Abstract
The pathway of sterol biosynthesis is highly conserved in all eucaryotic cells. We demonstrated structural and functional conservation of the rate-limiting enzyme of the mammalian pathway, 3-hydroxy-3-methyl-glutaryl coenzyme A reductase (HMG-CoA reductase), between the yeast Saccharomyces cerevisiae and humans. The amino acid sequence of the two yeast HMG-CoA reductase isozymes was deduced from DNA sequence analysis of the HMG1 and HMG2 genes. Extensive sequence similarity existed between the region of the mammalian enzyme encoding the active site and the corresponding region of the two yeast isozymes. Moreover, each of the yeast isozymes, like the mammalian enzyme, contained seven potential membrane-spanning domains in the NH2-terminal region of the protein. Expression of cDNA clones encoding either hamster or human HMG-CoA reductase rescued the viability of hmg1 hmg2 yeast cells lacking this enzyme. Thus, mammalian HMG-CoA reductase can provide sufficient catalytic function to replace both yeast isozymes in vivo. The availability of yeast cells whose growth depends on human HMG-CoA reductase may provide a microbial screen to identify new drugs that can modulate cholesterol biosynthesis.
Full text
PDF





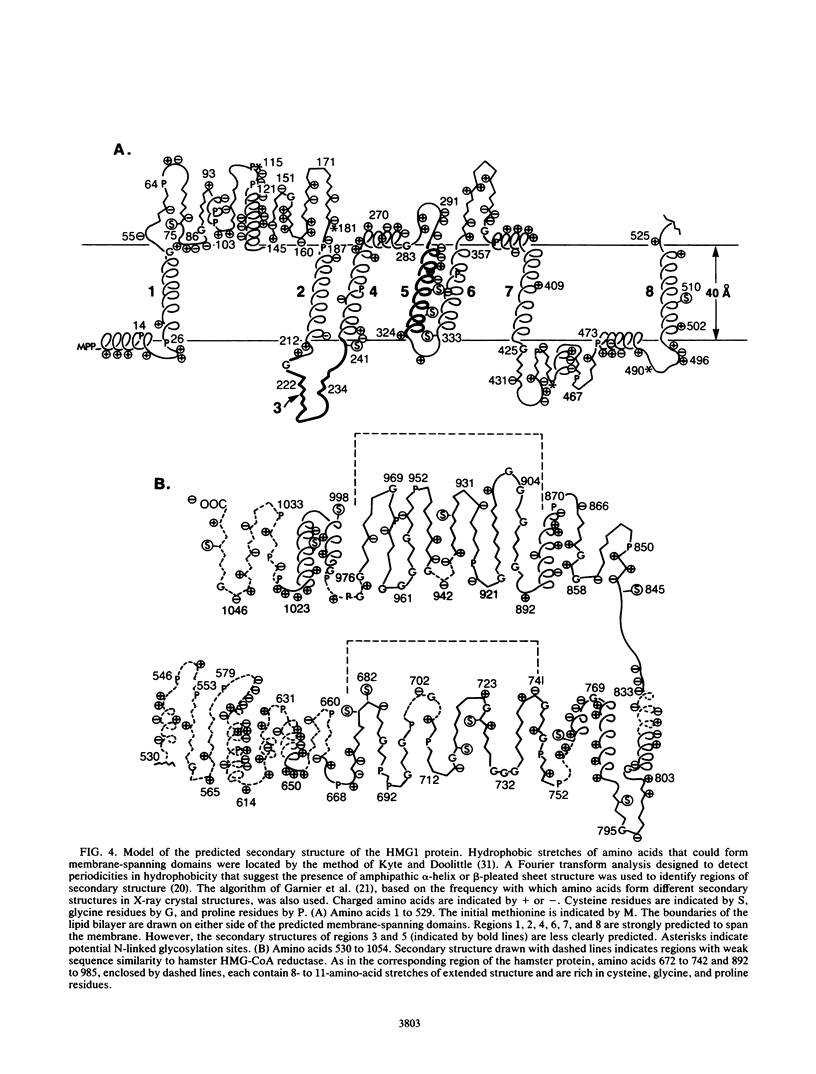
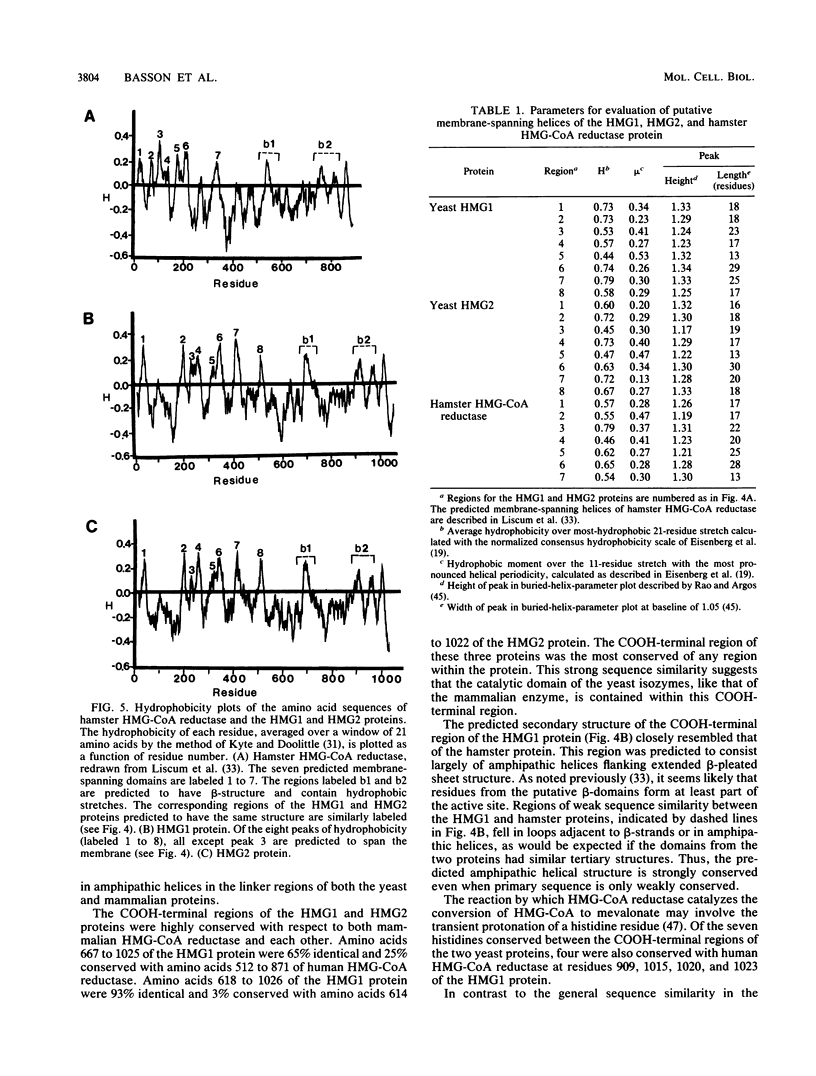
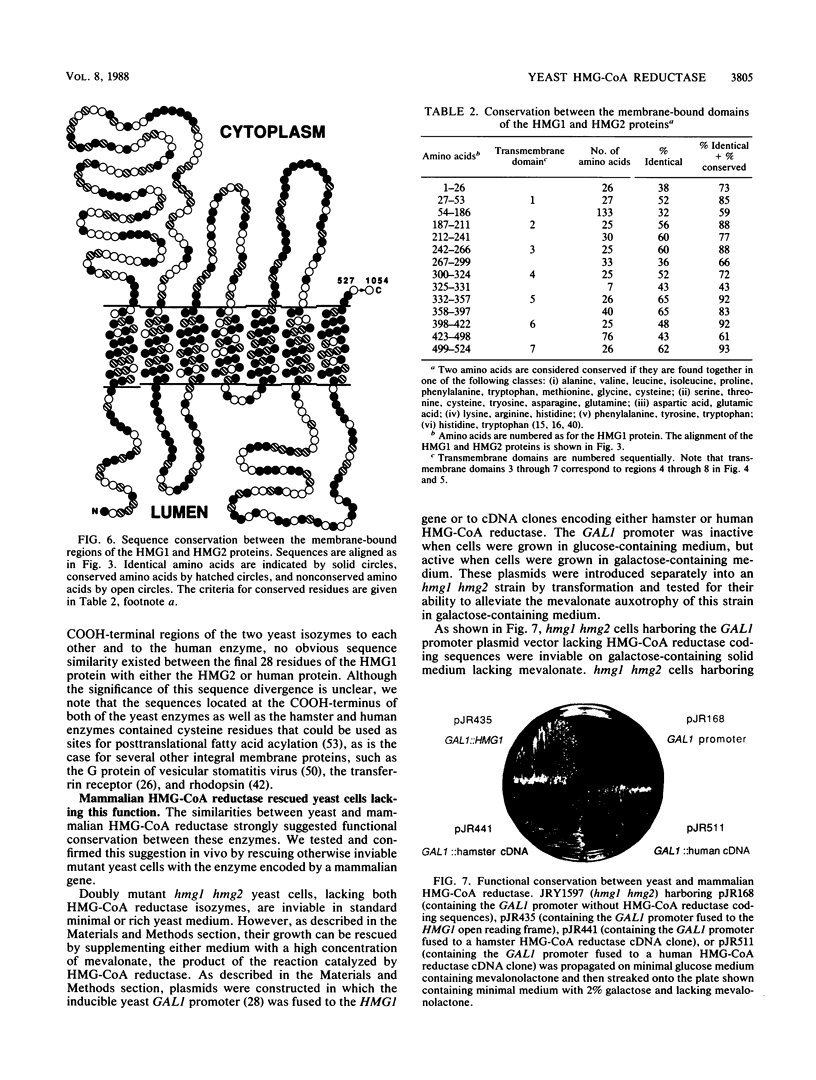



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abovich N., Rosbash M. Two genes for ribosomal protein 51 of Saccharomyces cerevisiae complement and contribute to the ribosomes. Mol Cell Biol. 1984 Sep;4(9):1871–1879. doi: 10.1128/mcb.4.9.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. G., Orci L., Brown M. S., Garcia-Segura L. M., Goldstein J. L. Ultrastructural analysis of crystalloid endoplasmic reticulum in UT-1 cells and its disappearance in response to cholesterol. J Cell Sci. 1983 Sep;63:1–20. doi: 10.1242/jcs.63.1.1. [DOI] [PubMed] [Google Scholar]
- Barnes G., Hansen W. J., Holcomb C. L., Rine J. Asparagine-linked glycosylation in Saccharomyces cerevisiae: genetic analysis of an early step. Mol Cell Biol. 1984 Nov;4(11):2381–2388. doi: 10.1128/mcb.4.11.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson M. E., Moore R. L., O'Rear J., Rine J. Identifying mutations in duplicated functions in Saccharomyces cerevisiae: recessive mutations in HMG-CoA reductase genes. Genetics. 1987 Dec;117(4):645–655. doi: 10.1093/genetics/117.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson M. E., Thorsness M., Rine J. Saccharomyces cerevisiae contains two functional genes encoding 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5563–5567. doi: 10.1073/pnas.83.15.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg Z. H., Stonik J. A., Brewer H. B., Jr Modulation of the enzymic activity of 3-hydroxy-3-methylglutaryl coenzyme A reductase by multiple kinase systems involving reversible phosphorylation: a review. Metabolism. 1987 Sep;36(9):900–917. doi: 10.1016/0026-0495(87)90101-6. [DOI] [PubMed] [Google Scholar]
- Bloch K. The biological synthesis of cholesterol. Science. 1965 Oct 1;150(3692):19–28. doi: 10.1126/science.150.3692.19. [DOI] [PubMed] [Google Scholar]
- Brown D. A., Simoni R. D. Biogenesis of 3-hydroxy-3-methylglutaryl-coenzyme A reductase, an integral glycoprotein of the endoplasmic reticulum. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1674–1678. doi: 10.1073/pnas.81.6.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. S., Goldstein J. L. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. 1980 Jul;21(5):505–517. [PubMed] [Google Scholar]
- Chin D. J., Gil G., Faust J. R., Goldstein J. L., Brown M. S., Luskey K. L. Sterols accelerate degradation of hamster 3-hydroxy-3-methylglutaryl coenzyme A reductase encoded by a constitutively expressed cDNA. Mol Cell Biol. 1985 Apr;5(4):634–641. doi: 10.1128/mcb.5.4.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin D. J., Gil G., Russell D. W., Liscum L., Luskey K. L., Basu S. K., Okayama H., Berg P., Goldstein J. L., Brown M. S. Nucleotide sequence of 3-hydroxy-3-methyl-glutaryl coenzyme A reductase, a glycoprotein of endoplasmic reticulum. Nature. 1984 Apr 12;308(5960):613–617. doi: 10.1038/308613a0. [DOI] [PubMed] [Google Scholar]
- Chin D. J., Luskey K. L., Anderson R. G., Faust J. R., Goldstein J. L., Brown M. S. Appearance of crystalloid endoplasmic reticulum in compactin-resistant Chinese hamster cells with a 500-fold increase in 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1185–1189. doi: 10.1073/pnas.79.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J., Kolodrubetz D., Grunstein M. The two yeast histone H2A genes encode similar protein subtypes. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1484–1487. doi: 10.1073/pnas.79.5.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke C. F., Fogelman A. M., Edwards P. A. Transcriptional regulation of the 3-hydroxy-3-methylglutaryl coenzyme A reductase gene in rat liver. J Biol Chem. 1985 Nov 15;260(26):14363–14367. [PubMed] [Google Scholar]
- Dayhoff M. O., Barker W. C., Hunt L. T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards P. A., Lan S. F., Fogelman A. M. Alterations in the rates of synthesis and degradation of rat liver 3-hydroxy-3-methylglutaryl coenzyme A reductase produced by cholestyramine and mevinolin. J Biol Chem. 1983 Sep 10;258(17):10219–10222. [PubMed] [Google Scholar]
- Eisenberg D., Schwarz E., Komaromy M., Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984 Oct 15;179(1):125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Finer-Moore J., Stroud R. M. Amphipathic analysis and possible formation of the ion channel in an acetylcholine receptor. Proc Natl Acad Sci U S A. 1984 Jan;81(1):155–159. doi: 10.1073/pnas.81.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Gil G., Faust J. R., Chin D. J., Goldstein J. L., Brown M. S. Membrane-bound domain of HMG CoA reductase is required for sterol-enhanced degradation of the enzyme. Cell. 1985 May;41(1):249–258. doi: 10.1016/0092-8674(85)90078-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing S. Q., Trowbridge I. S. Identification of the intermolecular disulfide bonds of the human transferrin receptor and its lipid-attachment site. EMBO J. 1987 Feb;6(2):327–331. doi: 10.1002/j.1460-2075.1987.tb04758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jingami H., Brown M. S., Goldstein J. L., Anderson R. G., Luskey K. L. Partial deletion of membrane-bound domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase eliminates sterol-enhanced degradation and prevents formation of crystalloid endoplasmic reticulum. J Cell Biol. 1987 Jun;104(6):1693–1704. doi: 10.1083/jcb.104.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G. A., Pazirandeh M., Krisans S. 3-Hydroxy-3-methylglutaryl coenzyme A reductase localization in rat liver peroxisomes and microsomes of control and cholestyramine-treated animals: quantitative biochemical and immunoelectron microscopical analyses. J Cell Biol. 1986 Sep;103(3):875–886. doi: 10.1083/jcb.103.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn L. A., Leigh J. S., Jr A statistical technique for predicting membrane protein structure. Biochim Biophys Acta. 1985 Apr 29;828(3):351–361. doi: 10.1016/0167-4838(85)90316-4. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Liscum L., Cummings R. D., Anderson R. G., DeMartino G. N., Goldstein J. L., Brown M. S. 3-Hydroxy-3-methylglutaryl-CoA reductase: a transmembrane glycoprotein of the endoplasmic reticulum with N-linked "high-mannose" oligosaccharides. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7165–7169. doi: 10.1073/pnas.80.23.7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum L., Luskey K. L., Chin D. J., Ho Y. K., Goldstein J. L., Brown M. S. Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase and its mRNA in rat liver as studied with a monoclonal antibody and a cDNA probe. J Biol Chem. 1983 Jul 10;258(13):8450–8455. [PubMed] [Google Scholar]
- Luskey K. L. Conservation of promoter sequence but not complex intron splicing pattern in human and hamster genes for 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol Cell Biol. 1987 May;7(5):1881–1893. doi: 10.1128/mcb.7.5.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luskey K. L., Stevens B. Human 3-hydroxy-3-methylglutaryl coenzyme A reductase. Conserved domains responsible for catalytic activity and sterol-regulated degradation. J Biol Chem. 1985 Aug 25;260(18):10271–10277. [PubMed] [Google Scholar]
- Michel H., Weyer K. A., Gruenberg H., Dunger I., Oesterhelt D., Lottspeich F. The 'light' and 'medium' subunits of the photosynthetic reaction centre from Rhodopseudomonas viridis: isolation of the genes, nucleotide and amino acid sequence. EMBO J. 1986 Jun;5(6):1149–1158. doi: 10.1002/j.1460-2075.1986.tb04340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohana Rao J. K., Argos P. A conformational preference parameter to predict helices in integral membrane proteins. Biochim Biophys Acta. 1986 Jan 30;869(2):197–214. doi: 10.1016/0167-4838(86)90295-5. [DOI] [PubMed] [Google Scholar]
- Nathans J., Thomas D., Hogness D. S. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986 Apr 11;232(4747):193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- O'Brien P. J., Zatz M. Acylation of bovine rhodopsin by [3H]palmitic acid. J Biol Chem. 1984 Apr 25;259(8):5054–5057. [PubMed] [Google Scholar]
- Pathak R. K., Luskey K. L., Anderson R. G. Biogenesis of the crystalloid endoplasmic reticulum in UT-1 cells: evidence that newly formed endoplasmic reticulum emerges from the nuclear envelope. J Cell Biol. 1986 Jun;102(6):2158–2168. doi: 10.1083/jcb.102.6.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peffley D., Sinensky M. Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase synthesis by a non-sterol mevalonate-derived product in Mev-1 cells. Apparent translational control. J Biol Chem. 1985 Aug 25;260(18):9949–9952. [PubMed] [Google Scholar]
- Reynolds G. A., Basu S. K., Osborne T. F., Chin D. J., Gil G., Brown M. S., Goldstein J. L., Luskey K. L. HMG CoA reductase: a negatively regulated gene with unusual promoter and 5' untranslated regions. Cell. 1984 Aug;38(1):275–285. doi: 10.1016/0092-8674(84)90549-x. [DOI] [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986 Oct 17;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Roitelman J., Shechter I. Regulation of rat liver 3-hydroxy-3-methylglutaryl coenzyme A reductase. Evidence for thiol-dependent allosteric modulation of enzyme activity. J Biol Chem. 1984 Jan 25;259(2):870–877. [PubMed] [Google Scholar]
- Rose J. K., Adams G. A., Gallione C. J. The presence of cysteine in the cytoplasmic domain of the vesicular stomatitis virus glycoprotein is required for palmitate addition. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2050–2054. doi: 10.1073/pnas.81.7.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz P. J., Pillus L., Grisafi P., Solomon F., Botstein D. Two functional alpha-tubulin genes of the yeast Saccharomyces cerevisiae encode divergent proteins. Mol Cell Biol. 1986 Nov;6(11):3711–3721. doi: 10.1128/mcb.6.11.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefton B. M., Buss J. E. The covalent modification of eukaryotic proteins with lipid. J Cell Biol. 1987 Jun;104(6):1449–1453. doi: 10.1083/jcb.104.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman L. S., Krummel B. M., Wilson A. C. Evolutionary shift in the site of cleavage of prelysozyme. J Biol Chem. 1986 Feb 15;261(5):2309–2313. [PubMed] [Google Scholar]
- Zammit V. A., Easom R. A. Regulation of hepatic HMG-CoA reductase in vivo by reversible phosphorylation. Biochim Biophys Acta. 1987 Feb 18;927(2):223–228. doi: 10.1016/0167-4889(87)90138-8. [DOI] [PubMed] [Google Scholar]



