Abstract
In the final steps of yeast ribosome synthesis, immature translation-incompetent pre-40S particles that contain 20S pre-rRNA are converted to the mature translation-competent subunits containing the 18S rRNA. An assay for 20S pre-rRNA cleavage in purified pre-40S particles showed that cleavage by the PIN domain endonuclease Nob1 was strongly stimulated by the GTPase activity of the cytoplasmic translation initiation factor eIF5b/Fun12. Cleavage of the 20S pre-rRNA was also inhibited in vivo and in vitro by blocking binding of Fun12 to the 25S rRNA through specific methylation of its binding site. Cleavage competent pre-40S particles stably associate with Fun12 and form 80S complexes with 60S ribosomal subunits. We propose that recruitment of 60S subunits promotes GTP-hydrolysis by Fun12, leading to structural rearrangements within the pre-40S particle that bring Nob1 and the pre-rRNA cleavage site together.
Keywords: pre-rRNA processing, nuclease, PIN domain, yeast, ribosome synthesis, GTPase
INTRODUCTION
The maturation of pre-ribosomal particles to produce the functional small and large ribosomal subunits is a complex, multi-step process, in which the processing of the pre-ribosomal RNAs (pre-rRNAs) must be coordinated with other assembly steps. During this maturation process, the 20S pre-rRNA is cleaved to 18S rRNA only within late, cytoplasmic pre-40S particles, raising the question of how the timing of cleavage is related to 40S subunit maturation.
Nucleotide tri-phosphatases form an important class of ribosome synthesis factors. Many ATPases and GTPases use the energy of NTP hydrolysis to drive or regulate structural changes within the pre-ribosomal particle. The RNA-stimulated ATPases, or RNA helicases, are a prominent group of ribosome synthesis factors. In yeast, 18 putative helicases play essential roles throughout the synthesis pathway but, of these, only Prp43 is known to participate in the late, cytoplasmic steps of 40S subunit synthesis. GTPases are also required at multiple steps in the synthesis pathway. While none had yet been clearly shown to participate directly in late steps of 40S synthesis in yeast, the GTPases Era and RsgA/YjeQ are required for 30S subunit synthesis in E. coli 1. Yeast Tsr1 has homology to GTPases, is required for 40S synthesis and occupies a very similar binding site to Era 2. In addition, a high-throughput analysis indicated that loss of the translation initiation factor and GTPase eIF5b, designated Fun12 (Function unknown 12) in yeast, results in 20S pre-rRNA accumulation 3, suggesting a potential role in proof-reading the maturation of 40S subunits towards translation competence. Notably, the bacterial translation initiation factor IF2, which is homologous to eIF5b/Fun12 is a high-copy suppressor of RsgA/YjeQ mutations 4, suggesting that it might have a conserved function in small-subunit synthesis.
The nuclear steps in ribosome synthesis are subject to very active quality control systems. In contrast, surveillance of the late cytoplasmic steps in ribosome synthesis appears to be less active, and 3′ unprocessed forms of the 18S and 5.8S rRNAs accumulate to high levels in strains with processing defects. Previous analyses mapped the locations of ribosome synthesis factors within the pre-40S and pre-60S particles, revealing that several proteins occupy sites that are required for key functional interactions during translation initiation and elongation 2,5-9. The clear conclusion was that immature pre-ribosomes are actively prevented from interacting non-productively with the translation machinery. In apparent contrast, other data indicated that late pre-40S particles, which were accumulated in strains lacking processing factors, could associate with 60S subunits. This suggested that the last steps in pre-40S maturation might involve functional interactions with the translation machinery 10.
The final RNA processing step during 40S subunit maturation is the cleavage of the 3′ end of the mature 18S rRNA by Nob1, a PIN domain endonuclease 11-15. Nob1 binds to early, nuclear pre-ribosomes that are exported to the cytoplasm prior to pre-rRNA cleavage, indicating that the timing of Nob1 cleavage is regulated and linked to 40S subunit maturation. Genetic analyses implicated Prp43 in this process, acting together with its cofactor Pfa1 11,16.
To better understand the late steps in 40S subunit maturation, we have reproduced 18S rRNA 3′ cleavage within pre-40S particles and investigated the factors involved in the presumed structural reorganization that is required for the acquisition of cleavage competence.
RESULTS
Prior cleavage at site A2 does not stimulate cleavage at site D by Nob1 in vitro
During 40S subunit maturation, Nob1 associates with early, 90S pre-ribosomes in the nucleus and is exported with the pre-40S particles to the cytoplasm, where it is active in pre-rRNA cleavage. This strongly indicates that the nuclease activity of Nob1 is regulated and linked to other steps in pre-40S maturation.
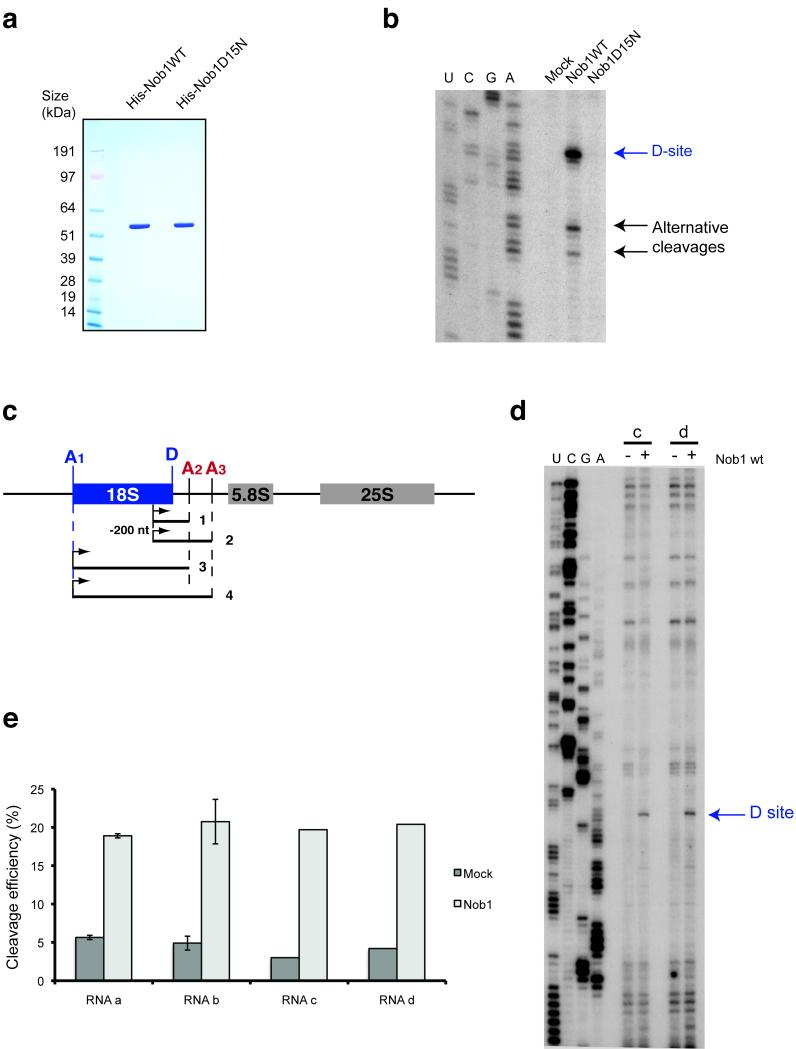
Our previous analyses of the in vitro activity of Nob1 utilized protein purified from yeast extracts 11. However, the lethal phenotypes of Nob1 mutants complicated functional analyses in vitro. We therefore purified recombinant wild-type and mutant forms of Nob1 expressed in E. coli. (Fig. 1a). Wild-type Nob1 showed approximately 20% cleavage efficiency (Fig. 1b) on an in vitro transcribed RNA substrate that contained site D (substrate a in Fig. 1c). Lower levels of cleavage were detected at alternative sites with sequences related to site D, as previously observed for Nob1 purified from yeast 11. These may arise only in vitro because the positioning of Nob1 in vivo is constrained by the pre-40S structure. The mutation in the PIN domain (D15N) abolished cleavage, confirming that this activity is solely due to Nob1 activity (Fig. 1b).
Figure 1.
In vitro RNA cleavage by Nob1 is not affected by sequences 3′ to site A2. (a) Recombinant forms of wild-type and mutant Nob1 were purified over nickel resin followed by gel filtration. 2 μg of each protein was loaded on an SDS-gel and Coomassie stained. (b) Cleavage assay on in vitro transcribed RNA. In vitro transcribed RNA was incubated with wild-type or mutant Nob1 and analyzed by primer extension. The major cleavage at site D is indicated, as are alternative cleavage sites, which were previously observed in vitro at sequences similar to site D 11. (c) Structures of long RNA substrates, mimicking the pre-rRNA before A2 cleavage (substrates 2 and 4) and after A2 cleavage (substrates 1 and 3). (d,e) The cleavage efficiency was determined by primer extension on the two longer RNA substrates (3 and 4) (d) and quantified using northern hybridization data (e). The ratio between cleaved fragment (D-A2) and intact RNA is presented as a histogram. These experiments were repeated for the shorter RNAs (substrates 1 and 2) but were performed only once for substrates 3 and 4 (with 5′ ends at site A1) due to the difficulties in purifying these long RNA species.
A model was proposed for the regulation of the timing of cleavage at site D, in which prior cleavage at A2 and release of the 3′ region of ITS1 triggers an RNA conformational switch that activates D site cleavage 17. To test this model we compared four sets of RNA substrates using in vitro endonuclease cleavage assays (Fig. 1c). Transcripts 1 and 2 have 5′ ends within the 18S rRNA 200nt 5′ to site D, while substrates 3 and 4 extend to site A1 at the 5′ end of 18S. Substrates 1 and 3 have 3′ ends at site A2, whereas 2 and 4 extend to site A3, mimicking a pre-rRNA prior to A2 cleavage that is not predicted to act as a cleavage substrate.
For all four substrates, the cleavage site was analyzed by primer extension and the efficiency was quantified by Northern hybridization (Figs. 1d, 1e and data not shown). On all substrates tested, similar cleavage efficiencies were observed (~20%), with no clear differences in the presence or absence of the A2-A3 region.
We also sought to test the pre-rRNA refolding model by extensive phylogenetic comparison of yeast ITS1 sequences and their secondary structures. This confirmed previously published and empirically tested models 18-20 and showed that the secondary structure of the A2-A3 region was conserved. The primary sequence of the ITS1 region situated between A2 and A3 was variable, whereas the pre-rRNA refolding model postulates that this region interacts with a very highly conserved sequence in the 3′ end of 18S. In contrast, the actual A2 and A3 processing sites were highly conserved at sequence level, as was a stem loop structure located just upstream of A3. Furthermore, no significant similarity could be found between Saccharomyces ITS1 sequences or predicted secondary structures and those of more distant fungi (Schizosaccharomyces or Pezizomycotina) (Supplementary Fig. 1 and Supplementary Note).
While these data do not demonstrate that the pre-rRNA refolding model is incorrect, they indicate that this is not sufficient to explain Nob1 activation in the cytoplasm and is unlikely to be conserved in organisms that are not closely related to S. cerevisiae.
In vitro cleavage by Nob1 is stimulated by ATP and GTP addition
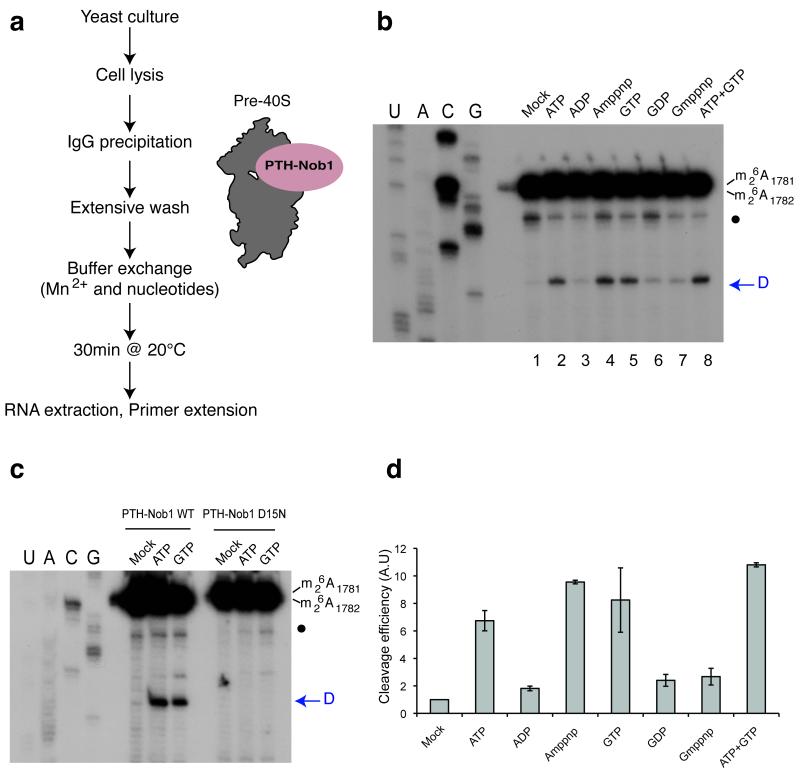
Since the in vitro cleavage data and phylogenetic comparisons failed to provide clear support for the role of RNA structure reorganization in the regulation of 20S pre-rRNA processing, we attempted to assess the roles of protein factors. To facilitate these analyses, we developed an in vitro cleavage assay using purified pre-40S particles. To ensure that late, cleavage-competent pre-40S particles were present in the precipitate, tagged Nob1 was used as the bait for purification (Fig. 2). The N-terminally tagged Protein A-TEV-His6 (PTH)-Nob1 fusion used was previously shown to support Nob1 function in vivo 2.
Figure 2.
In vitro cleavage in pre-40S particles is stimulated by ATP or GTP addition. (a) Diagram representing steps in the assay for in vitro 40S subunit maturation. (b) Primer extension analysis showing the activation of in vitro site D cleavage in pre-40S particles by nucleotide addition. The strong upper stops result from termination at the sites of 18S rRNA base-methylation at A1779 and A1780. These modifications precede site D cleavage in vivo. The blue arrow indicates site D. A closed circle indicates an additional primer extension stop seen in some experiments, which corresponds to a run of G-C base-pairs at the foot of H45 in the 18S rRNA. The nucleotides indicated were added at 1mM. (c) Comparison of cleavage stimulation observed with pre-40S particles purified using wild-type (PTH-Nob1 WT) or the catalytically inactive PTH-Nob1D15N protein. (d) Variation of cleavage efficiencies in relation to nucleotide addition. Signals obtained for cleavage at site D in panel (b) were quantified, corrected for RNA loading (using a common stop in the ITS1 as a standard) and normalized to the mock-treated sample (set to 1). The average of three independent experiments is shown, with the standard deviation indicated on top of the histogram.
Like other PIN-domain nucleases, the in vitro cleavage activity of Nob1 is strongly stimulated by Mn2+ 11. Pre-ribosomes were therefore affinity-purified using PTH-Nob1 on an IgG column and extensively washed in buffer containing 5mM Mg2+ but lacking Mn2+(Fig. 2a). In vitro cleavage was then activated by buffer exchange to 5mM Mn2+, 2mM Mg2+, with or without NTPs. Following incubation of the particles for 30 min at 20°C, cleavage was assessed by primer extension (Fig. 2b and 2c; arrows indicate the location of site D) and quantified (Fig. 2d).
Addition of Mn2+ without nucleotides induced only a very low level of site D cleavage in the pre-40S particles (Fig. 2b, lane 1). Inclusion of either ATP or GTP at 1mM final concentration enhanced cleavage (~7 fold and 8 fold, respectively) (Fig. 2b, lanes 2 and 5; Fig. 2c). In contrast, neither ADP nor GDP conferred clear cleavage stimulation (Fig. 2b, lanes 3 and 6). The effects of addition of both ATP and GTP were not synergistic (Fig. 2b, lane 8), but were modestly additive when analyzed over multiple experiments (Fig. 2d). Since Nob1 is bound to both early and late pre-ribosomes, it would be anticipated that only a fraction of the co-purified pre-ribosomes are cleavage competent. Moreover, it is very likely processing will have occurred that in a fraction of the cleavage competent particles pre-40S during the extended incubations needed for purification.
To assess the requirement for ATP and GTP hydrolysis, the non-hydrolysable analogues AMP-PNP and GMP-PNP were also tested. Cleavage was stimulated ~9 fold by AMP-PNP (Fig. 2b, lane 4), but was not stimulated by GMP-PNP (Fig. 2b, lane 7). Many NTPases are able to bind and hydrolyze either ATP or GTP, but this result clearly demonstrates that these are distinct activities in 40S processing.
To fully confirm that the observed cleavage was due to the activity of Nob1, pre-40S particles were also purified using the catalytically inactive PTH-Nob1D15N mutant protein (Fig. 2c). No cleavage activity was associated with pre-40S particles containing Nob1D15N, in the absence or presence of ATP or GTP.
We conclude that two different factors activate Nob1 cleavage in pre-40S particles, which possess ATP-binding and GTPase activity, respectively.
Fun12 (eIF5b) is a GTPase involved in 20S processing
We initially sought to confirm the identity of the GTPase(s) involved. The inventory of late pre-40S components suggested an obvious candidate factor in Tsr1 21, which acts to block premature association of pre-40S particles with 60S subunits 8. Tsr1 has apparent sequence homology to GTPases and occupies a binding site on the 18S rRNA that overlaps that of the E.coli GTPase Era 1,2,8,21. However, despite this prima facie evidence, we were unable to detect GTP-binding or GTPase activity associated with recombinant Tsr1 (Supplementary Fig. 2, and data not shown). A point mutation (K44A) was made in the Tsr1 GTGK motif, which is similar to the typical GSGK p-loop motif that is involved in nucleotide binding. In other GTPases, Lys44 would be strongly predicted to lie in the active site and mutation of this residue should eliminate the putative GTPase activity and disrupt Tsr1 function. Tellingly, when expressed in a P GAL-TSR1 strain on glucose medium, the Tsr1K44A protein supported normal growth and 40S synthesis (Supplementary Fig. 2e). We conclude that Tsr1 is unlikely to provide the GTPase activity that stimulates in vitro 20S pre-rRNA cleavage.
An alternative candidate was the translation initiation factor eIF5b (Fun12), a GTPase that was proposed to play a role in 20S processing in vivo 3. During translation initiation, Fun12 (eIF5b) promotes binding of tRNAfMet and subunit joining 22,23.
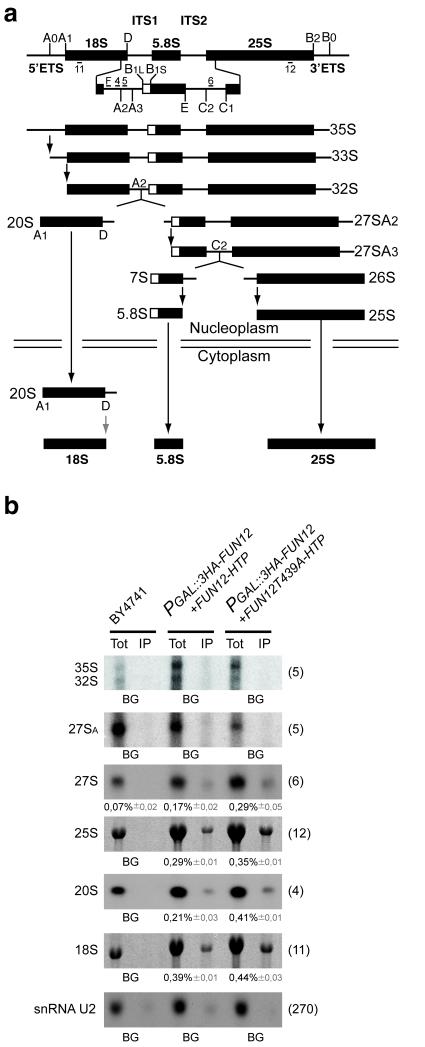
To assess whether Fun12 is directly involved in 20S processing, its association with pre-40S particles was tested by immunoprecipitation using Fun12 fused to a C-terminal His6-TEV-Protein A (HTP) tag. In addition, the immunoprecipitation efficiency of wild-type Fun12 was compared to a mutant (T439A), which retains GTP-binding but lacks hydrolysis activity and is therefore predicted to show increased retention on its substrate. RNA species co-precipitated with Fun12 following purification on IgG sepharose were analyzed by northern hybridization (Fig. 3b). As expected, both wild-type and mutant Fun12 bound mature 18S and 25S rRNA, consistent with its function in translation, but not the snRNA U2 involved in pre-mRNA splicing. In addition, Fun12 interacted with 20S pre-rRNA and, to a lesser extent, with 27SA/B. The mutant Fun12T439A showed approximately 2 fold greater co-precipitation with 20S pre-rRNA, with lesser increases for the mature rRNAs and 27S pre-rRNA.
Figure 3.
Fun12 is associated with pre-rRNA.
(a) Pre-rRNA processing pathway and probe locations. Probe F was Cy3 labeled and used for the FISH analyses shown in Figures 4 and 6. (b) Co-precipitation of pre-rRNA and rRNA with Fun12-HTP and GTPase defective Fun12T439A-HTP. 1.5% of the input was loaded as total RNA (Tot). RNA was separated on a 1.2% agarose gel containing glyoxal and transferred to Hybond N+ prior to hybridization with the probes indicated in the left of the panels. The ratio between the two signals was quantified and the co-precipitation efficiency determined from three independent experiments for each pre-rRNA species is presented with the standard deviation shown in gray. Hybridization probes used are indicated on the left of the panels.
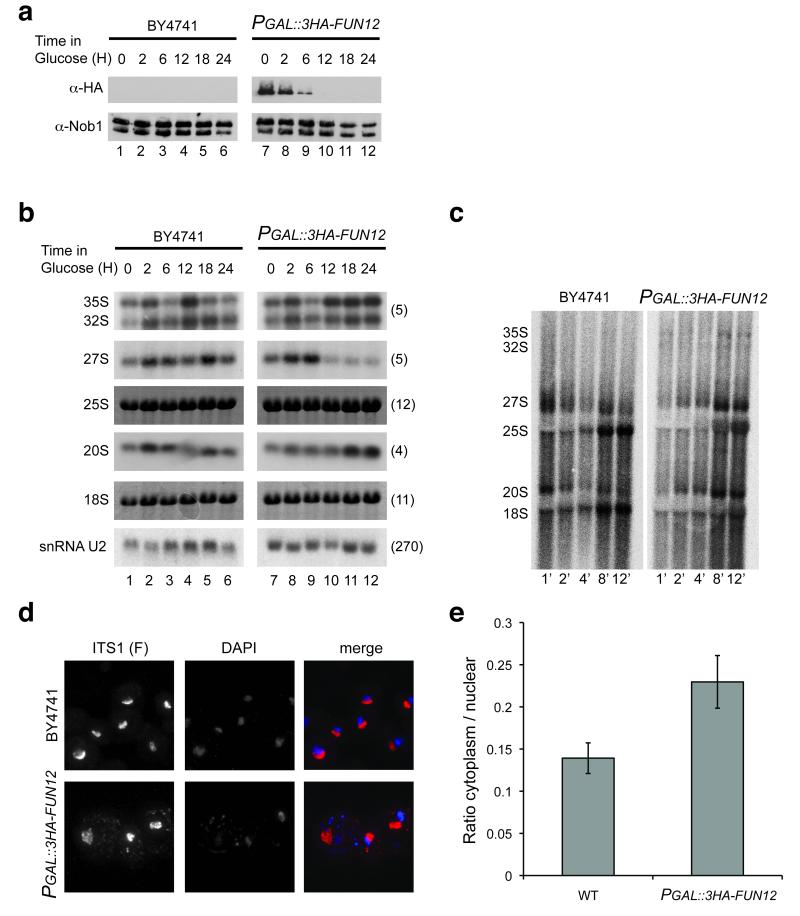
Yeast Fun12 is non-essential for viability, at least in some strain backgrounds, but its loss confers a substantial growth defect. To further test the involvement of Fun12 in pre-rRNA processing the FUN12 ORF was fused to an N-terminal triple HA-tag and placed under the control of a PGAL promoter (PGAL::3HA-FUN12). Western blotting (Fig. 4a) showed that Fun12 abundance was strongly reduced after 6 h in glucose medium, and was undetectable after 12 h depletion. Depletion of Fun12 did not strongly affect the abundance of Nob1 (Fig. 4a). Under these growth conditions, Nob1 migrates as two bands that we speculate reflect post-translational modification. However, the ability of recombinant Nob1 to cleave 20S pre-rRNA in vitro shows that this putative modification is not required for activity.
Figure 4.
Fun12 is required for efficient pre-20S processing.
(a) Western-blot analysis of Fun12 depletion using an anti-HA antibody. Effects of Fun12 depletion on general protein levels were determined using an anti-Nob1 antibody. Proteins were extracted from cells growing on galactose medium (0 h) and following transfer to glucose medium for the times indicated. (b) Northern hybridization analysis of pre-rRNA processing during Fun12 depletion. (c) Pulse-chase labeling of pre-rRNA processing in wild-type and P GAL::3HA-FUN12 strains. Following transfer to glucose medium for 20 h, cells were labeled with [3H] uracil for 2 min and chased with unlabeled uracil for the times indicated. Labeled RNA was visualized using a Fuji imager. (d) FISH analysis of wild-type and PGAL::3HA-FUN12 strains, following transfer to glucose medium for 20 h. Cells were hybridized with Cy3-labeled oligonucleotide F complementary to the 5′ region of ITS1, which is present in the 20S pre-rRNA and all earlier precursors. The nucleoplasm is labeled with DAPI. (e) Quantitation of FISH data. Signals were integrated over the nuclear and cytoplasmic areas of maximum projections. 15 individual cells were analyzed for each strain.
The effects of Fun12 depletion on pre-rRNA processing were determined by northern hybridization (Fig. 4b). After 12 h depletion, accumulation of the 35S primary transcript was detected, together with a reduction in the level of 27S pre-rRNA, indicating that cleavage at site A2 was impaired. This may reflect the inhibition of ribosomal protein synthesis, since depletion of several different ribosomal proteins leads to inhibition of these cleavages 24. Despite the reduction in 20S pre-rRNA synthesis implied by the reduction in A2 cleavage, clear 20S accumulation was observed after 18 h depletion. These results strongly indicate that depletion of Fun12 impairs 20S processing. Clear depletion of mature 18S rRNA relative to 25S was not observed, probably because, unlike the 20S pre-rRNA, the mature 18S rRNA is stable and depleted only by growth. Growth is inhibited by depletion of Fun12, but this is primarily due to the inhibition of translation. We predict that the residual 18S synthesis is sufficient to support the reduced requirement for ribosome synthesis.
To further assess the effects of Fun12 depletion, pulse-chase labeling was performed (Fig. 4c). The appearance of mature 18S rRNA was strongly retarded in the Fun12-depleted strain, showing that maturation of the entire 20S pre-rRNA population is delayed. Maturation of 27S pre-rRNA to 25S also showed some delay, although this was less marked than for 20S processing. It is currently unclear whether this reflects an additional involvement of Fun12 in 27S processing, or is a consequence of reduced protein synthesis in the Fun12-depleted strain.
The 20S pre-rRNA is processed to 18S rRNA after export of the pre-40S particles to the cytoplasm 25. The inhibition of processing is therefore expected to lead to the accumulation of cytoplasmic 20S pre-rRNA. To determine whether this is the case we monitored the level of dimethylation of nucleotides A1781 and A1782. These modifications are introduced into the 20S pre-rRNA by the Dim1 methyltransferase following export to the cytoplasm 26,27, and therefore allow a determination of whether the accumulated 20S pre-rRNA has indeed been exported. Dimethylation prevents base-pairing of the adenosines, and this in turn prevents oligonucleotide-directed, RNase H cleavage at this site 28. As shown in Supplementary Fig. 3, in vitro transcribed, unmodified RNAs were cleaved with >90% efficiency, whereas <10% of 20S pre-rRNA recovered from the fun12Δ strain was cleaved, showing it to be predominately cytoplasmic. This finding was confirmed by fluorescent hybridization (Fig. 4d) using a Cy3-labeled probe (oligo F in Fig. 3a). This is complementary to the 5′ region of ITS1, which is present in 20S pre-rRNA as well as all earlier precursors, including the nascent transcripts. As previously observed, the FISH signal is predominately nucleolar, but the cytoplasmic signal was clearly enhanced in Fun12-depleted strain. Quantification of the FISH data confirmed the increased ratio of the cytoplasmic to nuclear signal in the strain depleted of Fun12 (Fig. 4e).
These results indicate that Fun12 is directly associated with pre-40S particles and suggested that GTP hydrolysis by Fun12 might be important for cytoplasmic 20S pre-rRNA cleavage.
Fun12 activates Nob1 function in pre-40S particles in vitro
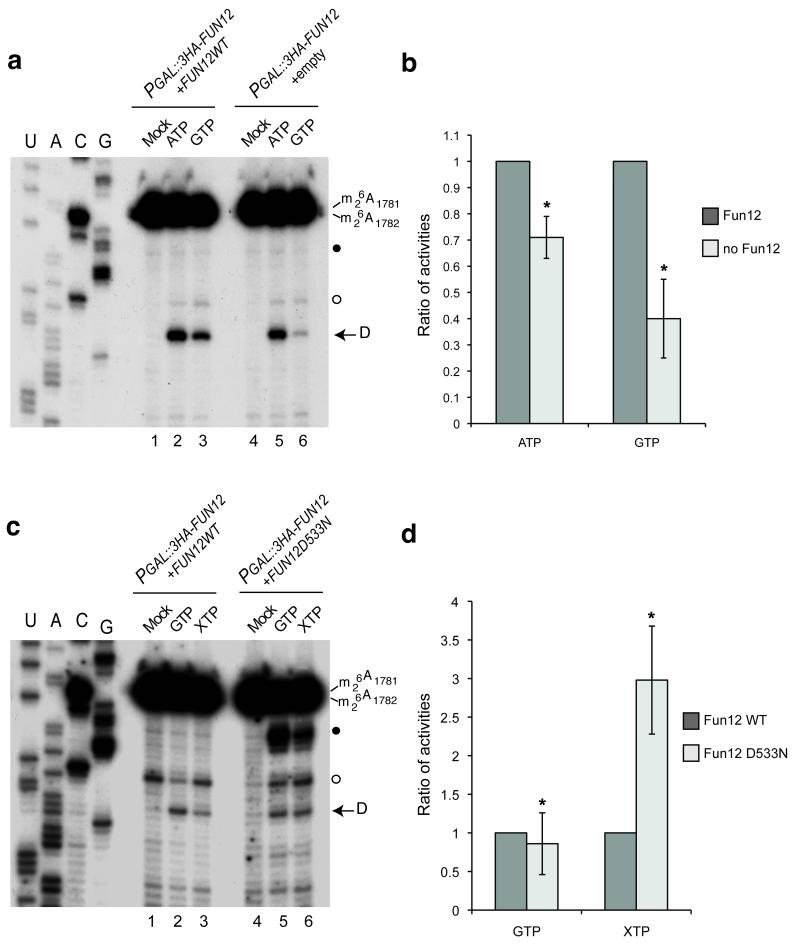
To test whether the activity of Fun12 is responsible for the GTP-dependent stimulation of in vitro pre-rRNA cleavage, this analysis was performed using pre-40S particles purified from P GAL::FUN12 strains genetically depleted of endogenous Fun12 for 12 h and carrying either an empty plasmid or a plasmid expressing wild-type Fun12 under the control of an ADH1 promoter. Depletion of Fun12 reduced GTP-stimulated cleavage ~2.5 fold (Figs. 5a and 5b). Some GTP stimulation was still observed but depletion of proteins under GAL-control is never complete and a reduced level of Fun12 will remain in the depleted extract. A modest but reproducible reduction in ATP-stimulated cleavage was also observed, possibly reflecting a reduced level of cleavage-competent pre-40S particles in the cell extract.
Figure 5.
Fun12 is responsible for GTP-stimulation of in vitro cleavage.
(a) Strain YSLD69 (PGAL::3HA-NOB1; PGAL::3HA-FUN12; pRS415-PTH-Nob1) was transformed with empty (empty) or PADH1::FUN12 plasmids. Following transfer to glucose medium for 12 h pre-40S particles were purified using PTH-Nob1 and cleavage at site D determined as in Fig. 2. (b) Cleavage efficiencies observed after addition of ATP or GTP were quantified relative to the control. The ratios of the activities observed from cell lysates expressing or depleted of Fun12 are presented (* P<0.2). (c) Site D cleavage in PTH-Nob1 pre-40S fractions isolated from strain YSLD69 carrying either a plasmid allowing low level expression of wild-type, non-tagged Fun12 under the control of a MET25 promoter or a plasmid expressing the Fun12D533N mutant that allows hydrolysis of XTP. Cells were transferred to medium containing 2% glucose and 8 mg l−1 methionine (2.5 fold lower than the normal concentration) for 10 h before pre-40S purification and analysis as in Fig. 2. (d) Activities induced by either GTP or XTP were quantified in comparison to the mock control. The ratio of the activities observed in cell lysates expressing Fun12 WT or mutant is presented (* P<0.2).
Specific mutations in the active sites of some GTPases allow the use of xanthine triphosphate (XTP) in addition to or instead of GTP. XTP does not occur naturally and is not normally a substrate for GTPases, so this allows specific determination of the roles played by an individual GTPase in vitro, even when other GTPases are present. Such mutations were previously described for human eIF5b and yeast Fun12, in which the active site mutation (D533N) allows XTP hydrolysis 22. Expression of Fun12D533N from a plasmid under the control of an ADH1 promoter had a strong dominant negative effect on cell growth even when P GAL::FUN12 was induced, presumably due to competition. We therefore used a much weaker MET25 promoter for this analysis. In cells expressing only wild-type Fun12, no significant stimulation of cleavage was seen following XTP addition (Figs. 5c and 5d). In contrast, XTP substantially stimulated cleavage in lysates from strains expressing Fun12D533N, demonstrating that the GTPase activity of Fun12 stimulates site D cleavage in vitro.
Stimulation by either GTP or XTP in Fig. 5c is lower than GTP stimulation in Fig. 5a, presumably reflecting the low level of Fun12 expression under control of the weak MET25promoter. In addition, the stability of the association of Fun12D533N with pre-40S particles was reduced in comparison with WT (data not shown).
The interaction of Fun12 with 60S subunits is required for efficient 20S processing
Fun12 is involved in 40S-60S subunit joining during translation initiation, suggesting the possibility that its role in stimulating 20S pre-rRNA processing within pre-40S particles might also involve interactions with 60S particles. Testing this hypothesis by depletion of 60S ribosomes is not feasible, so we sought to disrupt the interaction between Fun12 and the 25S rRNA. The 25S rRNA contact sites for Fun12 on the 25S rRNA have been identified (Fig. 6a) 29,30, but generating cells that contain only a mutant form of the 25S rRNA is also problematic, particularly if this impairs subunit function. However, previous analyses showed that 2′-O-methyl groups can be introduced at specific nucleotides using artificial box C/D snoRNAs, and at some positions these inhibit ribosome function 31. We therefore tested the effect of introducing an additional ectopic methylation at a Fun12 contact site (Gm2307) using a construct derived from snR75, which normally directs methylation only at G2288 (Gm2288).
Figure 6.
Fun12 binding to 25S rRNA is required for efficient 20S processing.
(a) 25S region involved in subunit joining. Fun12 binding sites are indicated by circled nucleotides. The snR75 interaction that directs methylation at Gm2288 is indicated in red. The accessory guide interaction is indicated in blue, along with the new target for snR75Mut2 at Gm2307. (b) Constructs expressing wild-type or mutant snR75. Red box; methylation guide. Blue box; accessory guide. In snR75Mut1, the accessory guide is non-functional. In snR75Mut2, the accessory guide is extended and confers ectopic methylation at Gm2307. The snoRNAs are expressed from the intron of the ACT1 gene flanked by PGAL and TADH, in a strain deleted for the SNR78-72 cluster. (c,d) Effects of expression of snR75Mut2 on pre-rRNA processing in vivo. (c) FISH analysis as in Figure 4d. (d) Northern analysis of pre-rRNAs using the probes indicated on the left. Mature rRNAs were visualized by EthBr staining. (e) Cleavage assay as in Figure 2 in lysates from cells expressing snR75 or snR75Mut2. Lanes 1 and 4, cleavage without added GTP. Lanes 2 and 5, with 1mM GTP. Lanes 3 and 6, with 1mM GTP plus 100pmol of purified ribosomal subunits (ribs). (f) Quantification of cleavage efficiency.
Plasmid-based constructs were designed for expression, under a PGAL1 promoter, of wild-type snR75, snR75Mut1, in which the accessory guide has been disrupted reducing methylation efficiency 28, and snR75Mut2 that directs methylation on G2288 and, via mutation of the accessory guide, an additional methylation at G2307 (Fig. 6b). These were expressed in a strain lacking the SNR78-72 gene cluster that includes snR75 28. Growth tests (Supplementary Fig. 4a) showed that expression of snR75Mut2 conferred a growth defect. Expression of snR75Mut1 decreased the level of Gm2288 methylation (Supplementary Fig. 4b), as shown previously for a snR75 mutant lacking extra base-pairing with 25S, confirming that methylation can be controlled by mutation of the accessory guide 28. Primer extension confirmed that the Gm2307 modification is detected only in cells expressing snR75Mut2 (Supplementary Fig. 5b).
The capacity of HA-Fun12 to precipitate pre-ribosomes and mature subunits was tested in strains expressing either wild-type snR75 or snR75Mut2. Western blotting revealed no difference in recovery of HA-Fun12 (Supplementary Fig. 4c). Northern hybridization showed greatly reduced co-precipitation of mature 25S and 18S rRNA, confirming disruption of Fun12 interactions during translation initiation (Supplementary Fig. 4d). In addition, the association of Fun12 with 20S pre-rRNA was reduced in the strain expressing snR75Mut2 (Supplementary Fig. 4d), indicating that this interaction also requires association of 60S with pre-40S. Some reduction in Fun12 association with 27S pre-rRNA was also observed on snR75Mut2 expression, consistent with specific interactions between Fun12 and pre-60S particles.
The effects of snR75Mut2 expression and consequent 25S methylation on 20S processing were tested by FISH analysis and northern hybridization. In Figure 6c, cells were hybridized with a Cy3-labeled oligonucleotide (probe F) complementary to the 5′ region of ITS1, which is present in the 20S pre-rRNA and all earlier precursors. The nucleoplasm is labeled with DAPI. Clear cytoplasmic accumulation of 20S pre-rRNA was observed when methylation is present on Gm2307. The wild-type cytoplasmic 20S signal in the wild-type is lower in Figure 6c than in Figure 4d, due to reduced ribosome synthesis on galactose compared to glucose medium. Northern hybridization (Fig. 6d) confirmed the accumulation of 20S pre-rRNA, whereas 35S and 27S pre-rRNA levels appeared unaffected. We conclude that the association of Fun12 with the 60S ribosomal subunit is required for efficient processing of cytoplasmic 20S in vivo.
To assess the involvement of 60S subunits in 20S pre-rRNA cleavage in vitro, pre-40S particles were prepared from lysates of strains expressing wild-type snR75 and snR75Mut2 (Figs. 6e and 6f). The wild-type strain showed approximately 2 fold more cleavage in the presence of GTP than the snR75Mut2 strain (Fig. 6e), consistent with a requirement for Fun12-25S association for efficient cleavage. To further test this, the purified pre-40S particles were supplemented with 60S subunits from wild-type lysates (Fig. 6e, lanes 3 and 6). Cleavage stimulation by 60S addition was substantially greater in the snR75Mut2 strain than in the wild-type (Fig. 6f). We interpret this as showing that the accumulated pre-40S particles in the snR75Mut2 strain identified by northern hybridization (Fig. 6d) can be activated for cleavage by association with exogenously added, unmodified 60S subunits, but that interaction with the endogenous subunits is blocked by 25S methylation.
Initial analyses were performed in strains deleted for the SNR78-72 cluster and complemented by expression of only snR75 (Fig. 6 and Supplementary Fig. 4). To confirm that the loss of other snoRNAs in the cluster did not contribute to the observed phenotypes, we constructed plasmids that included the entire cluster except SNR75, with snR75 expressed independently (Supplementary Fig. 5a). In these strains, northern analyses confirmed the specific accumulation of 20S pre-rRNA (Supplementary Fig. 5b) and RNase H resistance confirmed the specific methylation at Gm2307 (Supplementary Fig. 5c).
We conclude that both the GTPase activity of Fun12 and its functional association with the 60S subunit are required for efficient 3′ processing of the 18S rRNA.
Pre-40S particles associate with 60S subunits
To determine whether Fun12 shows stable or transient pre-ribosome association, its presence in pre-40S particles purified with HTP-Nob1 was assessed by western blotting (Fig. 7a). Around 2% of Fun12 was co-precipitated with pre-40S; a strikingly high figure considering that most of the Fun12 population is presumably engaged in translation initiation. This finding suggested that mature 60S subunits might also be stably associated with pre-40S particles. Ethidium bromide (EthBr) staining of RNA associated with the pre-40S particles showed a high level of mature 25S rRNA (Fig. 7b). This does not represent non-specific recovery as no corresponding signal was seen in the mock precipitation using a non-tagged strain. As noted above, it is likely that cleavage competent pre-40S particles undergo some level of cleavage during the extended incubation steps needed for purification, and this may contribute to the 18S signal observed in the Nob1 precipitate. If pre-40S cleavage normally occurs in the pre-40S/60S complex, we would expect this form to be increased in the pre-40S particles containing Nob1D15N, since these are specifically blocked at the cleavage step. As shown in Fig. 7b and quantified in Fig. 7c, this is indeed the case.
Figure 7.
Pre-40S particles stably associate with Fun12 and mature 60S prior to final 40S maturation.
(a) Pre-40S particles were purified from cells expressing Fun12-HA, using Nob1-HTP as bait. Control cells expressed Fun12-HA but not Nob1-HTP. Recovery of Nob1-HTP (α-Nob1) and association of Fun12-HA (α-HA) with pre-40S particles were tested by western blot. 2% of total extract (Tot) and 100% of the IPs (IP) were loaded. (b) 25S rRNA is specifically associated with PTH-Nob1 in vivo. RNAs associated with PTH-Nob1 or with catalytically inactive PTH-Nob1D15N were analyzed on an agarose gel (IP). 1.5% of the input was loaded as a total (Tot). Non-tagged cells were used as control. (c) 25S:20S ratios co-precipitated with PTH-Nob1 or PTH-Nob1D15N, quantified by EthBr staining and normalized for their relative lengths (* P<0.2). (d) Pre-40S particles purified using Nob1-HTP or PTH-Nob1D15N were analyzed by size exclusion. Comparison to a calibration curve (data not shown) revealed that the two major peaks correspond to 80S (“80S”) and 40S particles (pre-40S). The distribution of Nob1 was assessed by western blot (middle panel) and quantified using a Licor odyssey system (lower panel). (e) EM imaging with negative staining showing representative views of particles from the two peaks.
During translation initiation, Fun12 (eIF5b) promotes 40S/60S subunit joining. To assess whether Fun12 also facilitates pre-40S/60S interactions, snR75Mut2 was expressed in a strain expressing PTH-tagged Nob1 (Supplementary Fig. 5e). The introduction of the Gm2307 modification by snR75Mut2 significantly reduced recovery of 25S rRNA with Nob1 associated pre-40S particles, strongly indicating that Fun12 promotes pre-40S/60S joining.
To assess whether the pre-40S particles are assembled into discrete complexes of higher molecular weight, pre-40S particles associated with Nob1 and Nob1D15N were analyzed by gel filtration. In both cases the RNA distribution (Fig. 7d, upper panel) showed two major peaks. These were shown by comparison to a calibration curve to be approximately 80S and 40S in size. Western blotting revealed two matching peaks of Nob1 distribution (Fig. 7d middle panel; quantified in Fig. 7d, lower panel). Comparison of the pre-40S peaks associated with Nob1 and Nob1D15N (blue and red lines, respectively, in Fig. 7d upper panel), shows a marked increase in the 80S fraction for the D15N mutant, which is in good agreement with the RNA precipitation data shown in Figs. 7b and 7c.
These data strongly suggested that the pre-40S and mature 60S subunits form complexes resembling 80S ribosomes. This was verified by imaging fractions from the gel filtration complex by electron microscopy (Fig. 7e). The distinctive mature 60S subunit structure is readily visible in the pre-40S associated complexes present in the 80S fractions.
DISCUSSION
Four yeast pre-rRNA endonucleases, Rnt1, RNase MRP, Rcl1 and Nob1, have been shown to cleave their cognate target sites in vitro in naked RNA substrates 11,32-34. In vivo, however, the timing of cleavage by MRP and Nob1, at least, is regulated and correlated with other events in ribosome synthesis 11,17,35. Detailed understanding of the mechanisms involved has been elusive, since these processes have been refractory to in vitro analyses in minimal systems. Here we report the development of an assay for in vitro maturation of purified pre-40S particles by the Nob1 endonuclease that allows the dissection of factors and interactions responsible for the acquisition of cleavage competence. This assay did not provide support for the importance of a proposed RNA conformational switch in regulating cleavage 17, but revealed independent ATP- and GTP-stimulated activities that each promote site D cleavage by Nob1.
Multiple ATPases are associated with pre-40S particles, including the protein kinases Rio1, Rio2 and Hrr25, and the RNA helicase Prp43, and analyses of these factors are ongoing. Stimulation by GTP requires hydrolysis since substantially less activity was seen in the presence of GMP-PNP, and no stimulation was seen using GDP. We initially considered that Tsr1 might be the active GTPase due to sequence homology to known GTPases and the similarity between its binding site and that of the E.coli GTPase Era 2. However, we were unable to detect GTP-binding or GTPase activity associated with Tsr1, and mutation of the hypothetical active site did not confer any clear phenotype. An alternative candidate was the yeast eIF5b homologue Fun12; a translation initiation factor and bona fide GTPase that was previously implicated in 20S maturation 3. Genetic depletion of Fun12 was confirmed to inhibit site D cleavage in vivo and in vitro, leading to accumulation of cytoplasmic 20S pre-rRNA. Pulse-chase labeling showed a substantial delay in 18S maturation, showing that Fun12 is normally involved in the processing of all pre-20S, rather than affecting a subpopulation of pre-40S particles. Moreover, 20S pre-rRNA was co-precipitation of with tagged Fun12, indicating a direct interaction with pre-40S particles rather than indirect effects due to reduced protein synthesis. However, the clearest demonstration that the GTPase activity of Fun12 stimulates D site cleavage came from the finding that in vivo expression of a Fun12 mutant capable of hydrolyzing XTP conferred XTP-stimulation of in vitro cleavage on the pre-40S particle purified from the resulting cell lysate. Another translation initiation factor Hcr1/eIF3j was also reported to play a role in 20S processing in yeast 36. Hcr1 physically interacts with Fun12 37, suggesting that their functions in 20S processing are related. However, depletion of several other translation initiation factors did not affect 20S maturation 10,38,39, making it unlikely that the actual initiation of translation is required for processing.
Fun12 also interacted with 27S pre-rRNA and Fun12 depletion impaired 27S to 25S processing, albeit to a lesser extent than 20S processing. However, it remains unclear whether Fun12 also plays a direct role in pre-60S maturation.
During translation initiation, Fun12 binds both 40S and 60S subunits, suggesting the possibility that this might also be the case during pre-40S maturation. To test this hypothesis, we attempted to disrupt the interaction between Fun12 and 60S subunits, by introducing an ectopic site of 2′-O-methylation at a nucleotide (G2307) previously shown to be a contact site for Fun12 in the 25S rRNA 29,30. The presence of this single additional methyl group in 25S rRNA impaired 20S pre-rRNA processing in vivo and in vitro and disrupted Fun12 association with 40S and 60S subunits. This suggested that the functionally important interactions between Fun12 and pre-40S particles might take place in the context of a trimeric complex with mature 60S ribosomes.
Pre-40S particles containing 20S pre-rRNA that cannot be 3′ processed due to depletion of synthesis factors were previously reported to interact with 60S subunits, and indeed with polysomes 10. Analysis of RNA co-precipitated with HTP-tagged Nob1 by Ethidium bromide staining showed that the recovery of 25S rRNA was around 30-40% of the level of 20S pre-rRNA. The 25S rRNA was not detectable using a non-tagged strain, indicating that its presence does not represent contamination by mature subunits. A substantially higher level of 25S rRNA (around 80% of the abundance of 20S pre-rRNA) was recovered using a tagged form of the catalytically inactive Nob1D15N protein, despite the very substantial 20S accumulation in this strain 11-15.
To assess the nature of the particles that contain the 20S and 25S rRNAs, Nob1 complexes were affinity-purified and then subjected to gel filtration. In the column fractions, Nob1 and associated RNAs were found in two peaks, corresponding to 40S and 80S sized complexes. Visualization by electron microscopy revealed that the 80S complexes included structures resembling mature 60S subunits. Consistent with the RNA precipitation data, a substantially higher fraction of the complexes associated with Nob1D15N were in the 80S form. Nob1 is associated with 20S pre-rRNA in the nucleus and in early cytoplasmic pre-40S particles, as well as in late, cleavage competent particles. The Nob1D15N mutation lies in the PIN domain active site and does not affect Nob1 binding to the pre-rRNA 2, but leads to substantial accumulation of very late pre-40S particles. These data would therefore be consistent with 25S rRNA association and 80S complex formation only on late, cleavage competent pre-40S particles.
In contrast to the effects of the Nob1D15N mutation, expression of snR75Mut2 significantly reduced the recovery of 25S with pre-40S particles. In the presence of snR75Mut2 the interaction between Fun12 and the 25S rRNA is impaired by introduction of a single methyl group at G2307. Together with the functional data for the role of the 25S binding by Fun12 in cleavage, this indicates that final pre-40S maturation takes place in association with 60S particles and that this interaction is promoted or stabilized by Fun12.
The role of Fun12 in late 40S maturation
Fun12 (eIF5b) promotes tRNAfMet binding and subunit joining 22,23, and is an attractive candidate to combine functional proof-reading with late assembly steps in 40S synthesis (Fig. 8). 20S pre-rRNA cleavage is inhibited by depletion of Fun12 3, and also by mutation or depletion of r-proteins located along the mRNA-binding cleft 40. We speculate that the ITS1 5′ region, which forms an extended stem-loop structure, may associate with the mRNA binding cleft in the pre-40S particles 40. Placement of ITS1 in the cleft would pull site D away from Nob1, which is bound to H40 in the 18S rRNA 2,41. The 3′ terminal three nts of 18S rRNA are missing from the ribosome crystal structures, and are therefore not included in the model shown (Fig.8a) but are indicated with three green beads.
Figure 8.
Model for the role of Fun12 (eIF5b) in pre-40S processing.
(a,b) Side view of mature 50S particles 49. Panel (b) is a zoom of the region indicated by a square in panel (a). The head (cream) and body (green) are highlighted. Six nts at the 3′ end of 18S are represented in green as a surface (nt −6 to −4) or as beads (nt −3 to 3′). The Nob1 binding site is indicated in blue. Positions of ribosomal proteins Rps5 and Rps14, which are involved in site D cleavage, are highlighted in black. The path of the mRNA across the 40S subunit is indicated 49. (c) Model of steps potentially driving cleavage at site D. (c1) ITS1 is proposed to be located in the mRNA-binding cleft. (c2) Fun12 (purple) binds pre-40S particles together with the 60S subunit. (c3) GTP hydrolysis by Fun12 drives movement of the head domain and displaces ITS1 within the mRNA binding cleft, bringing site D towards Nob1. 40S head to body rotation is proposed to participate in bringing the Nob1 active site together with site D. (c4) Cleavage of ITS1 and release of Nob1 generates mature, translation-competent 40S subunits.
Detailed structural data on the role of eIF5b/Fun12 during eukaryotic translation initiation are currently lacking. However, the bacterial homologue IF2 has been shown to promote subunit joining and to promote small subunit rotation, leading to a translation competent conformation 42. Notably, while subunit joining is promoted by GMPPNP, subunit rotation requires GTP hydrolysis. The C-terminal domain of IF2 interacts with the 16S rRNA and rpS12, and is responsible for positioning tRNAfMet 43. Notably, prior to GTP hydrolysis by IF2, the interaction between the Shine-Dalgarno sequence at the 3′ end of the 16S rRNA and the anti-Shine-Dalgarno sequence in the mRNA holds the mRNA in a “standby” position, from which it must be moved in order for translation to initiate 43,44.
Attainment of a structure close to the mature 40S and/or nuclear export may allow Fun12 to bind the pre-40S particle in association with mature 60S subunits. The reported Fun12 binding site on 18S H5 is distant from Nob1, but close to Prp43 and to the mRNA binding cleft 30. During bacterial translation initiation, GTP hydrolysis by IF2 moves the Shine-Dalgarno/anti-Shine-Dalgarno duplex backwards (i.e. to the right in the orientation shown in Fig. 8) 43,44. We propose that a similar movement, possibly in combination with small subunit rotation 42, displaces ITS1 duplex from the mRNA-binding cleft and brings site D into proximity with the active site in the PIN domain of the endonuclease Nob1. On GTP binding, the effector domain of bacterial EF2 (a Fun12 (eIF5b) homologue) moves by some 37Å 45, consistent with estimates of the distance separating Nob1 from the 3′ end of 18S rRNA 2,8.
Links between translation competence and subunit maturation
Previous analyses have reported that multiple ribosome synthesis factors occupy sites in both the pre-40S and pre-60S particles that are required for key functional interactions of the mature subunits during translation (reviewed in 9). These are presumably important to ensure that immature pre-ribosomes do not engage non-productively with the translation machinery. Given the high rate of pre-ribosome production (around 2000 per minute in yeast) this would be a potentially significant problem. However, the data presented here support the model that at later steps in maturation at least one translation initiation factor and the 60S subunit interact with the pre-40S particle during final maturation. The obvious conclusion is that this represents a “proof-reading” step to ensure that the subunits are close to translation-competence.
In the slime mold Dictyostelium discoideum, pre-ribosomes that cannot engage with the translation machinery do not undergo late rRNA maturation steps 46. In Bacteria, the translation initiation factor IF2, which is homologous to eIF5b/Fun12 is implicated in 3′ maturation of 16S and 23S rRNA. Moreover, there are clear links between maturation of the bacterial 30S and 50S subunits and genetic interactions between IF2 and ribosome synthesis factors 4,47,48. Together these results indicate the existence of a conserved pathway, in which translation initiation factors and subunit interactions are involved in final steps of ribosome maturation. We postulate that functional proof-reading of pre-ribosomes prior to their release for translation is an ancient process that was likely already present in the last universal common ancestor of all extant organisms.
ONLINE MATERIALS AND METHODS
Strains, media, plasmids and cloning
Saccharomyces cerevisiae strain BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) was used as the parental strain 50. Strains with PGAL promoters and tags where generated by PCR as described 51. Strain genotypes are described in Supplementary Table 1. Strains were grown in YPD (1% yeast extract, 2% peptone, 2% dextrose), YPG (1% yeast extract, 2% peptone, 2% galactose), YPG/S (1% yeast extract, 2% peptone, 1% galactose, 1% sucrose) or yeast minimal medium (Formedium).
Oligonucleotides used are listed in Supplementary Table 2. The plasmid pRS-PTH-NOB1 was previously described 2. The plasmid pRS415-Fun12-HTP was constructed after PCR amplification of the FUN12 ORF and cloning into pRS415-HTP 52 using Xma1 restriction sites. The mutants Fun12D553N and Fun12T439A were generated by site-directed PCR mutagenesis. Wild-type and mutant FUN12 ORFs were subcloned into pRS416-MET25 or pRS426-ADH1 using Xma1 sites. The plasmid pSL64 allowing expression of Nob1-HIS was previously described 16. The variant pSL65 allowing expression of Nob1D15N was generated by site-directed PCR mutagenesis as described 11. The TSR1 ORF was PCR amplified and cloned into pCR4-TOPO. From this vector Tsr1 was subcloned into p415PL, generating p415-PL-HIS used for in vivo analyses or into pGEX4-T, generating GST-Tsr1-HIS used to express and purify this protein from E. coli. TSR1 mutations were generated by direct PCR mutagenesis. For intronic expression of snoRNAs from the PGAL1 promoter, oligonucleotides forming the genes for snR75 or its mutants were inserted in place of the artificial snoRNA in the actin/snoRNA cassette described previously 28. Plasmids were transformed into derivatives of YPH499 (MATa, ura3-52, lys2-801, ade2-101, leu2Δ1, his3-Δ200, trp1-Δ63) or BY4741 (Supplementary Table 1) deleted for the SNR78-72 gene cluster 28. For growth analysis, YPH499ΔsnR78-72 transformants were spotted as ten-fold serial dilutions on YPD or YPG plates and incubated at 30°C for two days.
In vitro RNA cleavage assay
The recombinant proteins Nob1-HIS and Nob1D15N-HIS were purified as described 16. Ribosomal RNA substrates were transcribed in vitro and pre-folded for 20 min at 55°C in the presence of 10 mM MgCl2 and 50 mM HEPES (pH 7.4) 53. Nuclease activity assays were performed at 30°C in 25 mM Tris/HCl (pH 7.6), 75 mM NaCl, 2 mM dithiothreitol, 100 μg ml−1 bovine serum albumin, 0.8 unit μl−1 RNasin, 4.5% glycerol, 0.05% Nonidet P-40, 0.3 μM E. coli tRNA and 5 mM MnCl2. 10 μl reactions containing 35 pmol of protein were pre-incubated for 5 min at 30°C. RNA substrate (0.1 pmol for RNAs c and d; 0.5 pmol for RNAs a and b) was added and incubated for 60 min at 30°C. After a proteinase K digestion for 30 min at 37°C, the RNA substrate was extracted, precipitated and analyzed by primer extension and northern hybridization. Primer extension used probe ITS1RT and products resolved on a denaturing 12% polyacrylamide, 8 M urea gel. For northern analysis RNA was separated on a 1% agarose gel containing glyoxal and transferred to Hybond N+. Cleavage efficiencies were quantified by northern hybridization using probe 004 that reveals both the cleaved and non-cleaved species.
In vitro pre-ribosome maturation assay
P GAL::NOB1, PTH-NOB1 cells were grown in 1 litre of YNB, glucose (2%) for 8 h to an OD600 of 0.5, collected and washed with phosphate-buffered saline. Extracts were prepared in 500 μl of Buffer A (50 mM Tris pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0,1% NP-40, 1 mM DTT and protease inhibitors (Roche)) using Zirconia beads as previously described 54. Aliquots corresponding to 12 mg of proteins were added to 50 μl of immunoglobulin G (IgG)-sepharose beads (GE healthcare) in a 1 ml final volume of buffer A. Immunoprecipitation was performed at 4°C for 1.5 h. Beads were then washed 3 times (5 min per wash) with 1 ml of buffer A at 4°C. Most of the supernatant was discarded and 50 μl of buffer X (50 mM Tris pH 7.5, 150 mM NaCl, 5 mM Mn2+, 0.1% NP-40, 1 mM DTT, 10% glycerol) added to the pellet (recovered by resuspension in 20 μl of remaining buffer A) to reach a final volume of 70 μl. Nucleotides were added when required at a final concentration of 1 mM. Reactions were incubated at 20°C for 30 min and RNAs were then immediately extracted as described 55.
Immunoprecipitation
Immunoprecipitation using the ProtA-tag on IgG sepharose beads was performed as described 55. HA-tagged proteins were precipitated using 50 μl of Gamma-bind matrix (GE healthcare) coated with 2 μg anti-HA probe sz (sc-7392 Santa Cruz Biotechnology).
RNA extraction and analysis
RNA extractions and northern blot analyses were performed as previously described 56. For primer extension used Superscript III (Invitrogen). For the sequencing ladder 2 pmol of in vitro transcribed ITS1 was used as template. Methylation activity was determined by reverse transcription in reduced nucleotide concentrations or by site-specific RNase H cleavage as described 28. Pulse-chase labeling of pre-rRNA was performed as described 55, using 100 *Ci [5,6-3H] uracil (PerkinElmer) per 40 OD600 of yeast cells.
Fluorescence In Situ Hybridization
Yeast cells were fixed for 30min with 4% paraformaldehyde in culture media, treated in Bsphero buffer (0,1 M Phosphate, 1.2 M Sorbitol, 0.2 mM PMSF, 2 mM VRC, 30 mM β-mercaptoethanol and 0.1 mg/ml) for 9 min at 30°C and incubated overnight in 70% Ethanol at −20°C. After rehydration, in 2x SSC, cells were hybridized overnight at 37°C in 40 *l of a mixture containing 10% dextran sulfate, 2 mM VRC, 0,02% RNase-free bovine serum albumin (BSA), 40 *g of E.coli tRNA, 2x SSC, 10% formamide and 30 ng of probe. Cells were washed twice for 30 min in 1x SSC, 10% formamide.
Ribosome purification
Ribosomal subunits were prepared as described 57. Two liters of BY4741 were cultivated in YPD, until OD600 of 0.6. After cell lysis (with a freezer mill 6870 from SPEX), the lysate was processed as described 57.
Separation of pre-ribosomal particles by size exclusion chromatography
36 liters of cells expressing PTH-Nob1-WT or 24 liters of cells expressing PTH-Nob1D15N were lysed in a freezer mill 6870 (SPEX). Lysates were diluted in Buffer A and loaded on an IgG-sepharose column. After overnight elution using TEV-protease, eluates were concentrated on Vivaspin 100 KDa cut-off columns. Concentrated eluates were loaded on a Superose 6 10/300 GL column (GE healthcare) using an Äkta prime system (GE healthcare) in Buffer A without Triton. Fractions corresponding to each peak were pooled and used for EM analysis.
Electron microscopy
For negative staining, 5μl samples were placed on a freshly glow-discharged, carbon-coated grid and then washed three times with water, 2 times with uranyl acetate (2% w/v) and stained with uranyl acetate for 5 min before drying. Micrographs shown were recorded with a Tecnai F20 electron microscope equipped with a TVIPS F816 digital camera and operating at 200 kV.
Model building and phylogenetic analysis
For the models presented in Fig. 8 PyMol software was used to manipulate crystal structures 3U5C, 3U5E and 1G7T 49,58. For phylogenetic comparisons ITS1 sequences were identified by BLAST using the 3′ and 5′ regions of 18S and 5.8S rRNAs, respectively, or retrieved from rDNA sequences accumulated previously 20,28. Alignments were assembled manually using the 4SALE sequence editor (http://4sale.bioapps.biozentrum.uni-wuerzburg.de/) and formatted with Jalview (http://www.jalview.org/). In the pile-up species are abbreviated as in 28; plus Torulaspora delbrueckii (Tdel), Hansenula wingei (Hwin; Pichia canadensis) 20, and Endomyces fibuliger (Efib; accession no. U10409).
Tsr1 purification and analysis
Tsr1-HTP expressed in S.cerevisiae was purified on IgG sepharose beads or on GTP agarose beads. 250 ml of cells were used for each IP. Cell extract was prepared in lysis buffer (50 mM Tris/HCl (pH 7.5), 2.5 mM MgCl2, 150 mM NaCl, 5 mM β-mercaptoethanol). Extract was incubated with IgG sepharose or GTP agarose beads for 1 h. After three washes in lysis buffer TEV cleavage (with 10 μg GST-TEV) was performed for 2 h at 18°C in the presence of 40U of RNasin. The TEV eluate was separated on a denaturing polyacrylamide gel. Recombinant proteins corresponding to either wild-type Tsr1 or the Tsr1K44A mutant were purified from E. coli using GST constructs. The K44A mutation is located in the Tsr1 GTGK domain, which is similar to the GSGK p-loop that is involved in nucleotide binding in active GTPases. Purification efficiencies were assessed on gel by Coomassie blue staining. Immunoprecipitation with purified GST-fusion proteins was performed using GTP-agarose beads. Proteins bound to the beads were visualized on Coomassie-stained gels. For the analysis of the effect of Tsr1K44A on pre-RNA processing, a chromosomal PGAL::3HA-TSR1-HTP strain was transformed an empty plasmid, a plasmid expressing wild-type Tsr1 (Tsr1-WT ) or a plasmid expressing mutant Tsr1K44A. Strains were transferred to glucose medium for 12 h, to deplete Tr1-HTP, RNA was extracted and resolved on 2% BPTE agarose gel. The EthBr signal intensity was measured using a Fuji imager.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the Wellcome Trust (SL, CS, SG, DT; 077248) and (RWvN, NJW), the Royal Society (CS), The Darwin Trust of Edinburgh (BB) and an EMBO LTF (SL). The EM-facility was supported by the Wellcome Trust and the Scottish Universities Life Science Alliance (SULSA). Work in the Wellcome Trust Centre for Cell Biology is supported by Wellcome Trust core funding [092076]. We thank Eléonore Fayet-Lebaron for her technical support and critical reading of the manuscript. We thank CTCB facilities for protein purification. Use of the CTCB Facilities was supported by The Wellcome Trust, the Scottish University Life Sciences Alliance (SULSA) and the BBSRC.
Footnotes
AUTHOR CONTRIBUTIONS SL, CS, RWvN, AS, SG, NJW, DT designed experiments; SL, CS, RWvN, AS, DW, BB, SG performed experiments; SL, CS, RWvN, AS, BB, SG, NJW, DT analyzed data; SL, CS, RWvN, AS, SG, NJW, DT wrote the paper.
REFERENCES
- 1.Inoue K, Alsina J, Chen J, Inouye M. Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli. Mol. Microbiol. 2003;48:1005–1016. doi: 10.1046/j.1365-2958.2003.03475.x. [DOI] [PubMed] [Google Scholar]
- 2.Granneman S, Petfalski E, Swiatkowska A, Tollervey D. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J. 2010 doi: 10.1038/emboj.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Z, et al. Rational Extension of the Ribosome Biogenesis Pathway Using Network-Guided Genetics. PLoS Biol. 2009;7:e1000213. doi: 10.1371/journal.pbio.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell TL, Brown ED. Genetic interaction screens with ordered overexpression and deletion clone sets implicate the Escherichia coli GTPase YjeQ in late ribosome biogenesis. J. Bacteriol. 2008;190:2537–2545. doi: 10.1128/JB.01744-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lo KY, et al. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell. 2010;39:196–208. doi: 10.1016/j.molcel.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sengupta J, et al. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J. Cell Biol. 2010;189:1079–1086. doi: 10.1083/jcb.201001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kemmler S, Occhipinti L, Veisu M, Panse VG. Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J. Cell Biol. 2009;186:863–880. doi: 10.1083/jcb.200904111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strunk BS, et al. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem. Sci. 2010;35:260–266. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soudet J, Gelugne JP, Belhabich-Baumas K, Caizergues-Ferrer M, Mougin A. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J. 2010;29:80–92. doi: 10.1038/emboj.2009.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pertschy B, et al. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem. 2009;284:35079–91. doi: 10.1074/jbc.M109.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fatica A, Oeffinger M, Dlakic M, Tollervey D. Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol. Cell. Biol. 2003;23:1798–807. doi: 10.1128/MCB.23.5.1798-1807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fatica A, Tollervey D, Dlakic M. The PIN domain of Nob1p is required for 20S pre-rRNA cleavage at site D. RNA. 2004;10:1698–1701. doi: 10.1261/rna.7123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamanna AC, Karbstein K. Nob1 binds the single-stranded cleavage site D at the 3′-end of 18S rRNA with its PIN domain. Proc Natl Acad Sci U S A. 2009;106:14259–64. doi: 10.1073/pnas.0905403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woolls HA, Lamanna AC, Karbstein K. Roles of Dim2 in ribosome assembly. J Biol Chem. 2011;286:2578–86. doi: 10.1074/jbc.M110.191494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lebaron S, et al. The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J. 2009;28:3808–19. doi: 10.1038/emboj.2009.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamanna AC, Karbstein K. An RNA conformational switch regulates pre-18S rRNA cleavage. J. Mol. Biol. 2011;405:3–17. doi: 10.1016/j.jmb.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allmang C, et al. Processing of the yeast pre-rRNA at sites A2 and A3 is linked. RNA. 1996;2:63–73. [PMC free article] [PubMed] [Google Scholar]
- 19.Yeh L-CC, Thweatt R, Lee JC. Internal transcribed spacer 1 of the yeast precursor ribosomal RNA. Higher order structure and common structural motifs. Biochemistry. 1990;29:5911–5918. doi: 10.1021/bi00477a005. [DOI] [PubMed] [Google Scholar]
- 20.Van Nues RW, et al. Separate structural elements within internal transcribed spacer 1 of Saccharomyces cerevisiae precursor ribosomal RNA direct the formation of 17S and 26S rRNA. Nucleic Acids Res. 1994;22:912–929. doi: 10.1093/nar/22.6.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelperin D, Horton L, Beckman J, Hensold J, Lemmon SK. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA. 2001;7:1268–1283. doi: 10.1017/s1355838201013073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JH, et al. Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc. Natl. Acad. Sci. USA. 2002;99:16689–16694. doi: 10.1073/pnas.262569399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pestova TV, et al. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- 24.Ferreira-Cerca S, et al. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol Cell. 2007;28:446–57. doi: 10.1016/j.molcel.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 25.Udem SA, Warner JR. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 1973;248:1412–1416. [PubMed] [Google Scholar]
- 26.Brand RC, Klootwijk J, van Steenbergen TJM, de Kok AJ, Planta RJ. Secondary methylation of yeast ribosomal precursor RNA. Eur. J. Biochem. 1977;75:311–318. doi: 10.1111/j.1432-1033.1977.tb11531.x. [DOI] [PubMed] [Google Scholar]
- 27.Lafontaine D, Delcour J, Glasser A-L, Desgrès J, Vandenhaute J. The DIM1 gene responsible for the conserved m26Am26A dimethylation in the 3′-terminal loop of 18S rRNA is essential in yeast. J. Mol. Biol. 1994;241:492–497. doi: 10.1006/jmbi.1994.1525. [DOI] [PubMed] [Google Scholar]
- 28.van Nues RW, et al. Box C/D snoRNP catalysed methylation is aided by additional pre-rRNA base-pairing. EMBO J. 2011;30:2420–2430. doi: 10.1038/emboj.2011.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Unbehaun A, et al. Position of eukaryotic initiation factor eIF5B on the 80S ribosome mapped by directed hydroxyl radical probing. EMBO J. 2007;26:3109–3123. doi: 10.1038/sj.emboj.7601751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin B-S, et al. rRNA Suppressor of a Eukaryotic Translation Initiation Factor 5B/Initiation Factor 2 Mutant Reveals a Binding Site for Translational GTPases on the Small Ribosomal Subunit. Molecular and Cellular Biology. 2009;29:808–821. doi: 10.1128/MCB.00896-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344–51. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 32.Lygerou Z, Allmang C, Tollervey D, Séraphin B. Accurate processing of a eukaryotic pre-rRNA by RNase MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 33.Abou Elela S, Igel H, Ares M., Jr. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 34.Horn DM, Mason SL, Karbstein K. Rcl1 protein, a novel nuclease for 18 S ribosomal RNA production. J. Biol. Chem. 2011;286:34082–34087. doi: 10.1074/jbc.M111.268649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allmang C, Tollervey D. The role of the 3′ external transcribed spacer in yeast pre-rRNA processing. J. Mol. Biol. 1998;278:67–78. doi: 10.1006/jmbi.1998.1693. [DOI] [PubMed] [Google Scholar]
- 36.Valasek L, Hasek J, Nielsen KH, Hinnebusch AG. Dual Function of eIF3j/Hcr1p in Processing 20 S Pre-rRNA and Translation Initiation. J. Biol. Chem. 2001;276:43351–43360. doi: 10.1074/jbc.M106887200. [DOI] [PubMed] [Google Scholar]
- 37.Collins SR, et al. Toward a Comprehensive Atlas of the Physical Interactome of Saccharomyces cerevisiae. Molec. Cell. Proteom. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Senger B, et al. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell. 2001;8:1363–1373. doi: 10.1016/s1097-2765(01)00403-8. [DOI] [PubMed] [Google Scholar]
- 39.Yarunin A, et al. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J. 2005;24:580–588. doi: 10.1038/sj.emboj.7600540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neueder A, et al. A local role for the small ribosomal subunit primary binder rpS5 in final 18S rRNA processing in yeast. PLoS One. 2010;5:e10194. doi: 10.1371/journal.pone.0010194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veith T, et al. Structural and functional analysis of the archaeal endonuclease Nob1. Nucleic Acids Res. 2012;40:3259–74. doi: 10.1093/nar/gkr1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marshall RA, Aitken CE, Puglisi JD. GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol. Cell. 2009;35:37–47. doi: 10.1016/j.molcel.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Julian P, et al. The Cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli. PLoS Biol. 2011;9:e1001095. doi: 10.1371/journal.pbio.1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.La Teana A, Gualerzi CO, Brimacombe R. From stand-by to decoding site. Adjustment of the mRNA on the 30S ribosomal subunit under the influence of the initiation factors. RNA. 1995;1:772–782. [PMC free article] [PubMed] [Google Scholar]
- 45.Valle M, et al. Locking and Unlocking of Ribosomal Motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 46.Mangiarotti G, Chiaberge S, Bulfone S. rRNA maturation as a “quality” control step in ribosomal subunit assembly in Dictyostelium discoideum. J. Biol. Chem. 1997;272:27818–27822. doi: 10.1074/jbc.272.44.27818. [DOI] [PubMed] [Google Scholar]
- 47.Kaczanowska M, Ryden-Aulin M. Ribosome Biogenesis and the Translation Process in Escherichia coli. Microbiol. Molec. Biol. Revs. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belotserkovsky JM, Dabbs ER, Isaksson LA. Mutations in 16S rRNA that suppress cold-sensitive initiation factor 1 affect ribosomal subunit association. FEBS J. 2011;278:3508–3517. doi: 10.1111/j.1742-4658.2011.08272.x. [DOI] [PubMed] [Google Scholar]
- 49.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 50.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 51.Petracek ME, Longtine MS. PCR-based engineering of yeast genome. Methods Enzymol. 2002;350:445–469. doi: 10.1016/s0076-6879(02)50978-2. [DOI] [PubMed] [Google Scholar]
- 52.Bohnsack MT, et al. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karbstein K, Jonas S, Doudna JA. An essential GTPase promotes assembly of preribosomal RNA processing complexes. Mol. Cell. 2005;20:633–643. doi: 10.1016/j.molcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 54.Granneman S, Kudla G, Petfalski E, Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high throughput analysis of cDNAs. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9613–9818. doi: 10.1073/pnas.0901997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lebaron S, et al. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol. Cell. Biol. 2005;25:9269–9282. doi: 10.1128/MCB.25.21.9269-9282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tollervey D. High level of complexity of small nuclear RNAs from fungi and plants. J. Mol. Biol. 1987;196:355–361. doi: 10.1016/0022-2836(87)90696-6. [DOI] [PubMed] [Google Scholar]
- 57.Leshin JA, Rakauskaite R, Dinman JD, Meskauskas A. Enhanced purity, activity and structural integrity of yeast ribosomes purified using a general chromatographic method. RNA Biol. 2010;7:354–360. doi: 10.4161/rna.7.3.11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roll-Mecak A, Cao C, Dever TE, Burley SK. X-Ray Structures of the Universal Translation Initiation Factor IF2/eIF5B: Conformational Changes on GDP and GTP Binding. Cell. 2000;103:781–792. doi: 10.1016/s0092-8674(00)00181-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.