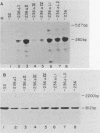
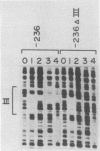
Abstract
The adenovirus early region 3 (E3) promoter is an early viral promoter which is strongly induced by the adenovirus transactivator protein E1A. DNase I footprinting with HeLa cell extracts has identified four factor-binding domains which appear to be involved in basal and E1A-induced transcriptional regulation. These binding domains may bind TATA region-binding factors (site I), the CREB/ATF protein (site II), the AP-1 protein (site III), and nuclear factor I/CTF (site IV). Recently, it has been shown that the DNA-binding domain of transcription factor AP-1 has homology with the yeast transcription factor GCN4 and that the yeast transactivator protein GAL4 is able to stimulate transcription in HeLa cells from promoters containing GAL4-binding sites. These results suggest an evolutionary conservation of both transcription factors and the mechanisms responsible for transcriptional activation in Saccharomyces cerevisiae and higher eucaryotic organisms. To determine whether similar patterns of transcriptional regulation were seen with the E3 promoter in HeLa and yeast cells, the E3 promoter fused to the chloramphenicol acetyltransferase (cat) gene was cloned into a high-copy-number plasmid and stably introduced into yeast cells. S1 analysis revealed that similar E3 promoter mRNA start sites were found in yeast and HeLa cells. DNase I footprinting with partially purified yeast extracts revealed that four regions of the E3 promoter were protected. Several of these regions were similar to binding sites determined by using HeLa cell extracts. Oligonucleotide mutagenesis of these binding domains indicated their importance in the transcriptional regulation of the E3 promoter in yeast cells. These results suggest that similar cellular transcription factor-binding sites may be involved in the regulation of promoters in both yeast and mammalian cells.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angel P., Allegretto E. A., Okino S. T., Hattori K., Boyle W. J., Hunter T., Karin M. Oncogene jun encodes a sequence-specific trans-activator similar to AP-1. Nature. 1988 Mar 10;332(6160):166–171. doi: 10.1038/332166a0. [DOI] [PubMed] [Google Scholar]
- Angel P., Imagawa M., Chiu R., Stein B., Imbra R. J., Rahmsdorf H. J., Jonat C., Herrlich P., Karin M. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987 Jun 19;49(6):729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- Arndt K., Fink G. R. GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5' TGACTC 3' sequences. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8516–8520. doi: 10.1073/pnas.83.22.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J. Adenovirus promoters and E1A transactivation. Annu Rev Genet. 1986;20:45–79. doi: 10.1146/annurev.ge.20.120186.000401. [DOI] [PubMed] [Google Scholar]
- Bohmann D., Bos T. J., Admon A., Nishimura T., Vogt P. K., Tjian R. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science. 1987 Dec 4;238(4832):1386–1392. doi: 10.1126/science.2825349. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Specific protein binding to far upstream activating sequences in polymerase II promoters. Proc Natl Acad Sci U S A. 1985 Jan;82(1):43–47. doi: 10.1073/pnas.82.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Lue N. F., Kornberg R. D. A GAL family of upstream activating sequences in yeast: roles in both induction and repression of transcription. EMBO J. 1986 Mar;5(3):603–608. doi: 10.1002/j.1460-2075.1986.tb04253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985 Dec;43(3 Pt 2):729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- Brent R. Repression of transcription in yeast. Cell. 1985 Aug;42(1):3–4. doi: 10.1016/s0092-8674(85)80091-x. [DOI] [PubMed] [Google Scholar]
- Briggs M. R., Kadonaga J. T., Bell S. P., Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986 Oct 3;234(4772):47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- Broach J. R. Construction of high copy yeast vectors using 2-microns circle sequences. Methods Enzymol. 1983;101:307–325. doi: 10.1016/0076-6879(83)01024-1. [DOI] [PubMed] [Google Scholar]
- Chen W., Struhl K. Yeast mRNA initiation sites are determined primarily by specific sequences, not by the distance from the TATA element. EMBO J. 1985 Dec 1;4(12):3273–3280. doi: 10.1002/j.1460-2075.1985.tb04077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh L. A., Carthew R. W., Sharp P. A. A single polypeptide possesses the binding and transcription activities of the adenovirus major late transcription factor. Mol Cell Biol. 1986 Dec;6(12):4723–4733. doi: 10.1128/mcb.6.12.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm M., Berg P. Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. J Mol Appl Genet. 1982;1(5):457–481. [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J., Wu F., Gaynor R. Upstream regulatory regions required to stabilize binding to the TATA sequence in an adenovirus early promoter. Nucleic Acids Res. 1987 Oct 26;15(20):8367–8385. doi: 10.1093/nar/15.20.8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the "TATA box". Proc Natl Acad Sci U S A. 1984 Dec;81(24):7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984 Feb;36(2):503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters: positive and negative elements. Cell. 1984 Apr;36(4):799–800. doi: 10.1016/0092-8674(84)90028-x. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Hoar E. T., Guarente L. Each of three "TATA elements" specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Chang M., Kim U. J., Grunstein M. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell. 1987 Feb 27;48(4):589–597. doi: 10.1016/0092-8674(87)90237-6. [DOI] [PubMed] [Google Scholar]
- Hill D. E., Hope I. A., Macke J. P., Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986 Oct 24;234(4775):451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Hurst H. C., Jones N. C. Identification of factors that interact with the E1A-inducible adenovirus E3 promoter. Genes Dev. 1987 Dec;1(10):1132–1146. doi: 10.1101/gad.1.10.1132. [DOI] [PubMed] [Google Scholar]
- Imagawa M., Chiu R., Karin M. Transcription factor AP-2 mediates induction by two different signal-transduction pathways: protein kinase C and cAMP. Cell. 1987 Oct 23;51(2):251–260. doi: 10.1016/0092-8674(87)90152-8. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Herskowitz I. A repressor (MAT alpha 2 Product) and its operator control expression of a set of cell type specific genes in yeast. Cell. 1985 Aug;42(1):237–247. doi: 10.1016/s0092-8674(85)80119-7. [DOI] [PubMed] [Google Scholar]
- Jones K. A., Kadonaga J. T., Rosenfeld P. J., Kelly T. J., Tjian R. A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell. 1987 Jan 16;48(1):79–89. doi: 10.1016/0092-8674(87)90358-8. [DOI] [PubMed] [Google Scholar]
- Kakidani H., Ptashne M. GAL4 activates gene expression in mammalian cells. Cell. 1988 Jan 29;52(2):161–167. doi: 10.1016/0092-8674(88)90504-1. [DOI] [PubMed] [Google Scholar]
- Keegan L., Gill G., Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986 Feb 14;231(4739):699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- Kovesdi I., Reichel R., Nevins J. R. Identification of a cellular transcription factor involved in E1A trans-activation. Cell. 1986 Apr 25;45(2):219–228. doi: 10.1016/0092-8674(86)90386-7. [DOI] [PubMed] [Google Scholar]
- Lalonde B., Arcangioli B., Guarente L. A single Saccharomyces cerevisiae upstream activation site (UAS1) has two distinct regions essential for its activity. Mol Cell Biol. 1986 Dec;6(12):4690–4696. doi: 10.1128/mcb.6.12.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lech K., Anderson K., Brent R. DNA-bound Fos proteins activate transcription in yeast. Cell. 1988 Jan 29;52(2):179–184. doi: 10.1016/0092-8674(88)90506-5. [DOI] [PubMed] [Google Scholar]
- Lee K. A., Hai T. Y., SivaRaman L., Thimmappaya B., Hurst H. C., Jones N. C., Green M. R. A cellular protein, activating transcription factor, activates transcription of multiple E1A-inducible adenovirus early promoters. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8355–8359. doi: 10.1073/pnas.84.23.8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. G., Nurse P. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature. 1987 May 7;327(6117):31–35. doi: 10.1038/327031a0. [DOI] [PubMed] [Google Scholar]
- Lee W., Mitchell P., Tjian R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 1987 Jun 19;49(6):741–752. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- Leff T., Corden J., Elkaim R., Sassone-Corsi P. Transcriptional analysis of the adenovirus-5 EIII promoter: absence of sequence specificity for stimulation by EIa gene products. Nucleic Acids Res. 1985 Feb 25;13(4):1209–1221. doi: 10.1093/nar/13.4.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Hereford L. Yeast chromatin is uniformly digested by DNase-I. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4285–4288. doi: 10.1073/pnas.76.9.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Maki Y., Bos T. J., Davis C., Starbuck M., Vogt P. K. Avian sarcoma virus 17 carries the jun oncogene. Proc Natl Acad Sci U S A. 1987 May;84(9):2848–2852. doi: 10.1073/pnas.84.9.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil J. B., Smith M. Transcription initiation of the Saccharomyces cerevisiae iso-1-cytochrome c gene. Multiple, independent T-A-T-A sequences. J Mol Biol. 1986 Feb 5;187(3):363–378. doi: 10.1016/0022-2836(86)90439-0. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Wang C., Tjian R. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell. 1987 Sep 11;50(6):847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- Montminy M. R., Bilezikjian L. M. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987 Jul 9;328(6126):175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- Nagawa F., Fink G. R. The relationship between the "TATA" sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R., Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks C. L., Banerjee S., Spector D. J. Organization of the transcriptional control region of the E1b gene of adenovirus type 5. J Virol. 1988 Jan;62(1):54–67. doi: 10.1128/jvi.62.1.54-67.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer K., Prezant T., Guarente L. Yeast HAP1 activator binds to two upstream activation sites of different sequence. Cell. 1987 Apr 10;49(1):19–27. doi: 10.1016/0092-8674(87)90751-3. [DOI] [PubMed] [Google Scholar]
- Rosenfeld P. J., Kelly T. J. Purification of nuclear factor I by DNA recognition site affinity chromatography. J Biol Chem. 1986 Jan 25;261(3):1398–1408. [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985 Nov;43(1):165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Tatchell K. Transcription and regulatory signals at the mating type locus in yeast. Cell. 1984 Jul;37(3):969–978. doi: 10.1016/0092-8674(84)90431-8. [DOI] [PubMed] [Google Scholar]
- Sorger P. K., Lewis M. J., Pelham H. R. Heat shock factor is regulated differently in yeast and HeLa cells. Nature. 1987 Sep 3;329(6134):81–84. doi: 10.1038/329081a0. [DOI] [PubMed] [Google Scholar]
- Sorger P. K., Pelham H. R. Purification and characterization of a heat-shock element binding protein from yeast. EMBO J. 1987 Oct;6(10):3035–3041. doi: 10.1002/j.1460-2075.1987.tb02609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg P. W., Stern M. J., Clark I., Herskowitz I. Activation of the yeast HO gene by release from multiple negative controls. Cell. 1987 Feb 27;48(4):567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- Struhl K. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol Cell Biol. 1986 Nov;6(11):3847–3853. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Struhl K. The DNA-binding domains of the jun oncoprotein and the yeast GCN4 transcriptional activator protein are functionally homologous. Cell. 1987 Sep 11;50(6):841–846. doi: 10.1016/0092-8674(87)90511-3. [DOI] [PubMed] [Google Scholar]
- Struhl K. The yeast his3 promoter contains at least two distinct elements. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7385–7389. doi: 10.1073/pnas.79.23.7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt P. K., Bos T. J., Doolittle R. F. Homology between the DNA-binding domain of the GCN4 regulatory protein of yeast and the carboxyl-terminal region of a protein coded for by the oncogene jun. Proc Natl Acad Sci U S A. 1987 May;84(10):3316–3319. doi: 10.1073/pnas.84.10.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N., Jin J. R., Green S., Hollis M., Chambon P. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL4 trans-activator. Cell. 1988 Jan 29;52(2):169–178. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]
- Weeks D. L., Jones N. C. Adenovirus E3-early promoter: sequences required for activation by E1A. Nucleic Acids Res. 1985 Jul 25;13(14):5389–5402. doi: 10.1093/nar/13.14.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederrecht G., Shuey D. J., Kibbe W. A., Parker C. S. The Saccharomyces and Drosophila heat shock transcription factors are identical in size and DNA binding properties. Cell. 1987 Feb 13;48(3):507–515. doi: 10.1016/0092-8674(87)90201-7. [DOI] [PubMed] [Google Scholar]
- Wildeman A. G., Zenke M., Schatz C., Wintzerith M., Grundström T., Matthes H., Takahashi K., Chambon P. Specific protein binding to the simian virus 40 enhancer in vitro. Mol Cell Biol. 1986 Jun;6(6):2098–2105. doi: 10.1128/mcb.6.6.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Rosser D. S., Schmidt M. C., Berk A. A TATA box implicated in E1A transcriptional activation of a simple adenovirus 2 promoter. Nature. 1987 Apr 2;326(6112):512–515. doi: 10.1038/326512a0. [DOI] [PubMed] [Google Scholar]
- Zenke M., Grundström T., Matthes H., Wintzerith M., Schatz C., Wildeman A., Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986 Feb;5(2):387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]