabbit polyclonal anti-phosphorylated histone H3 (H3P) (Upstate Biotechnology) antibody and the DeadEnd colorimetric detection kit (Promega), respectively (Miyake et al., 2005).
For cell counts, the stained embryos were embedded in Technovit 7100 (Heraeus Kulzer, Wehrheim) and cut into 4-µm serial sections. The sections were counterstained with hematoxylin.
Cyclopamine treatments
Cyclopamine (Toronto Chemical) (Incardona et al., 1998) was dissolved at 10 mM in 95% ethanol. Embryos, in their chorions, were incubated in cyclopamine diluted to 100 µM in Zebrafish Ringer's solution starting at the time points indicated. Control embryos were treated simultaneously with an equal volume of 0.95% ethanol (cyclopamine carrier) in Zebrafish Ringer's solution.
Hydroxyurea–Aphidicholin (HUA) treatments
Mid-gastrula embryos (80% epiboly) were incubated in Zebrafish Ringer's solution containing 20 mM Hydroxyurea (Sigma–Aldrich), 150 µM Aphidicholin (Sigma–Aldrich), and 4% dimethyl sulfoxide (DMSO). Control embryos were treated simultaneously with an equal volume of 4% DMSO (HUA carrier) in Zebrafish Ringer's solution.
Results
Identification and characterization of zebrafish fgf22
Among vertebrates, amino acid sequences of most orthologous Fgfs are highly conserved (Itoh and Ornitz, 2004). A BLAST-search of the zebrafish cDNA and genomic DNA sequences with the amino acid sequence of human FGF22 identified a zebrafish amino acid sequence (207 amino acids) closely related to human FGF22 and mouse Fgf22 (supplementary material Fig. S1A). We isolated the full-length cDNA encoding the amino acid sequence from 24 hpf zebrafish embryo cDNA. Human FGF22 is closely linked to the BSG, HCN2, and POLRMT genes on chromosome 19 at p13.3 (supplementary material Fig. S1B). Therefore, we have examined this gene's location in the zebrafish genome. The gene was also closely linked to the zebrafish bsg, hcn2, and polrmt genes on chromosome 22 (supplementary material Fig. S1B). Thus, this gene was identified as zebrafish fgf22.
Expression pattern of fgf22
The temporal expression of fgf22 during embryonic development was examined by RT-PCR. As shown in Fig. 1A, fgf22 expression was first detected at low levels at 12 hpf. Subsequently, the expression gradually increased and was detected at least until 36 hpf.
Fig. 1. Expression of fgf22 in zebrafish embryos.
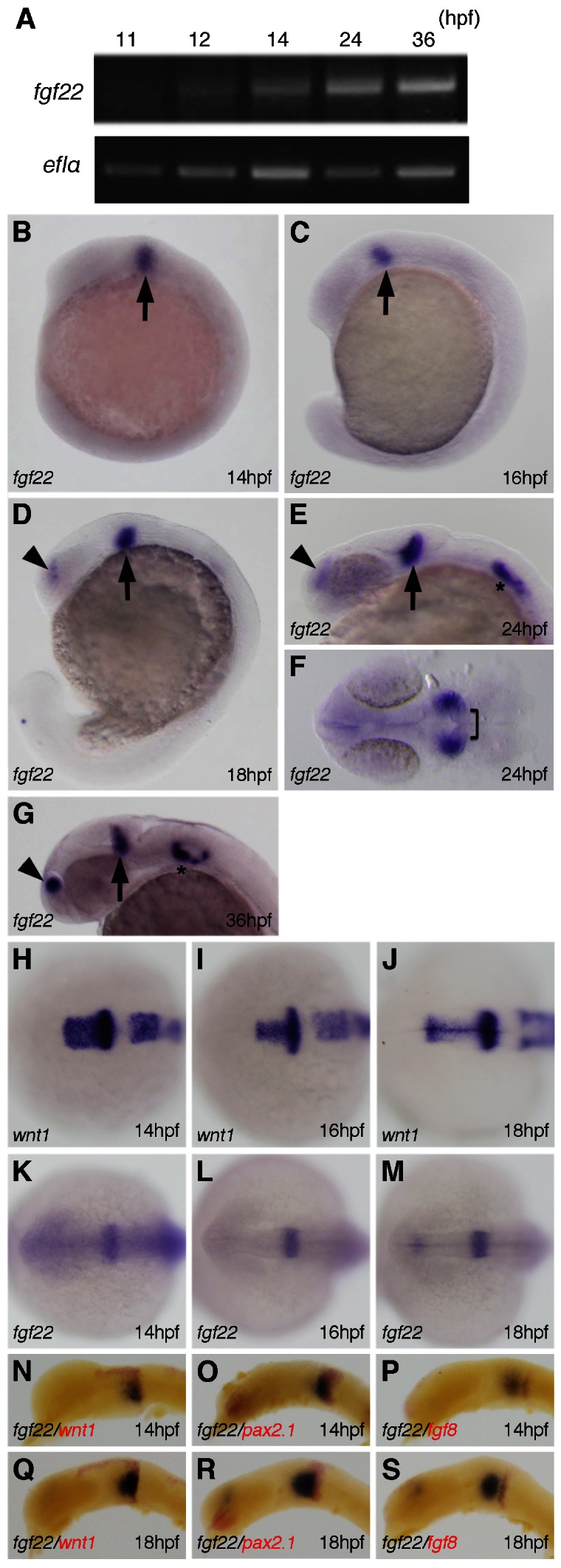
(A) Amplification of fgf22 by RT-PCR at the indicated stages. The lower panel shows results for ef1α as a control. (B–S) Expression pattern of fgf22 (B–G,K–S) and wnt1 (H–J) in zebrafish embryos at the indicated stages detected by whole-mount in situ hybridization. Embryos were double-labeled for wnt1 (red) (N,Q), pax2.1 (red) (O,R), or fgf8 (red) (P,S). Lateral views with anterior to the left and dorsal to the top (B–D,G,N–S). Dorsal views with anterior to the top (F,H–M). Arrowheads and asterisks indicate the telencephalon and otic vesicles, respectively.
We then investigated the spatiotemporal expression pattern of fgf22 by whole mount in situ hybridization. At 14 and 16 hpf, fgf22 was expressed near the posterior midbrain primordium (arrow), whereas fgf22 expression was not detected in the most dorsal part (Fig. 1B,C,H,I,K,L). To examine the spatial expression pattern of fgf22 in detail, the expression of fgf22 at 14 hpf was compared with those of wnt1, pax2.1 and fgf8, all of which are expressed persistently in the midbrain–hindbrain region (Lun and Brand, 1998). A comparison between fgf22 and wnt1 expression showed that the caudal boundary of fgf22 expression was nearly identical to the caudal boundary of the wnt1 expression domain (Fig. 1H,I,K,L,N). The caudal boundary of fgf22 expression was located posteriorly to the rostral boundary of pax2.1 expression and the posterior domain of fgf22 expression overlapped with the anterior domain of pax2.1 expression (Fig. 1O). On the other hand, the fgf8 expression domain was located at a distance from the fgf22 expression domain, since fgf8 was expressed in the posterior region contiguous to the pax2.1 expression domain (Fig. 1P). These observations indicate that fgf22 is expressed in both ventral and dorsal domains except most dorsal domain in the posterior midbrain and anterior MHB primordium. By 18 hpf, fgf22 expression had intensified in the ventral domain in the posterior midbrain and anterior MHB (Fig. 1D,J,M,Q–S). At 24 hpf, fgf22 expression was still detectable in the posterior midbrain (arrow), but no longer found in the anterior MHB (bracket) (Fig. 1E,F). The expression in the posterior midbrain (arrow) continued at least until 36 hpf (Fig. 1G). In addition, fgf22 was expressed in the telencephalon and otic vesicles at 18 and 24 hpf, respectively (Fig. 1D,E). At 36 hpf, fgf22 expression had intensified in both the telencephalon and the otic vesicles (Fig. 1G).
Inhibition of fgf22 functions results in defects in formation of the brain
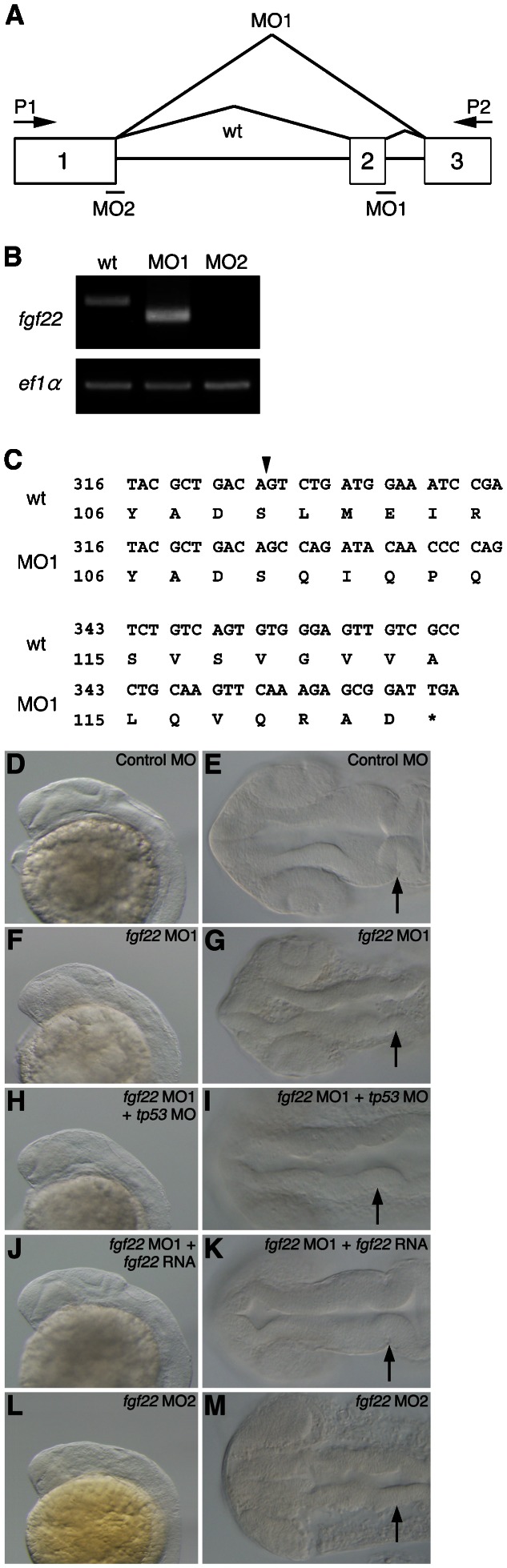
To examine the roles of fgf22 in zebrafish development, we performed knockdown experiments with MOs. We injected two independent splice-site-targeted MOs (MO1 and MO2) for fgf22 into 2-cell embryos and examined whether MOs could efficiently block the splicing of the fgf22 mRNA precursor in zebrafish embryos (Fig. 2A). Although the wild-type cDNA was subjected to normal splicing, the amplified cDNA from fgf22 MO1-injected embryos, which was shorter than the wild-type cDNA was subjected to abnormal splicing, resulting in a truncated translation product (Fig. 2B,C). In addition, the expression of mature fgf22 mRNA was greatly decreased in fgf22 MO2-injected embryos (Fig. 2B). These results indicate that both of the non-overlapping MOs effectively blocked the maturation of fgf22 mRNA.
Fig. 2. Inhibition of fgf22 functions in zebrafish embryos.
(A) The coding region of fgf22 is divided by two introns. Open boxes and black lines indicate exons and introns, respectively. MO indicates the target position of fgf22 MO. (B) fgf22 cDNA was amplified from wild-type or fgf22 MO-injected embryonic cDNA by RT-PCR using P1 and P2 primers, the positions of which are indicated by arrows (A). ef1α cDNA was also amplified as a control. (C) The nucleotide sequences of fgf22 cDNAs described above were determined. Numbers for the nucleotide sequence of the coding region and the amino acid sequence are shown. Arrowheads indicate splice-sites between exons one and two. (D–M) Lateral views (D,F,H,J,L) and dorsal views (E,G,I,K,M) of control MO-injected (D,E), fgf22 MO1-injected (F,G), fgf22 MO1- and tp53 MO-injected (H,I), fgf22 MO1- and fgf22 RNA-injected (J,K), and fgf22 MO2-injected (L,M) embryos at 24 hpf are shown.
The fgf22 morphants were morphologically defective in formation of the MHB constriction and exhibited a failure of the midbrain to evaginate laterally at 24 hpf (MO1, n = 407/476 and MO2, n = 79/99) (Fig. 2F,G,L,M). In addition, the fgf22 morphants showed morphological abnormality in the forebrain at 24 hpf (Fig. 2F,G,L,M). On the other hand, the control MO-injected embryos developed normally during embryogenesis (n = 25/25) (Fig. 2D,E). MOs might elicit undesirable off-target effects, which are rescued by co-knockdown of tp53 (Gerety and Wilkinson, 2011). We examined whether the co-injection of tp53 MO with fgf22 MO1 could rescue the phenotype of fgf22 MO1-injected embryos at 24 hpf. The co-injection of tp53 MO with fgf22 MO1 did not prevent the impaired neural development caused by fgf22 MO1 (n = 73/79) (Fig. 2H,I). Furthermore, the phenotype was also confirmed by RNA rescue experiments. The co-injection of fgf22 RNA with fgf22 MO1 rescued the defects in the brain caused by fgf22 MO1 (n = 37/51) (Fig. 2J,K). These results suggest that fgf22 is required for the formation of the MHB constriction, and normal development of the forebrain and midbrain during neurogenesis.
Cell proliferation in the midbrain is reduced in fgf22 morphants
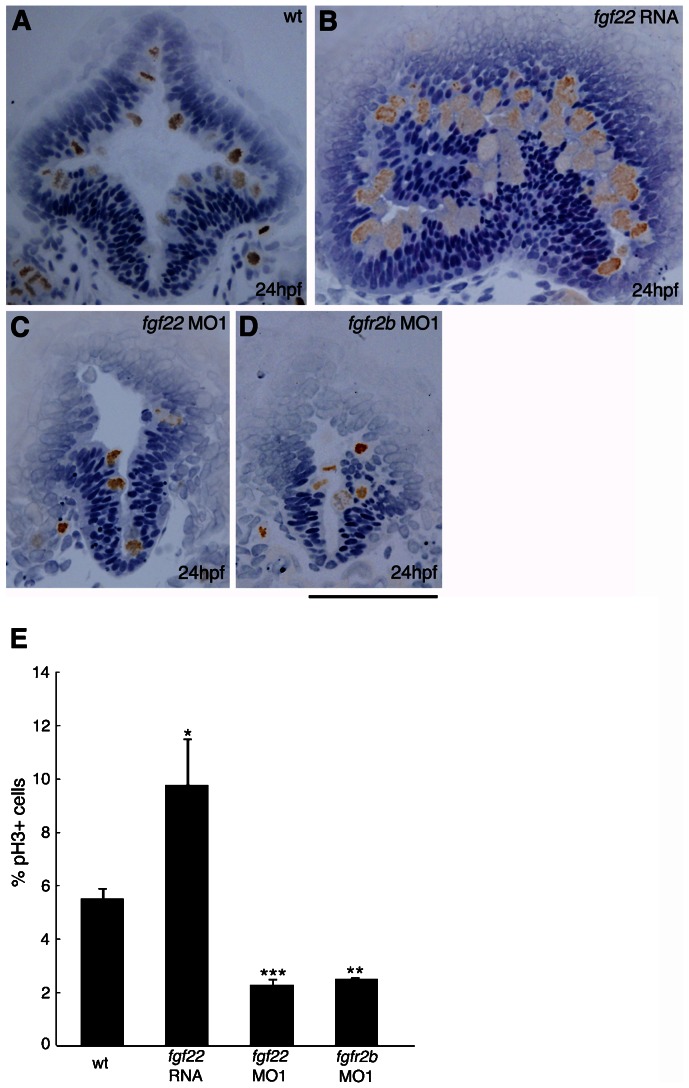
In mice, Fgf signaling regulates cell proliferation and cell survival in the midbrain (Xu et al., 2000; Chi et al., 2003; Trokovic et al., 2003). Therefore, we examined whether a defect in cell proliferation and/or cell survival could account for the observed morphological abnormality in the midbrain of fgf22 morphants. Phosphorylated histone H3 (pH3) was specifically detected in proliferating cells (Hendzel et al., 1997). We identified proliferating cells as pH3-positive cells. The rate of pH3-positive cells in the midbrain of fgf22 morphants was significantly decreased in comparison with that in wild-type embryos at 24 hpf (Fig. 3A,C,E). Conversely, the rate of pH3-positive cells in the midbrain was significantly increased in fgf22 RNA-injected embryos (Fig. 3A,B,E). These results suggest that fgf22 stimulates proliferation in the midbrain. Furthermore, fgf22 morphants were assayed for apoptotic cells via TUNEL labeling at 24 hpf. The number of apoptotic cells in the midbrain of fgf22 morphants was slightly increased in comparison with that in the wild-type embryos (n = 16/17) (supplementary material Fig. S2A,B).
Fig. 3. Comparison of cell proliferation in the midbrain of embryos injected with fgf22 RNA, fgf22 MO1, or fgfr2b MO1.
(A–D) Wild-type embryos (A) and embryos injected with fgf22 RNA (B), fgf22 MO1 (C), or fgfr2b MO1 (D) were stained using an anti-pH3 antibody. Panels show representative transverse sections of the midbrain at 24 hpf. Scale bar: 100 µm. (E) The percentage of proliferating cells labelled with anti-pH3 antibody in the midbrain of wild-type embryos and embryos injected with fgf22 RNA, fgf22 MO1, or fgfr2b MO1. Results are the mean ± S.D. for three independent sections from three embryos. The statistical significance of differences in mean values was assessed with the Student's t-test. Asterisks indicate statistical significance compared with the wild type (*P<0.05; **P<0.01; ***P<0.001).
Expression of roof plate marker genes is expanded in the midbrain of fgf22 morphants
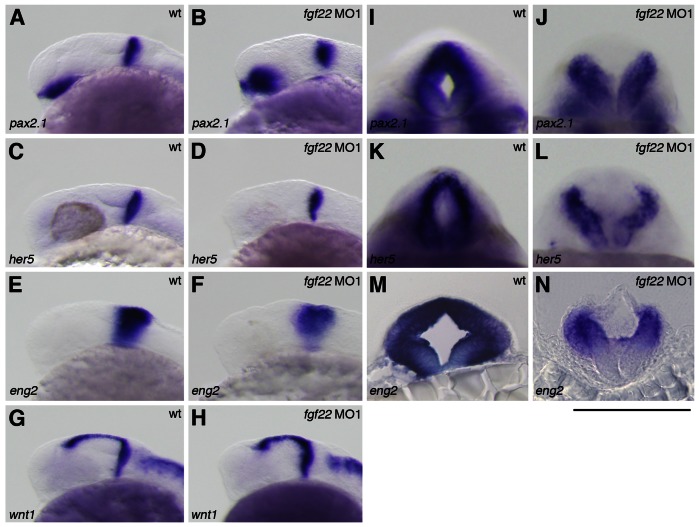
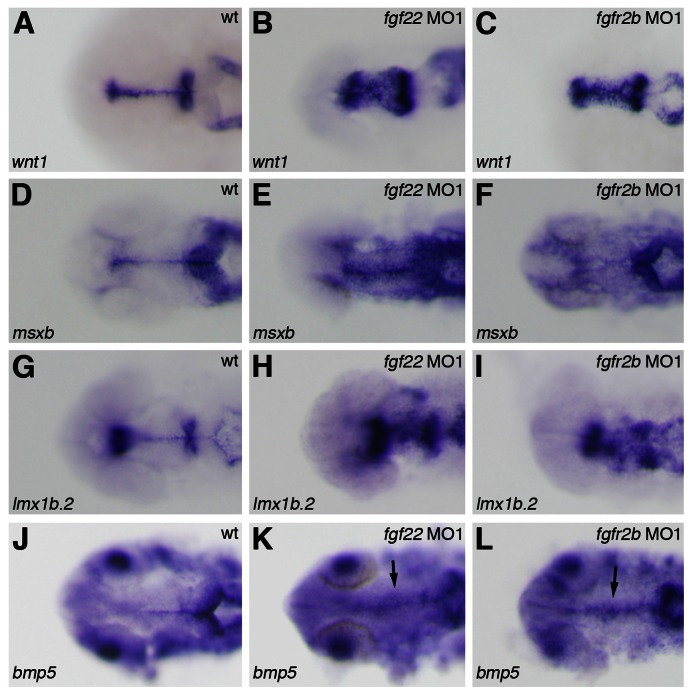
The fgf22 morphants showed morphological abnormality in the MHB constriction. Therefore, to investigate whether fgf22 is implicated in MHB development, we examined the expression of genes related with MHB patterning in fgf22 morphants at 24 hpf. In fgf22 morphants, the expression of pax2.1, her5, and eng2a was detected in the MHB (n = 27/27, n = 33/33, and n = 23/23, respectively) (Fig. 4A–F). However, optical cross-sections showed that the expression of pax2.1, her5, and eng2a in the dorsal domain of the MHB was eliminated or reduced in fgf22 morphants (n = 19/27, n = 24/33, and n = 17/23, respectively) (Fig. 4I–L; data not shown). On the other hand, the expression of wnt1 was detected in both the dorsal and ventral domains of the MHB in fgf22 morphants (n = 24/24) (Fig. 4G,H). These results indicate that loss of fgf22 function disrupts normal specification of the dorsal domain in the MHB. wnt1 is also expressed in the dorsal midline of the midbrain at 24 hpf (Fig. 5A). In fgf22 morphants, the lateral expansion of wnt1 expression was detected in the dorsal domain of the midbrain (MO1, n = 24/24 and MO2, n = 21/21) (Fig. 5A,B; supplementary material Fig. S3B). Furthermore, the expression of msxb, lmx1b.2, and bmp5, markers for the midbrain roof plate, in fgf22 morphants was up-regulated in the dorsal midbrain and their expression domains were expanded at 24 hpf (n = 31/32, n = 39/49, n = 15/16), respectively (Fig. 5D,E,G,H,J,K). Conversely, eng2a expression was eliminated in the dorsal domain of the posterior midbrain in fgf22 morphants (n = 17/23) (Fig. 4M,N). An analysis of transverse sections through the posterior midbrain showed that in fgf22 morphants, the roof plate, which is characteristically thin and marks the dorsal midline of the neural tube, was similarly thin but much wider than normal (Fig. 4M,N). These results suggest that Fgf22 signaling suppresses the roof plate fate in the midbrain.
Fig. 4. Gene expression in the MHB of the fgf22 morphants.
(A–N) The expression of pax2.1 (A,B,I,J), her5 (C,D,K,L), eng2 (E,F,M,N), and wnt1 (G,H) in wild-type embryos (A,C,E,G,I,K,M) and fgf22 morphants (B,D,F,H,J,L,N) at 24 hpf. A–H are lateral views, anterior to the left; I–L are optical cross-sections of the MHB; M,N are midbrain transverse sections. (I–N) Scale bar: 100 µm.
Fig. 5. Gene expression in the midbrain roof plate of the fgf22 and fgfr2b morphants.
The expression of wnt1 (A–C), msxb (D–F), lmx1b.2 (G–I), and bmp5 (J–L) in wild-type embryos (A,D,G,J) and fgf22 (B,E,H,K) and fgfr2b (C,F,I,L) morphants at 24 hpf.
D/V pattern forms incorrectly in the midbrain of fgf22 morphants
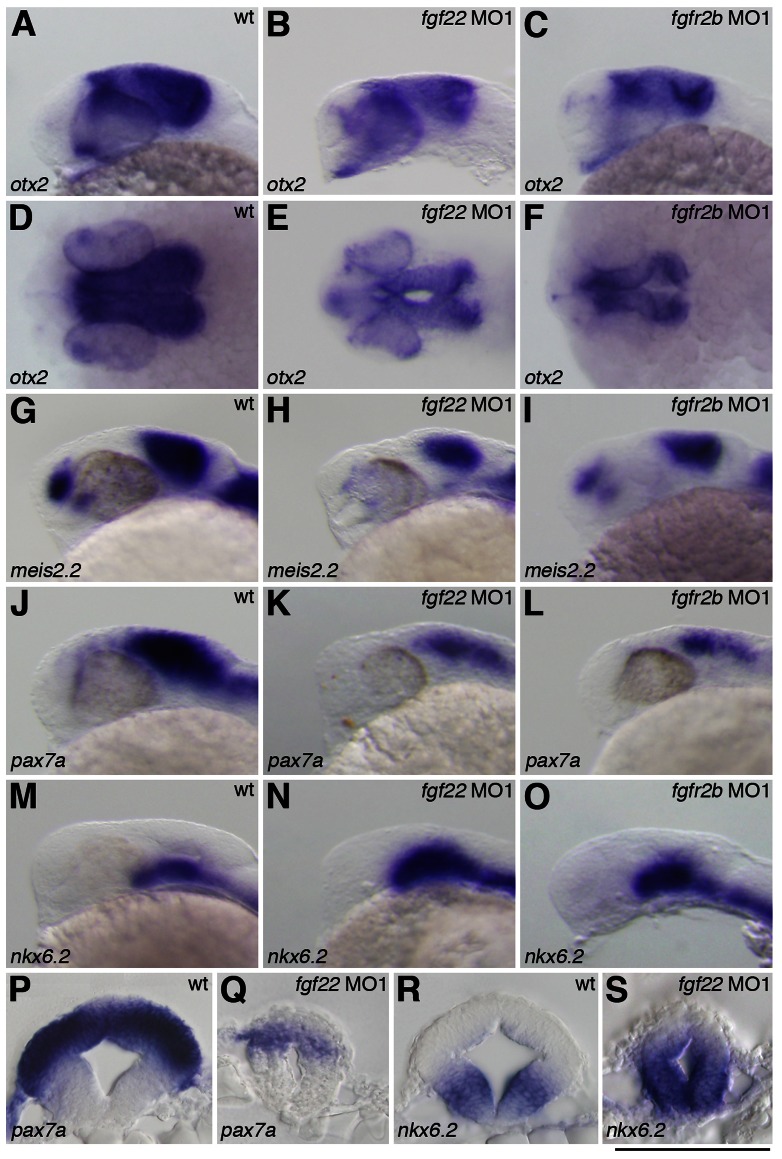
As mesencephalic morphology was altered following fgf22 knockdown, we investigated whether fgf22 was involved in specification of the midbrain. otx2 expressed in the midbrain is involved in midbrain patterning (Katahira et al., 2000). In fgf22 morphants, otx2 expression was down-regulated in the midbrain at 24 hpf (n = 28/30) (Fig. 6A,B). In particular, otx2 expression in the most dorsal domain of the tectum was completely eliminated in fgf22 morphants (n = 28/30) (Fig. 6D,E). In mice, Otx2 is also an important player in the regulation of midbrain D/V patterning (Alexandre and Wassef, 2003). Therefore, we investigated whether fgf22 is implicated in tectal fate specification. The expression of meis2.2 and pax7a was reduced in the most dorsal domain and the ventral domain of the tectum in fgf22 morphants at 24 hpf (MO1, n = 14/14, MO1, n = 15/15, and MO2, n = 13/14), respectively (Fig. 6G,H,J,K,P,Q; supplementary material Fig. S3D). Next, we investigated whether the reduction of tectal marker gene expression in fgf22 morphants was accompanied by the alteration of ventral marker gene expression. In fgf22 morphants, nkx6.2 expression was partially expanded into the dorsal region of the midbrain at 24 hpf (MO1, n = 21/23 and MO2, n = 16/17) (Fig. 6M,N,R,S; supplementary material Fig. S3F). Taken together, these results demonstrate that Fgf22 is required for normal tectal and tegumental development.
Fig. 6. Gene expression in the midbrain of the fgf22 and fgfr2b morphants.
The expression of otx2 (A–F), meis2.2 (G–I), pax7a (J–L,P,Q) and nkx6.2 (M–O,R,S) in wild-type embryos (A,D,G,J,M,P,R) and fgf22 (B,E,H,K,N,Q,S) and fgfr2b (C,F,I,L,O) morphants at 24 hpf. A–C,G–O are lateral views, anterior to the left; D–F are dorsal views; P–S are midbrain transverse sections. (P–S) Scale bar: 100 µm.
Inhibition of fgfr2b functions disrupts formation of both the dorsal and ventral midbrain
Fgfr genes contain an extracellular ligand-binding domain with three immunoglobulin-like domains (Ι, ΙΙ and ΙΙΙ), a transmembrane domain, and a split intercellular tyrosine kinase domain (Itoh and Ornitz, 2004). Among them, immunoglobulin-like domain ΙΙΙ is involved in the determination of ligand-binding specificity and Fgfr1-Fgfr3 encode two major versions of the domain (ΙΙΙb and ΙΙΙc) generated by alternative splicing (Itoh and Ornitz, 2004). Human FGF22 specifically bound to human FGFR2b in vitro and zebrafish fgfr2 are expressed in the midbrain during somitogenesis (Zhang et al., 2006; Ota et al., 2010). These findings suggest fgfr2b to be involved in the roles of fgf22 in the midbrain; therefore, we injected two splice-site-targeted MOs (MO1 and MO2) for fgfr2b into 2-cell embryos to investigate the role of fgfr2b in midbrain development. In embryos injected with fgfr2b MOs, the expression of mature fgfr2b mRNA was greatly decreased, whereas the expression of mature fgfr2c mRNA was unaffected (supplementary material Fig. S3A). As fgfr2b MOs could efficiently block the splicing of fgfr2b mRNA in embryos, we examined gene expression in the midbrain of fgfr2b morphants at 24 hpf. wnt1 expression was expanded laterally (MO1, n = 23/24 and MO2, n = 19/20) (Fig. 5C; supplementary material Fig. S3C). The expression of msxb, lmx1b.2, and bmp5 was also up-regulated in the dorsal midbrain (n = 43/43, n = 26/28, and n = 16/20, respectively) (Fig. 5F,I,L). On the other hand, pax7a expression was reduced in both the dorsal and ventral regions of the tectum (MO1, n = 20/21 and MO2, n = 14/18) (Fig. 6L; supplementary material Fig. S3E). The expression of otx2 and meis2.2 was also reduced in the tectum (n = 22/22 and n = 20/22, respectively) (Fig. 6C,F,I). Conversely, nkx6.2 expression in the tegmentum was expanded dorsally (MO1, n = 11/11 and MO2, n = 13/15) (Fig. 6O; supplementary material Fig. S3G). These results indicate that fgfr2b is involved in normal tectal and tegumental development.
Next, we examined proliferating cells in fgfr2b morphants at 24 hpf. The rate of pH3-positive cells in the midbrain was significantly decreased compared with that in wild-type embryos (Fig. 3D,E). These results suggest that fgfr2b is involved in cell proliferation. Thus, the phenotype of fgfr2b morphants was essentially similar to that of fgf22 morphants.
Phenotype of fgf22 knockdown in the midbrain differs from that caused by inhibition of Hh signaling
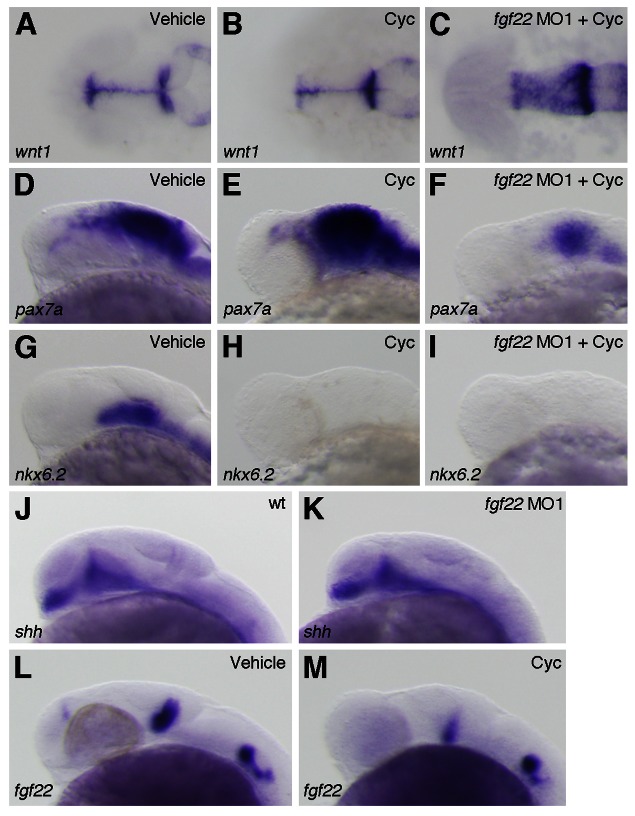
Hh molecules produced in the floor plate function in D/V midbrain patterning. The misexpression of Shh in the midbrain transforms cell fate specification, from dorsal to ventral (Agarwala et al., 2001; Bayly et al., 2007). Conversely, no ventral cells remain and markers for dorsal cells are extended ventrally in the midbrain of Shh null mutants (Fedtsova and Turner, 2001; Fogel et al., 2008). As the alkaloid cyclopamine completely blocked Hh signaling at the level of Smoothened, which transduces hedgehog signals, in zebrafish (Taipale et al., 2000; Miyake et al., 2005), we analyzed the D/V midbrain patterning in embryos treated with cyclopamine. The embryos treated with cyclopamine from 5 hpf onwards showed a normal expression of wnt1 in the dorsal midbrain at 24 hpf, whereas they showed a ventral expansion of pax7a expression and a loss of nkx6.2 expression in the midbrain (n = 13/16, n = 24/24, and n = 22/22, respectively) (Fig. 7A,B,D,E,G,H). This result was consistent with that for shh null mutants, whereas the phenotype of fgf22 morphants differed from that of the embryos treated with cyclopamine. Therefore, we examined whether shh expression was affected by inhibition of fgf22. shh expression was not affected in fgf22 morphants at 24 hpf (n = 33/33) (Fig. 7J,K). Furthermore, we examined whether fgf22 expression was responsive to Hh signaling. Surprisingly, fgf22 expression was reduced in the posterior midbrain of the embryos treated with cyclopamine at 24 hpf (n = 12/12) (Fig. 7L,M). However, fgf22 expression in the posterior midbrain was still detected in cyclopamine-treated embryos. Next, we investigated whether a dorsalization of the midbrain caused by blocking Hh signaling was affected by fgf22 knockdown at 24 hpf. In the embryos injected with fgf22 MO1 and treated with cyclopamine, wnt1 expression was expanded laterally compared with that in the embryos treated with cyclopamine (n = 18/19) (Fig. 7C). This result suggests that wnt1 is regulated by fgf22 but not by Hh signaling in the midbrain. On the other hand, pax7a expression was strongly reduced in the embryos injected with fgf22 MO1 and treated with cyclopamine compared with the embryos treated with cyclopamine (n = 44/49) (Fig. 7F). This result suggests that an expansion of pax7a expression in the midbrain caused by inhibition of Hh signaling is suppressed by inhibition of fgf22. A loss of nkx6.2 expression in the midbrain caused by blocking Hh signaling was unaffected by fgf22 knockdown (n = 14/14) (Fig. 7I). This result indicates that inhibition of fgf22 does not rescue a loss of nkx6.2 expression caused by inhibition of Hh signaling in the ventral midbrain.
Fig. 7. Interactions between fgf22 and Hh signaling in the midbrain.
(A–I) The expression of wnt1 (A–C), pax7a (D–F) and nkx6.2 (G–I) at 24 hpf in wild-type embryos treated with 0.95% ethanol (A,D,G) or cyclopamine (B,E,H) and fgf22 morphants treated with cyclopamine (C,F,I). (J–M) The expression of shh (J,K) and fgf22 (L,M) at 24 hpf in wild-type embryos (J), fgf22 morphants (K), and wild-type embryos treated with 0.95% ethanol (L) or cyclopamine (M).
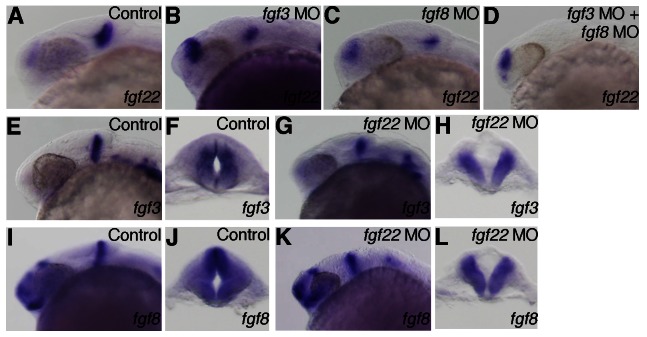
fgf22 expression in the midbrain is lost in fgf3/fgf8 double morphant embryos
Transplantation and ablation experiments in avian embryos have indicated that the isthmic organizer is involved in the positioning and development of the midbrain roof plate (Alexandre and Wassef, 2003). Fgf8 induces the isthmic node and participates in the formation of the MHB and midbrain roof plate in avian embryos (Bally-Cuif and Wassef, 1994; Crossley et al., 1996; Alexandre et al., 2006). In zebrafish, fgf3 and fgf8 are expressed in the MHB (Reifers et al., 1998; Kwak et al., 2006). Therefore, we examined whether the expression of roof plate marker genes was affected by inhibition of Fgf3 and Fgf8 signaling. The embryos co-injected with fgf3 MO and fgf8 MO showed a lateral expansion of wnt1 expression in the midbrain at 24 hpf (n = 22/23) (Fig. 9A). Furthermore, they showed a reduction of pax7a expression and ventral expansion of nkx6.2 expression in the midbrain (n = 36/45 and n = 15/20, respectively) (Fig. 9C,E). This phenotype is similar to that of fgf22 morphants. In zebrafish, fgf3 and fgf8 are expressed in the MHB primordium at earlier stages than fgf22 expression in the posterior midbrain primordium. Therefore, we examined whether fgf22 expression in the midbrain was affected by inhibition of Fgf3 and Fgf8 signaling. Although fgf22 expression was reduced in the posterior midbrain of the embryos injected with either fgf3 MO or fgf8 MO at 24 hpf, it was still detected (n = 12/12 and n = 12/13, respectively) (Fig. 8A–C). On the other hand, fgf22 expression was completely lost in the posterior midbrain of the embryos co-injected with fgf3 MO and fgf8 MO (n = 19/19) (Fig. 8D). In fgf22 morphants, the expression of fgf3 and fgf8 was detected in the MHB (n = 17/17 and n = 22/22, respectively) (Fig. 8E,G,I,K). The analysis of optical cross-sections showed that the expression of fgf3 and fgf8 was eliminated or reduced in the dorsal domain of the MHB in fgf22 morphants (n = 17/27 and n = 22/22, respectively) (Fig. 8F,H,J,L). This is possibly due to expansion of the roof plate, where fgf3 and fgf8 are not expressed. These results suggest that a combinatorial function of fgf3 and fgf8 is involved in the regulation of fgf22 expression in the posterior midbrain but fgf22 may not regulate fgf3 and fgf8 expression in the MHB.
Fig. 9. Rescue of midbrain region-specific marker loss in fgf3/fgf8 morphants by fgf22 RNA.
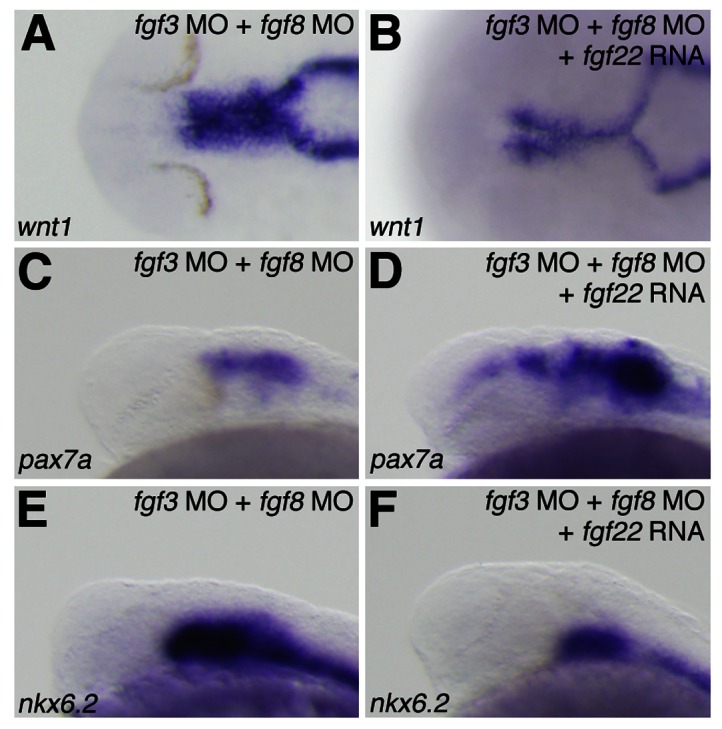
The expression of wnt1 (A,B), pax7a (C,D), and nkx6.2 (E,F) at 24 hpf in fgf3/fgf8 morphants (A,C,E) and fgf22 RNA-injected fgf3/fgf8 morphants (B,D,F).
Fig. 8. Interactions between fgf3, fgf8 and fgf22.
(A–D) The expression of fgf22 at 24 hpf in embryos injected with control MO (A), fgf3 MO (B), fgf8 MO (C), and fgf3 MO and fgf8 MO (D). (E–L) The expression of fgf3 (E–H) and fgf8 (I–L) at 24 hpf in embryos injected with control MO (E,F,I,J) and fgf22 MO (G,H,K,L). A–D,E,G,I,K are lateral views, anterior to the left; F,H,J,L are optical cross-sections of the MHB.
Next, to investigate whether fgf3- and fgf8-mediated loss of fgf22 function leads to defects in dorsal midbrain specification, we injected fgf3/fgf8 double morphants with fgf22 RNA. The injection depressed an expansion of wnt1 and nkx6.2 expression in the dorsal midbrain caused by co-injection of fgf3 MO and fgf8 MO (n = 24/26 and n = 30/33, respectively) (Fig. 9B,F). Furthermore, pax7a expression was up-regulated in the dorsal midbrain of fgf3/fgf8 double morphants injected with fgf22 RNA (n = 25/32) (Fig. 9D). These results indicate that fgf22 partially rescues the phenotype caused by inhibition of fgf3 and fgf8 function in the specification of the dorsal midbrain.
Blocking proliferation does not contribute to specification of the dorsal midbrain
As the decreased proliferation of tectal precursors might contribute to the reduction in the pax7a expression domain of the dorsal midbrain, we addressed whether decreases in cellular proliferation can secondarily cause patterning defects. To block proliferation, wild-type embryos were treated with hydroxyurea and aphidicholin (HUA), which have been used previously in zebrafish to reduce proliferation (Ikegami et al., 1999; Lyons et al., 2005). We applied HUA to embryos at 8 hpf, and analyzed midbrain development at 24 hpf. HUA treatment effectively inhibited proliferation, as evidenced by a reduction in the number of pH3-positive cells in the midbrain (n = 7/7) (supplementary material Fig. S4A,B). However, the domain of pax7a expression appeared relatively normal (n = 22/23) (supplementary material Fig. S4E,F). In addition, we observed no shift of wnt1 and nkx6.2 into the dorsal domain (n = 23/23 and n = 26/26, respectively) (supplementary material Fig. S4C,D,G,H). These results argue that the midbrain patterning defects that arise when Fgf signaling is disrupted are not due to decreases in localized proliferation.
Discussion
fgf22 controls cell proliferation in the midbrain
Fgf signaling regulates the proliferation and differentiation of specific neuronal cell types in the midbrain (Ye et al., 1998; Xu et al., 2000; Trokovic et al., 2005). Among the fgf family, fgf22 showed an unique expression pattern in the midbrain and MHB primordia. fgf22 morphants showed a decrease in tectal volume. fgf22 knockdown significantly inhibited cell proliferation in the midbrain. However, the knockdown did not strongly stimulate apoptosis in the midbrain. In addition, overexpression of fgf22 resulted increased cell proliferation in the midbrain. These results indicate that the reduction of tectum volume was not due to apoptosis rather due to the decreased cell proliferation in fgf22 morphants. fgfr2b knockdown also resulted in decreased cell proliferation in the midbrain and fgfr2b morphants showed very similar morphological defects to those obtained by fgf22 knockdown. Thus, it is suggested that Fgf22 signaling is mediated through Fgfr2b during cell proliferation in the midbrain.
fgf22 is involved in formation of the roof plate
Roof plate cells are induced to form by Bmp signals from the epidermal ectoderm and develop at the dorsal midline of the neural tube (Liem et al., 1995). Members of the Msx family have been implicated as downstream targets of Bmps and are induced to express in regions where Bmp signaling is active (Furuta et al., 1997; Graham et al., 1994; Liem et al., 1995; Shimeld et al., 1996; Timmer et al., 2002). Bmp signaling can be inhibited by Fgf signaling in the forebrain and midbrain (Storm et al., 2003; Alexandre et al., 2006). In zebrafish, bmp5 and msxb are expressed in the midbrain roof plate (Miyake et al., 2012) and the expression of bmp5 and msxb was increased in fgf22 morphants. This suggests that Fgf22 regulates Bmp signaling in the midbrain. Overexpression of Msx1 induces the ectopic expression of Lmx1 and Wnt1 (Liu et al., 2004). Lmx1b is sufficient to form a functional roof plate in the hindbrain and spinal cord (Chizhikov and Millen, 2004; Mishima et al., 2009). The increased expression of wnt1 and lmx1b.2 in the midbrain of fgf22 morphants may be due to an expansion of msxb expression. Therefore, Fgf22 may function to suppress the mediolateral extent of Bmp signaling from the center of the roof plate in the midbrain. On the other hand, loss of Fgf22 function led to a loss of MHB markers in the dorsal MHB region. This result suggests Fgf22 to be involved in the specification of the dorsal MHB region. However, the defect in the dorsal domain of the MHB might be due to the lateral expansion of the midbrain roof plate in fgf22 morphants. Furthermore, fgfr2b knockdown resulted in the expanded expression of roof plate markers and fgfr2b morphants showed very similar dorsal patterning defects to those observed after fgf22 knockdown. Thus, it is suggested that Fgf22 signaling suppresses the roof plate fate in the midbrain and it is mediated through Fgfr2b.
fgf22 is required for specification of the tectum
Otx2 is essential for the formation of all forebrain- and midbrain-derived structures (Acampora et al., 1995). Meis2 is both necessary and sufficient for tectal fate specification (Agoston and Schulte, 2009). Meis2 acts downstream of Otx2 and is a direct partner of Otx2 in the tectum (Agoston and Schulte, 2009). In fgf22 morphants, the expression of otx2 and meis2.2 was reduced in the midbrain. In addition, pax7a expression in the tectum was reduced in fgf22 morphants. fgf22 knockdown resulted in decreased proliferation and fgf22 morphants showed a decrease in tectal volume. However, decreased proliferation is not sufficient to cause patterning defects in the midbrain, as pax7a expression in the alar plate was not reduced in the midbrain in embryos treated with HUA in spite of decreased proliferation in this domain. Thus, reduced proliferation is not a major mechanism contributing to the reduction of tectal cell fate in fgf22 morphants. These results indicate that fgf22 is required for the specification of the tectum.
The roof plate is an important signaling center that controls dorsal CNS patterning and specification through secretion of the Bmp and Wnt signaling molecules. In fgf22 morphants, dorsal pax7a expression was reduced and the roof plate markers shifted into the domain where pax7a expression was absent. Because pax7a is not expressed in the roof plate, the decreased expression of pax7a in the tectum might cause the expanded expression of the roof plate markers in fgf22 morphants. Ventral nkx6.2 expression also shifted into the dorsal domain in the midbrain of fgf22 morphants, whereas fgf22 knockdown did not induce the expression of nkx6.2 in embryos with blocked Hh signaling. On the other hand, fgf22 knockdown strongly suppressed the up-regulation of pax7a expression caused by blocking Hh signaling. These results indicate that fgf22 is not involved in specification of the tegmentum and the increased expression of nkx6.2 in fgf22 morphants may be due to a reduction of pax7a expression. Thus, it is suggested that fgf22 is involved in specification of the tectum by controlling pax7a expression. Furthermore, fgfr2b knockdown resulted in the decreased expression of pax7a and the increased expression of nkx6.2. The loss of the dorsal midbrain in the morphants might secondarily induce expansion of the most dorsal tissues in the midbrain. Therefore, Fgf22 signaling is suggested to be mediated through Fgfr2b in the specification of the tectum.
fgf3 and fgf8 are required for fgf22 expression in the posterior midbrain
Hh signaling is involved in D/V patterning of the midbrain. Cross talk between Fgf and Hh signaling is critical for brain development (Brewster et al., 2000). fgf22 expression in the posterior midbrain was reduced in embryos with blocked Hh signaling, whereas shh expression was unaffected in fgf22 morphants. However, we speculate that fgf22 expression in the posterior midbrain is reduced by a secondary effect of dorsalization of the midbrain in embryos with blocked Hh signaling, because the phenotype of fgf22 morphants was opposite to that of embryos with blocked Hh signaling. Thus, the function of fgf22 differed from that of Hh signaling in the development of the midbrain roof plate and the specification of the tectum.
The isthmic organizer is implicated in the formation of the caudal roof plate in the midbrain and is crucial for the growth and patterning of the midbrain (Martínez, 2001; Rhinn and Brand, 2001; Wurst and Bally-Cuif, 2001; Alexandre and Wassef, 2003). When the isthmic tissue is ablated, the roof plate of the caudal midbrain fails to develop (Alexandre and Wassef, 2005). Fgf8 is a key component of the isthmic organizer and Fgf8 bead implantation experiments have demonstrated that isthmic organizer signals, in particular Fgf8, are involved in the positioning and differentiation of the midbrain roof plate (Alexandre and Wassef, 2005). In zebrafish, fgf3 and fgf8 are expressed in the MHB (Reifers et al., 1998; Kwak et al., 2006) and at earlier stages than fgf22. In the fgf3/fgf8 double morphant embryos, fgf22 expression was completely lost in the posterior midbrain. This result indicates that fgf22 expression in the posterior midbrain is regulated by Fgf3 and Fgf8 signaling from the MHB territory. The phenotype of the fgf3/fgf8 double morphant embryos was similar to that of fgf22 morphants. Furthermore, injection of fgf22 RNA into fgf3/fgf8 double morphant embryos rescued the reduction of pax7a expression and the expansion of wnt1 and nkx6.2 expression caused by co-injection of fgf3 MO and fgf8 MO. Thus, fgf22 is implicated in the development of the midbrain roof plate and the specification of the tectum as a downstream factor of Fgf3 and Fgf8 signaling.
The present study indicates that Fgf22 is involved in cell proliferation, roof plate formation, and tectum specification through Fgfr2b in the zebrafish midbrain. Furthermore, fgf22 is implicated in midbrain development as a factor downstream of fgf3 and fgf8 expressed in the MHB but not of Hh expressed in the floor plate. The present findings should provide new insights into roles of Fgf signaling in midbrain development in zebrafish. However, phenotypes of fgf22 knockdown in zebrafish differ from those of Fgf22-deficient mice. In Fgf22-deficient mice, the differentiation of excitatory nerve terminals on dendrites of CA3 pyramidal neurons in the hippocampus and the development of retinal terminals in the dorsal lateral geniculate nucleus are impaired (Terauchi et al., 2010; Singh et al., 2012). As no distinct expression of Fgf22 was observed in mouse embryos (Yaylaoglu et al., 2005), the different phenotypes may be due to different expression patterns in zebrafish and mice. In mice, other Fgfs expressed in the midbrain may be involved in cell proliferation, roof plate formation, and tectum specification in midbrain. This will be addressed in a future study.
Supplementary Material
Acknowledgments
We wish to thank Y. Nakagawa and T. Mido for technical assistance. This work was in part supported by Grant-in-Aid for Exploratory Research No. 23659035 (to N.I.) and for Young Scientists (B) No. 21790075 (to A.M.) from the Japan Society for the Promotion of Science.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Acampora D., Mazan S., Lallemand Y., Avantaggiato V., Maury M., Simeone A., Brûlet P. (1995). Forebrain and midbrain regions are deleted in Otx2-/- mutants due to a defective anterior neuroectoderm specification during gastrulation. Development 121, 3279–3290. [DOI] [PubMed] [Google Scholar]
- Agarwala S., Sanders T. A., Ragsdale C. W. (2001). Sonic hedgehog control of size and shape in midbrain pattern formation. Science 291, 2147–2150 10.1126/science.1058624 [DOI] [PubMed] [Google Scholar]
- Agoston Z., Schulte D. (2009). Meis2 competes with the Groucho co-repressor Tle4 for binding to Otx2 and specifies tectal fate without induction of a secondary midbrain-hindbrain boundary organizer. Development 136, 3311–3322 10.1242/dev.037770 [DOI] [PubMed] [Google Scholar]
- Alexandre P., Wassef M. (2003). The isthmic organizer links anteroposterior and dorsoventral patterning in the mid/hindbrain by generating roof plate structures. Development 130, 5331–5338 10.1242/dev.00756 [DOI] [PubMed] [Google Scholar]
- Alexandre P., Wassef M. (2005). Does the isthmic organizer influence D/V patterning of the midbrain? Brain Res. Brain Res. Rev. 49, 127–133 10.1016/j.brainresrev.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Alexandre P., Bachy I., Marcou M., Wassef M. (2006). Positive and negative regulations by FGF8 contribute to midbrain roof plate developmental plasticity. Development 133, 2905–2913 10.1242/dev.02460 [DOI] [PubMed] [Google Scholar]
- Altmann C. R., Brivanlou A. H. (2001). Neural patterning in the vertebrate embryo. Int. Rev. Cytol. 203, 447–482 10.1016/S0074-7696(01)03013-3 [DOI] [PubMed] [Google Scholar]
- Bally–Cuif L., Wassef M. (1994). Ectopic induction and reorganization of Wnt-1 expression in quail/chick chimeras. Development 120, 3379–3394. [DOI] [PubMed] [Google Scholar]
- Bayly R. D., Ngo M., Aglyamova G. V., Agarwala S. (2007). Regulation of ventral midbrain patterning by Hedgehog signaling. Development 134, 2115–2124 10.1242/dev.02850 [DOI] [PubMed] [Google Scholar]
- Brewster R., Mullor J. L., Ruiz i Altaba A. (2000). Gli2 functions in FGF signaling during antero-posterior patterning. Development 127, 4395–4405. [DOI] [PubMed] [Google Scholar]
- Briscoe J., Ericson J. (2001). Specification of neuronal fates in the ventral neural tube. Curr. Opin. Neurobiol. 11, 43–49 10.1016/S0959-4388(00)00172-0 [DOI] [PubMed] [Google Scholar]
- Chi C. L., Martinez S., Wurst W., Martin G. R. (2003). The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development 130, 2633–2644 10.1242/dev.00487 [DOI] [PubMed] [Google Scholar]
- Chiang C., Litingtung Y., Lee E., Young K. E., Corden J. L., Westphal H., Beachy P. A. (1996). Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383, 407–413 10.1038/383407a0 [DOI] [PubMed] [Google Scholar]
- Chizhikov V. V., Millen K. J. (2004). Control of roof plate development and signaling by Lmx1b in the caudal vertebrate CNS. J. Neurosci. 24, 5694–5703 10.1523/JNEUROSCI.0758-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley P. H., Martinez S., Martin G. R. (1996). Midbrain development induced by FGF8 in the chick embryo. Nature 380, 66–68 10.1038/380066a0 [DOI] [PubMed] [Google Scholar]
- Ekker M., Wegner J., Akimenko M. A., Westerfield M. (1992). Coordinate embryonic expression of three zebrafish engrailed genes. Development 116, 1001–1010. [DOI] [PubMed] [Google Scholar]
- Ekker M., Akimenko M. A., Allende M. L., Smith R., Drouin G., Langille R. M., Weinberg E. S., Westerfield M. (1997). Relationships among msx gene structure and function in zebrafish and other vertebrates. Mol. Biol. Evol. 14, 1008–1022 10.1093/oxfordjournals.molbev.a025707 [DOI] [PubMed] [Google Scholar]
- Elsen G. E., Choi L. Y., Millen K. J., Grinblat Y., Prince V. E. (2008). Zic1 and Zic4 regulate zebrafish roof plate specification and hindbrain ventricle morphogenesis. Dev. Biol. 314, 376–392 10.1016/j.ydbio.2007.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedtsova N., Turner E. E. (2001). Signals from the ventral midline and isthmus regulate the development of Brn3.0-expressing neurons in the midbrain. Mech. Dev. 105, 129–144 10.1016/S0925-4773(01)00399-9 [DOI] [PubMed] [Google Scholar]
- Fogel J. L., Chiang C., Huang X., Agarwala S. (2008). Ventral specification and perturbed boundary formation in the mouse midbrain in the absence of Hedgehog signaling. Dev. Dyn. 237, 1359–1372 10.1002/dvdy.21536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y., Piston D. W., Hogan B. L. (1997). Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development 124, 2203–2212. [DOI] [PubMed] [Google Scholar]
- Gerety S. S., Wilkinson D. G. (2011). Morpholino artifacts provide pitfalls and reveal a novel role for pro-apoptotic genes in hindbrain boundary development. Dev. Biol. 350, 279–289 10.1016/j.ydbio.2010.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A., Francis–West P., Brickell P., Lumsden A. (1994). The signalling molecule BMP4 mediates apoptosis in the rhombencephalic neural crest. Nature 372, 684–686 10.1038/372684a0 [DOI] [PubMed] [Google Scholar]
- Guner B., Karlstrom R. O. (2007). Cloning of zebrafish nkx6.2 and a comprehensive analysis of the conserved transcriptional response to Hedgehog/Gli signaling in the zebrafish neural tube. Gene Expr. Patterns 7, 596–605 10.1016/j.modgep.2007.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel M. J., Wei Y., Mancini M. A., Van Hooser A., Ranalli T., Brinkley B. R., Bazett–Jones D. P., Allis C. D. (1997). Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106, 348–360 10.1007/s004120050256 [DOI] [PubMed] [Google Scholar]
- Holzschuh J., Wada N., Wada C., Schaffer A., Javidan Y., Tallafuss A., Bally–Cuif L., Schilling T. F. (2005). Requirements for endoderm and BMP signaling in sensory neurogenesis in zebrafish. Development 132, 3731–3742 10.1242/dev.01936 [DOI] [PubMed] [Google Scholar]
- Ikegami R., Hunter P., Yager T. D. (1999). Developmental activation of the capability to undergo checkpoint-induced apoptosis in the early zebrafish embryo. Dev. Biol. 209, 409–433 10.1006/dbio.1999.9243 [DOI] [PubMed] [Google Scholar]
- Incardona J. P., Gaffield W., Kapur R. P., Roelink H. (1998). The teratogenic Veratrum alkaloid cyclopamine inhibits sonic hedgehog signal transduction. Development 125, 3553–3562. [DOI] [PubMed] [Google Scholar]
- Itoh N., Ornitz D. M. (2004). Evolution of the Fgf and Fgfr gene families. Trends Genet. 20, 563–569 10.1016/j.tig.2004.08.007 [DOI] [PubMed] [Google Scholar]
- Katahira T., Sato T., Sugiyama S., Okafuji T., Araki I., Funahashi J., Nakamura H. (2000). Interaction between Otx2 and Gbx2 defines the organizing center for the optic tectum. Mech. Dev. 91, 43–52 10.1016/S0925-4773(99)00262-2 [DOI] [PubMed] [Google Scholar]
- Kelly G. M., Moon R. T. (1995). Involvement of wnt1 and pax2 in the formation of the midbrain-hindbrain boundary in the zebrafish gastrula. Dev. Genet. 17, 129–140 10.1002/dvg.1020170205 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Krauss S., Johansen T., Korzh V., Fjose A. (1991). Expression of the zebrafish paired box gene pax[zf-b] during early neurogenesis. Development 113, 1193–1206. [DOI] [PubMed] [Google Scholar]
- Kwak S. J., Vemaraju S., Moorman S. J., Zeddies D., Popper A. N., Riley B. B. (2006). Zebrafish pax5 regulates development of the utricular macula and vestibular function. Dev. Dyn. 235, 3026–3038 10.1002/dvdy.20961 [DOI] [PubMed] [Google Scholar]
- Liem K. F., Jr, Tremml G., Roelink H., Jessell T. M. (1995). Dorsal differentiation of neural plate cells induced by BMP-mediated signals from epidermal ectoderm. Cell 82, 969–979 10.1016/0092-8674(95)90276-7 [DOI] [PubMed] [Google Scholar]
- Liu A., Joyner A. L. (2001). EN and GBX2 play essential roles downstream of FGF8 in patterning the mouse mid/hindbrain region. Development 128, 181–191. [DOI] [PubMed] [Google Scholar]
- Liu Y., Helms A. W., Johnson J. E. (2004). Distinct activities of Msx1 and Msx3 in dorsal neural tube development. Development 131, 1017–1028 10.1242/dev.00994 [DOI] [PubMed] [Google Scholar]
- Lun K., Brand M. (1998). A series of no isthmus (noi) alleles of the zebrafish pax2.1 gene reveals multiple signaling events in development of the midbrain-hindbrain boundary. Development 125, 3049–3062. [DOI] [PubMed] [Google Scholar]
- Lyons D. A., Pogoda H. M., Voas M. G., Woods I. G., Diamond B., Nix R., Arana N., Jacobs J., Talbot W. S. (2005). erbb3 and erbb2 are essential for schwann cell migration and myelination in zebrafish. Curr. Biol. 15, 513–524 10.1016/j.cub.2005.02.030 [DOI] [PubMed] [Google Scholar]
- Maroon H., Walshe J., Mahmood R., Kiefer P., Dickson C., Mason I. (2002). Fgf3 and Fgf8 are required together for formation of the otic placode and vesicle. Development 129, 2099–2108. [DOI] [PubMed] [Google Scholar]
- Martínez S. (2001). The isthmic organizer and brain regionalization. Int. J. Dev. Biol. 45, 367–371. [PubMed] [Google Scholar]
- Mishima Y., Lindgren A. G., Chizhikov V. V., Johnson R. L., Millen K. J. (2009). Overlapping function of Lmx1a and Lmx1b in anterior hindbrain roof plate formation and cerebellar growth. J. Neurosci. 29, 11377–11384 10.1523/JNEUROSCI.0969-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake A., Nakayama Y., Konishi M., Itoh N. (2005). Fgf19 regulated by Hh signaling is required for zebrafish forebrain development. Dev. Biol. 288, 259–275 10.1016/j.ydbio.2005.09.042 [DOI] [PubMed] [Google Scholar]
- Miyake A., Nihno S., Murakoshi Y., Satsuka A., Nakayama Y., Itoh N. (2012). Neucrin, a novel secreted antagonist of canonical Wnt signaling, plays roles in developing neural tissues in zebrafish. Mech. Dev. 128, 577–590 10.1016/j.mod.2012.01.001 [DOI] [PubMed] [Google Scholar]
- Mori H., Miyazaki Y., Morita T., Nitta H., Mishina M. (1994). Different spatio-temporal expressions of three otx homeoprotein transcripts during zebrafish embryogenesis. Brain Res. Mol. Brain Res. 27, 221–231 10.1016/0169-328X(94)90004-3 [DOI] [PubMed] [Google Scholar]
- Müller M., von Weizsäcker E., Campos–Ortega J. A. (1996). Transcription of a zebrafish gene of the hairy-Enhancer of split family delineates the midbrain anlage in the neural plate. Dev. Genes Evol. 206, 153–160 10.1007/s004270050041 [DOI] [PubMed] [Google Scholar]
- Nasevicius A., Ekker S. C. (2000). Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26, 216–220 10.1038/79951 [DOI] [PubMed] [Google Scholar]
- Ota S., Tonou–Fujimori N., Tonou–Fujimori N., Nakayama Y., Ito Y., Kawamura A., Yamasu K. (2010). FGF receptor gene expression and its regulation by FGF signaling during early zebrafish development. Genesis 48, 707–716 10.1002/dvg.20682 [DOI] [PubMed] [Google Scholar]
- Phillips B. T., Bolding K., Riley B. B. (2001). Zebrafish fgf3 and fgf8 encode redundant functions required for otic placode induction. Dev. Biol. 235, 351–365 10.1006/dbio.2001.0297 [DOI] [PubMed] [Google Scholar]
- Reifers F., Böhli H., Walsh E. C., Crossley P. H., Stainier D. Y., Brand M. (1998). Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development 125, 2381–2395. [DOI] [PubMed] [Google Scholar]
- Rhinn M., Brand M. (2001). The midbrain–hindbrain boundary organizer. Curr. Opin. Neurobiol. 11, 34–42 10.1016/S0959-4388(00)00171-9 [DOI] [PubMed] [Google Scholar]
- Seo H. C., Saetre B. O., Håvik B., Ellingsen S., Fjose A. (1998). The zebrafish Pax3 and Pax7 homologues are highly conserved, encode multiple isoforms and show dynamic segment-like expression in the developing brain. Mech. Dev. 70, 49–63 10.1016/S0925-4773(97)00175-5 [DOI] [PubMed] [Google Scholar]
- Shimeld S. M., McKay I. J., Sharpe P. T. (1996). The murine homeobox gene Msx-3 shows highly restricted expression in the developing neural tube. Mech. Dev. 55, 201–210 10.1016/0925-4773(96)00505-9 [DOI] [PubMed] [Google Scholar]
- Simeone A. (2002). Towards the comprehension of genetic mechanisms controlling brain morphogenesis. Trends Neurosci. 25, 119–121 10.1016/S0166-2236(00)02095-6 [DOI] [PubMed] [Google Scholar]
- Singh R., Su J., Brooks J., Terauchi A., Umemori H., Fox M. A. (2012). Fibroblast growth factor 22 contributes to the development of retinal nerve terminals in the dorsal lateral geniculate nucleus. Front. Mol. Neurosci 4, 61 10.3389/fnmol.2011.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm E. E., Rubenstein J. L., Martin G. R. (2003). Dosage of Fgf8 determines whether cell survival is positively or negatively regulated in the developing forebrain. Proc. Natl. Acad. Sci. USA 100, 1757–1762 10.1073/pnas.0337736100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J., Chen J. K., Cooper M. K., Wang B., Mann R. K., Milenkovic L., Scott M. P., Beachy P. A. (2000). Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature 406, 1005–1009 10.1038/35023008 [DOI] [PubMed] [Google Scholar]
- Terauchi A., Johnson–Venkatesh E. M., Toth A. B., Javed D., Sutton M. A., Umemori H. (2010). Distinct FGFs promote differentiation of excitatory and inhibitory synapses. Nature 465, 783–787 10.1038/nature09041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer J. R., Wang C., Niswander L. (2002). BMP signaling patterns the dorsal and intermediate neural tube via regulation of homeobox and helix-loop-helix transcription factors. Development 129, 2459–2472. [DOI] [PubMed] [Google Scholar]
- Trokovic R., Trokovic N., Hernesniemi S., Pirvola U., Vogt Weisenhorn D. M., Rossant J., McMahon A. P., Wurst W., Partanen J. (2003). FGFR1 is independently required in both developing mid- and hindbrain for sustained response to isthmic signals. EMBO J. 22, 1811–1823 10.1093/emboj/cdg169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trokovic R., Jukkola T., Saarimäki J., Peltopuro P., Naserke T., Weisenhorn D. M., Trokovic N., Wurst W., Partanen J. (2005). Fgfr1-dependent boundary cells between developing mid- and hindbrain. Dev. Biol. 278, 428–439 10.1016/j.ydbio.2004.11.024 [DOI] [PubMed] [Google Scholar]
- Turner D. L., Weintraub H. (1994). Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 8, 1434–1447 10.1101/gad.8.12.1434 [DOI] [PubMed] [Google Scholar]
- Waskiewicz A. J., Rikhof H. A., Hernandez R. E., Moens C. B. (2001). Zebrafish Meis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development 128, 4139–4151. [DOI] [PubMed] [Google Scholar]
- Westerfield M. (1995). The Zebrafish Book: A Guide For The Laboratory Use Of Zebrafish (Brachydanio Rerio). Eugene, OR: M. Westerfield. [Google Scholar]
- Wilson S. W., Brand M., Eisen J. S. (2002). Patterning the zebrafish central nervous system. Results Probl. Cell Differ. 40, 181–215. [DOI] [PubMed] [Google Scholar]
- Wurst W., Bally–Cuif L. (2001). Neural plate patterning: upstream and downstream of the isthmic organizer. Nat. Rev. Neurosci. 2, 99–108 10.1038/35053516 [DOI] [PubMed] [Google Scholar]
- Xu J., Liu Z., Ornitz D. M. (2000). Temporal and spatial gradients of Fgf8 and Fgf17 regulate proliferation and differentiation of midline cerebellar structures. Development 127, 1833–1843. [DOI] [PubMed] [Google Scholar]
- Yaylaoglu M. B., Titmus A., Visel A., Alvarez–Bolado G., Thaller C., Eichele G. (2005). Comprehensive expression atlas of fibroblast growth factors and their receptors generated by a novel robotic in situ hybridization platform. Dev. Dyn. 234, 371–386 10.1002/dvdy.20441 [DOI] [PubMed] [Google Scholar]
- Ye W., Shimamura K., Rubenstein J. L., Hynes M. A., Rosenthal A. (1998). FGF and Shh signals control dopaminergic and serotonergic cell fate in the anterior neural plate. Cell 93, 755–766 10.1016/S0092-8674(00)81437-3 [DOI] [PubMed] [Google Scholar]
- Zhang X., Ibrahimi O. A., Olsen S. K., Umemori H., Mohammadi M., Ornitz D. M. (2006). Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem. 281, 15694–15700 10.1074/jbc.M601252200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.