Abstract
In barley (Hordeum vulgare L.), chiasmata (the physical sites of genetic crossovers) are skewed towards the distal ends of chromosomes, effectively consigning a large proportion of genes to recombination coldspots. This has the effect of limiting potential genetic variability, and of reducing the efficiency of map-based cloning and breeding approaches for this crop. Shifting the sites of recombination to more proximal chromosome regions by forward and reverse genetic means may be profitable in terms of realizing the genetic potential of the species, but is predicated upon a better understanding of the mechanisms governing the sites of these events, and upon the ability to recognize real changes in recombination patterns. The barley MutL Homologue (HvMLH3), a marker for class I interfering crossovers, has been isolated and a specific antibody has been raised. Immunolocalization of HvMLH3 along with the synaptonemal complex transverse filament protein ZYP1, used in conjunction with fluorescence in situ hybridization (FISH) tagging of specific barley chromosomes, has enabled access to the physical recombination landscape of the barley cultivars Morex and Bowman. Consistent distal localization of HvMLH3 foci throughout the genome, and similar patterns of HvMLH3 foci within bivalents 2H and 3H have been observed. A difference in total numbers of HvMLH3 foci between these two cultivars has been quantified, which is interpreted as representing genotypic variation in class I crossover frequency. Discrepancies between the frequencies of HvMLH3 foci and crossover frequencies derived from linkage analysis point to the existence of at least two crossover pathways in barley. It is also shown that interference of HvMLH3 foci is relatively weak compared with other plant species.
Key words: Barley, chiasma, crossover, Hordeum vulgare, interference, MLH3.
Introduction
Meiosis is an integral part of sexual reproduction, since it halves the zygotic chromosome number during gametogenesis, and enables the recombination of genes through crossing over and the independent assortment of maternal and paternal chromosomes. Crossovers (COs) not only drive genetic variability by shuffling the maternal and paternal genomes, but also ensure the regular disjunction of homologues at anaphase I. In other words, each pair of homologues in the majority of sexually reproducing organisms must form at least one CO (the obligate CO) in order to maintain the mechanical efficiency of meiosis (Jones, 1984). Notwithstanding the stabilizing effect of COs, in most organisms their numbers are capped by positive interference, whereby one CO reduces the probability of a second simultaneous CO occurring in its vicinity (Muller, 1916; Haldane, 1931). Longer bivalents commonly have more COs than shorter bivalents, but positive interference ensures that COs are not clustered (Kleckner et al., 2003). COs may also be confined to particular regions of the genome, resulting in ‘hot’ regions where the frequency of COs is disproportionately high and ‘cold’ regions where COs are relatively suppressed (de Massy, 2003; Youds and Boulton, 2011). Several genomic features that influence recombination localization have been documented, such as gene density (Jensen-Seaman et al., 2004), histone modification (Baudat et al., 2010), and chromosomal territories (Obeso and Dawson, 2010).
Accessing the recombination landscape in agronomically important temperate cereals and grasses, such as wheat, rye, barley, and ryegrass, has been accomplished to date largely by the cytological scoring of chiasmata or through genetic linkage analysis (Bennett et al., 1973; King et al., 2007; Saintenac et al., 2009; Li et al., 2010). Physical and genetic mapping have been instrumental in determining the genetic potential of these crops, and have enabled the discovery of important genes and quantitative trait loci (QTLs). Many of these studies have shown that COs are not distributed evenly along the chromosome, but are predominantly localized in distal regions (Lukaszewski and Curtis, 1993; Künzel et al., 2000; Akhunov et al., 2003; Erayman et al., 2004; King et al., 2007). The result is that large tracts of interstitial and proximal regions recombine much less frequently, thereby limiting the potential genetic variability of the species and reducing the effectiveness of map-based cloning and breeding approaches. Shifting the sites of recombination may be profitable in terms of realizing the full genetic potential of crops, but is predicated upon better understanding of the mechanisms governing the distribution of these events, and upon the ability to assay accurately changes in recombination patterns resulting from interventions.
Visual assays of the number and distribution of COs are conducted in one of three ways: scoring chiasmata, scoring late nodules (LNs), or scoring immunodetected LN components. The cytological counterparts of mature CO events (chiasmata) can be scored by conventional bright-field microscopy at diakinesis or metaphase I in amenable organisms. While this may provide a rapid and effective means of estimating CO frequency in organisms possessing long chromosomes in which chiasmata can be easily discerned, it is however considerably less reliable in species with small, compact bivalents in which a proportion of the chiasmata cannot be resolved unambiguously by light microscopy. Alternatively, a different physical manifestation of mature CO events, LNs, can be observed by resolving synaptonemal complexes (SCs) at pachytene by electron microscopy (Sherman et al., 1992; Anderson and Stack, 2005). The scoring of LNs on SCs at pachytene provides a high resolution physical map of CO events, which corresponds well in some organisms to patterns of chiasmata at metaphase I (Anderson and Stack, 2005; Falque et al., 2009). Unfortunately, technical difficulties in preserving and resolving LNs have detracted from the widespread use of this approach. Additionally, a fluorescence-based assay using tetrad pollen has been used to determine the level of COs and gene conversion events at specific loci in Arabidopsis (Berchowitz and Copenhaver, 2009). Mutants that produce tetrad pollen have yet to be identified in the Poaceae, so comparable analyses are not yet possible in this important family of plants.
The discovery of the proteins involved in homologous recombination has enabled the development of an immunological method to score CO number and distribution. The most widely used approach is to immunolocalize the MLH1/MLH3 protein heterodimer during pachytene using fluorescence microscopy, in conjunction with antibodies that highlight the SC (Anderson et al., 1999; Moens et al., 2002; Lhuissier et al., 2007; Chelysheva et al., 2010). The MLH1 and MLH3 proteins are orthologous to the MutL DNA mismatch repair protein of Escherichia coli, and are crucial to the final stages of homologous recombination, since mutations in these key genes result in a severe reduction in CO frequencies (Baker et al., 1996; Wang et al., 1999; Jackson et al., 2006). To date, this heterodimer has been found to mark only sites of interhomologue CO events and not intersister events (Wijeratne and Ma, 2007). The MLH3 protein contains a highly conserved metal-binding domain [DQHA(X)2E(X)4E] which is predicted to endow endonuclease activity on the heterodimer for joint molecule resolution (Nishant et al., 2008). Immunogold labelling has shown that these two proteins are components of a subset of interfering LNs, and therefore provide an accurate and reliable mark for interfering (class I) CO events at pachytene (Moens et al., 2002; Lhuissier et al., 2007). The MLH1 protein persists at sites of chiasmata at diakinesis in Arabidopsis, unlike in other organisms where the MLH1 foci are lost prior to mature chiasma formation (Chelysheva et al., 2010). In order to map the distribution of the MLH1/3 foci along the length of bivalents, the SC is simultaneously immunodetected using antibodies to SC components, such as the Zip1 protein of the central element of Saccharomyces cerevisiae (Sym et al., 1993), or its orthologue in other species such as AtZYP1 in Arabidopsis (Higgins et al., 2005). The success of this method is to a large extent dependent upon the accuracy by which individual SCs can be traced from slides; to date, only a small number of organisms such as tomato, mouse and human have been amenable to this approach (Anderson et al., 1999; Sun et al., 2006; Lhuissier et al., 2007).
In order to overcome these limitations, Phillips et al. (2010) detailed a novel experimental approach to physically map CO events in barley at high resolution. This present study describes the isolation and utility of the barley orthologue of MLH3 (HvMLH3), and shows how the immunolocalization of HvMLH3 foci and the transverse filament protein ZYP1, together with fluorescence in situ hybridization (FISH) tagging of specific barley chromosomes, can be integrated to access the recombination landscape of this crop. It shows that the antibody to HvMLH3 protein reliably detects foci at pachytene that are likely to correspond to class I CO events, and reveals surprising differences in numbers of foci between two contrasting cultivars. It also shows consistent distal localization of class I COs in bivalents 2H and 3H of the two cultivars, and relatively weak CO interference compared with other plant species.
Materials and methods
Plant material
Barley [Hordeum vulgare cv. Morex, Bowman, and Optic (2n=2x=14)] was grown to maturity under 16h days with 60 μmol s–1 m–2 illumination at a constant 20 °C in standard greenhouse conditions.
In silico search for barley sequences and full-length cDNA cloning
The protein sequence of Arabidopsis MLH3 (AT4G35520) was used in an NCBI blastp search (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins), and the top hits for monocots were selected and their corresponding nucleotide sequences were downloaded. The retrieved nucleotide sequences were then used to find the barley homologues in the barley expressed sequence tag (EST) database HarvEST#35 assembly (http://www.harvest-web.org/hweb/bin/wc.dll?hwebProcess~hmain~&versid=5). Two unigenes (h35_29479 and h35_08952) were found and their sequences used to design oligonucleotides for 5’ and 3’ rapid amplification of cDNA ends (RACE) (Supplementary Table S1 available at JXB online).
Total RNA was extracted from barley (H. vulgare cv. Optic) inflorescences at meiosis (0.5–1.5mm anthers) using an RNAqueous® kit (Ambion). First-strand cDNA was synthesized with a GeneRacer kit (Invitrogen). The 5’ and 3’ ends of MLH3 cDNA were amplified by PCR according to the GeneRacer kit protocol. Briefly, 1 μl of the cDNA library was used as a template in the first PCR to amplify the 5’ or 3’ end of the cDNA in the presence of Phusion High-Fidelity DNA Polymerase (New England Biolabs). A 1 μl aliquot of this reaction was used as a template in the second PCR with specific nested oligonucleotides. The PCR fragments were cloned into pGEM-Teasy vector (Promega) and sequenced. These 5’ and 3’ sequences were used to design primers for full-length cDNA amplification from the cDNA using Expand High Fidelity Taq polymerase (Roche) (Supplementary Table S1 at JXB online). The full-length HvMLH3 cDNA sequence was deposited in GenBank (accession no. JQ855501).
RNA-seq
The barley Morex genome assembly database (International Barley Genome Sequencing Consortium, 2012) contains next-generation sequencing of total RNA extracted from three replicates each of eight different samples (see the Results) representing different organs and developmental stages of barley, as described by the authors in their supplementary information (http://www.nature.com/nature/journal/v491/n7426/extref/nature11543-s1.pdf). The Illumina RNA-seq reads (ftp://ftpmips.helmholtz-muenchen.de/plants/barley/public_data/) were assembled to generate a barley RNA-seq database that was BLAST searched with the HvMLH3 cDNA sequence in order to assay the relative levels of expression of this gene.
Multiple alignment and phylogenetic analysis
The plant genomes platform Plaza 2.5 [http://bioinformatics.psb.ugent.be/plaza/ (Van Bel et al., 2012)] was searched for sequences similar to the Arabidopsis MutL homologue 3 (AT4G35520). The full-length sequences were analysed with Pfam 26.0 (http://pfam.janelia.org/) and their conserved domains extracted with the EMBOSS 6.3.1 tool (http://mobyle.pasteur.fr/cgi-bin/portal.py#forms::extractseq). The sequences were aligned using the Clustal Omega protein multiple alignment program (Sievers et al., 2011), and reliably aligned alignment positions were selected using the Gblocks program (Talavera and Castresana, 2007) with the three least stringent options chosen. This resulted in an 18 sequence alignment of length 592 amino acids.
Model selection was carried out using the TOPALi (Milne et al., 2009) joint estimation of the evolutionary model and the phylogenetic tree. A JTT+G model (alpha shape parameter=1.118) was selected using the BIC criterion. A maximum likelihood tree was then estimated using the PhyML program (Guindon and Gascuel, 2003) launched from TOPALi, with bootstrapping (100 replicates). Phylogenetic tree diagrams were produced using Dendroscope (Huson et al., 2007), with Physcomitrella patens a natural choice for the outgroup.
The observed (single locus MLH3) and expected (multilocus) tree topologies were statistically tested for congruence using the Kishino–Hasegawa–Templeton (KHT) test (Kishino and Hasegawa, 1989). This was carried out by inputting the tree topologies (with zero branch lengths) into the PHYLIP PROML program along with the 15 sequence MLH3 alignment. The branch lengths for both trees were then re-estimated for the MLH3 data using the JTT+G model (the alpha shape parameter, 1.118, was obtained from the TOPALI model selection output and converted to a coefficient of variation of 0.945756 for use with PROML). The difference in the log likelihood of the trees (10.04) was not significantly different (P > 0.05) based on the KHT test, with a log likelihood (observed tree) of –9106.46 compared with –9116.49 for the expected tree, and a standard deviation of log likelihoods of 10.4771.
HvMLH3 antibody production
For antibody production, a fragment of 329 amino acids (residues 863–1191) at the C-terminus of the protein was used. Briefly, the corresponding sequence (2905–3895bp) was amplified from the full-length cDNA using specific primers (Supplementary Table S1 at JXB online) containing NdeI and XhoI restriction sites, respectively. The resulting fragment was digested and ligated into pET21b and then introduced into E. coli BL21 DE3 cells (Novagen) for recombinant protein expression. Recombinant protein induction, inclusion body purification, and protein refolding were performed as described by Kakeda et al. (1998). Rabbit polyclonal antiserum was produced against the recombinant protein by Biogenes (Germany).
Sequential FISH and immunolocalization
Morex and Bowman meiocytes at prophase I were gathered from inflorescences collected from several plants and embedded in polyacrylamide according to Phillips et al. (2010). Subsequent immunolocalization was carried out as detailed in Phillips et al. (2010) with the following modifications. The meiotic nuclei embedded in the polyacrylamide pads were incubated with anti-HvHML3 antibody and anti-AtZYP1 antibody (Higgins et al., 2005) both diluted to 1:150 in blocking buffer and incubated for 36h at 4 °C. FISH with centromere and 5S rDNA probes was performed according to Phillips et al. (2010). Briefly, 5S rDNA (Gerlach and Dyer, 1980) was labelled by PCR with biotin-16-dUTP (Roche) and a centromeric repeat (Nasuda et al., 2005) labelled with tetramethyl-rhodamine-5-dUTP (Roche). Chromosomes were denatured for 8min at 75 °C, incubated overnight at 37 °C, followed by two stringent washes in 0.1× SSC at 37 °C for 30min each. Pads subjected solely to immunolocalization were prepared as detailed in Phillips et al. (2012). The control experiment with HvMLH3 pre-immune serum did not show any specific binding during pachytene (Supplementary Fig. S1A, B at JXB online).
Chiasma counts of Morex and Bowman meiocytes at metaphase I were obtained from bright-field microscopy of standard squash preparations stained with 1% aceto-carmine.
Image capture and analysis
Nuclei were optically sectioned using a Leica TCS SP5 II confocal laser scanning microscope (CLSM) controlled by Leica LAS-AF software. Z-stacks were deconvolved using AutoQuant X2 (Media Cybernetics) and analysed using Imaris 7.3 (Bitplane). Imaris allows the Z-stacks to be rendered in 3D and in this space surfaces were manually added to trace each of the bivalents and mark and measure each of the HvMLH3 foci (Phillips et al., 2012). The selection of HvMHL3 foci was performed automatically using the ‘Spot detection’ wizard in Imaris 7.1, and the results were subsequently manually edited to remove any foci that lay away from the axes or to add in foci that the automatic detection occasionally failed to identify.
Results
Isolation and characterization of HvMLH3
HvMLH3 cDNA encodes a protein with conserved domains at the N- and C-termini. The full-length cDNA of 4186bp was cloned from meiotic inflorescences of barley, and the coding sequence is flanked by a 321bp GC-rich (74.9%) 5’-untranslated region (UTR) and 236bp 3’-UTR. The predicted protein product is 1208 amino acids long with a pI of 6.36 and mol. wt of 135.935kDa. Based on the 3’-RACE-PCR results and using the EST database HarvEST 35, it is concluded that HvMLH3 protein is encoded by a single gene in barley, as is the case for most of the plant species in the Plaza 2.5 database.
The alignment of full-length MLH3 protein sequences reveals conserved domains only at the N- and C-termini connected by a variable linker sequence. Pfam analysis of these sequences detected three conserved domains, namely the HATPase_c_3 domain and DNA_mis_repair family at the N-terminus and the MutL_C domain at the C-terminus. These three conserved domains were present in the barley sequence and extracted from all available plant MLH3 sequences. The alignment of these domains reveals several conserved blocks, including a putative endonuclease domain at the C-terminal-interacting domain (Supplementary Fig. S2 at JXB online).
The amino acid sequences of plant MLH3 proteins were used to estimate a maximum likelihood phylogenetic tree, with P. patens as the outgroup (Fig. 1A). If the sequences are orthologous, then the resulting 18 sequence tree is an estimate of the plant species phylogeny. However, this is based on only a single locus and is unlikely to resolve the species relationships fully; it should therefore be expected that the single locus tree would not be identical in topology to a multilocus tree, but would be not significantly different.
Fig. 1.
Plant MLH3 phylogeny and HvMlh3 expression. (A) Maximum likelihood phylogenetic tree based on 18 sequences. The positions of the five species labelled (***) do not agree with the species tree based on the multilocus analysis of Lee et al. (2011), and are associated with low bootstrap support. The three species labelled (>>) were not included in the multilocus tree of Lee et al. (2011). (B) The graph shows MLH3 expression levels in different organs of barley cv. Morex. Error bars represent the SD (n=3). FPKM, fragments per kb of transcript per million mapped reads; EMB, 4-day-old embryos dissected from germinating grains; LEA, shoots from seedlings (10cm shoot stage); INF1, young developing inflorescences (5mm); INF2, developing inflorescences (1–1.5cm); ROO, roots from seedlings (10cm shoot stage); NOD, developing tillers at the six-leaf stage, third internode; CAR5, developing grain, bracts removed (5 d post-anthesis); CAR15, developing grain, bracts removed (15 d post-anthesis).
A tree based on 15 of these 18 sequences was thus compared with the relationships among 15 shared species in the multilocus plant phylogenetic tree (22,833 sets of orthologues from nuclear genomes of 101 plant species) estimated by Lee et al. (2011). The single locus ‘observed’ MLH3 tree was compared with an expected tree constrained to have the same topology as 15 shared species in the Lee et al. (2011) multilocus tree using the KHT test. The test was not significant (P > 0.05) and it can therefore be concluded that the MLH3 tree is consistent with the species relationships and that the sequences can be considered orthologous.
The expression of the MLH3 gene was assayed using RNA-seq data assembled in the genomic database (International Barley Genome Sequencing Consortium, 2012) that was BLAST searched using the HvMLH3 cDNA sequence. The RNA-seq expression profiles revealed high levels of HvMLH3 transcript in inflorescences, moderate levels in young leaves and immature seeds at 5 days post-anthesis (dpa), and low abundance in embryos, stem internodes, roots, and immature seeds at 15 dpa (Fig. 1B).
Immunolocalization of HvMLH3 and the numbers of foci at pachytene
In order to determine the numbers of HvMLH3 foci during pachytene in barley bivalents, meiotic prophase cells of barley were embedded in polyacrylamide, and HvMLH3 and SC transverse filament protein ZYP1 were detected using immunofluorescence. HvMLH3 first appears during zygotene as discrete foci on synapsed axes marked by the AtZYP1 antibody (Fig. 2A, B). At pachytene (Fig. 2C), individual foci at the ends of bivalents (Fig. 2D) and in relatively close proximity (Fig. 2E) are readily resolved. Foci are lost from the axes during diplotene, with no foci remaining by the end of this stage (Fig. 2F).
Fig. 2.
Immunolocalization of HvMLH3 protein. (A) A zygotene nucleus showing polarized ZYP1 (grey) and HvMLH3 (green); enlargement of the pink box (B) shows an HvMLH3 focus at the end of the bivalent. (C) A pachytene nucleus containing ZYP1 (grey) and HvMLH3 (green); enlargement of the red box (D) shows HvMLH3 foci at the end of the SC, and enlargement of the yellow box (E) shows two HvMLH3 foci in close proximity. (F) A diplotene nucleus showing fragmented and spiralled ZYP1 cores (grey) and no HvMLH3 foci (green). (G) A pachytene nucleus showing ZYP1 (grey), HvMLH3 (green), centromeres (red), and 5S rDNA (green); enlargement of the green box (H) shows an HvMLH3 focus. The pachytene nucleus shown in (G) with its 2H bivalent (I) and 3H bivalent (J) isolated using Imaris. (K) The same two bivalents (I and J) with the position of the HvMLH3 foci highlighted by green arrows. (L) Plot of the number of HvMLH3 foci on 93 2H and 3H bivalents against SC length. (A–J) Images of nuclei embedded in polyacrylamide are deconvolved maximum projections of z-stacks captured using CLSM.
In order to ascertain if different cultivars of barley have different CO frequencies, 140 pachytene bivalents (from 20 nuclei) of six-row Morex and 70 pachytene bivalents (from 10 nuclei) of two-row Bowman were isolated using Imaris software, and the number of HvMLH3 foci in each was recorded (Table 1). A total of 384 foci was counted in Morex, giving a mean of 19.2 foci per nucleus [compared with a chiasma count of 13.65 at metaphase I (SD=0.67; n=20)] and a mean of 2.74 foci per bivalent. A total of 142 foci was counted in Bowman, which equates to a mean of 14.2 foci per nucleus [compared with a chiasma count of 13.72 at metaphase I (SD=0.44; n=20)] and a mean of 2.03 foci per bivalent (Table 1). Interestingly, this result clearly demonstrates that Bowman has significantly (t-test; P=0.0019) fewer HvMLH3 foci than Morex, indicating potential variation in CO frequencies even between cultivars. It is not possible to ascertain to what extent this variation is attributable to differences in numbers of foci between plants of each cultivar, as the meiotic material had to be pooled from several plants in order to prepare acrylamide pads.
Table 1.
The observed numbers and mean frequencies of HvMLH3 foci in bivalents of Morex and Bowman. Long arm (a) covers the region from the telomere to the 5S rDNA locus, and long arm (b) covers the 5S rDNA locus to the centromere. Data relating to the 2H and 3H bivalents are from Morex. The expected numbers of foci were calculated on the basis of the Poisson distribution, and tested against observed numbers using a χ2 goodness of fit test (*P-value ≤0.05).
| Total no. bivalents | Total no foci | Mean no. (SD) foci/nucleus | Mean no. (SD) foci/bivalent | No. of foci/bivalent | Observed | Expected | |
|---|---|---|---|---|---|---|---|
| Morex | 140 | 384 | 19.2 (4.29) | 2.74 (0.16) | 0 | 3 | 9 |
| 1 | 13 | 24.8 | |||||
| 2 | 48 | 33.9 | |||||
| 3 | 42 | 31 | |||||
| 4 | 27 | 21.2 | |||||
| ≥5 | 7 | 20 | |||||
| P-value | <0.001* | ||||||
| 2H | 93 | 201 | – | 2.16 (1.21) | 0 | 5 | 11.5 |
| Short arm | 95 | 1 | 28 | 24.0 | |||
| Long arm (a) | 72 | 2 | 23 | 26.9 | |||
| Long arm (b) | 34 | 3 | 24 | 19.4 | |||
| Long arm sum | 106 | 4 | 10 | 10.5 | |||
| 5 | 4 | 6.8 | |||||
| P-value | 0.1602 | ||||||
| 3H | 93 | 178 | – | 1.91 (1.18) | 0 | 6 | 14.7 |
| Short arm | 71 | 1 | 32 | 28.2 | |||
| Long arm (a) | 67 | 2 | 34 | 27.0 | |||
| Long arm (b) | 40 | 3 | 11 | 17.2 | |||
| Long arm sum | 107 | 4 | 7 | 8.2 | |||
| 5 | 4 | 4.5 | |||||
| P-value | 0.0508 | ||||||
| Bowman | 70 | 142 | 14.2 (2.35) | 2.03 (0.34) | 0 | 5 | 9.19 |
| 1 | 15 | 18.66 | |||||
| 2 | 26 | 18.94 | |||||
| 3 | 21 | 12.82 | |||||
| 4 | 3 | 6.51 | |||||
| 5 | 0 | 3.88 | |||||
| P-value | 0.0062* |
The observed numbers of bivalents with 0 to 5+ foci in both Morex and Bowman were compared using a χ2 goodness-of-fit test to the numbers expected on the basis of a Poisson distribution (Table 1). The difference is highly significant (P < 0.01), indicating that the foci are not randomly distributed between bivalents. The number of bivalents containing no foci is considerably lower than expected, as would be expected if all bivalents must have an obligate CO from the class I pathway as in Arabidopsis. Since Morex and Bowman are fully fertile and have no non-disjunction resulting from partial achiasmy, it is assumed that the antibody has simply not detected its epitope in the bivalents lacking foci.
Morex nuclei at pachytene were embedded in polyacrylamide, and ZYP1 and HvMLH3 were detected immunologically as above, but followed sequentially by FISH with centromeric and 5S rDNA probes (Fig. 2G). The centromeric probe delimits the two arms of the bivalent, and the 5S rDNA probe unambiguously identifies chromosomes 2H and 3H of the complement. The 2H bivalent is consistently longer than 3H and has a larger 5S signal. Despite the FISH step, HvMLH3 foci were still well preserved (Fig. 2H) and the 93 2H and 3H bivalents were readily reconstructed (Fig. 2I–K). Considerable variation in SC length was recorded for both bivalents (Supplementary Fig. S3, Supplementary Table S2 at JXB online), which could reflect the random sampling of bivalents undergoing normal condensation as pachytene progresses. Considerable variation was also noted in the chromosomal locations of the centromere and 5S rDNA locus (Supplementary Table S2). Clearly, the extent of condensation is not uniform along the length of the same bivalents from replicate nuclei. There is no correlation between the length of the bivalent and numbers of HvMLH3 foci (Fig. 2L; Supplementary Fig. S4A), inferring that foci are not lost or gained as pachytene progresses, and that foci faithfully mark the sites of COs.
The 2H bivalent has a mean of 2.16 HvMLH3 foci, compared with a mean of 1.91 for 3H (Table 1), a difference that is not significant (t-test; P=0.1623). The numbers of foci in the short arm of 2H are not significantly different from those in the long arm (t-test; P=0.3443), in contrast to 3H where the numbers of foci are significantly lower in the short arm (t-test; P=0.0009; Table 1). A significantly higher number of foci are present in the chromosome region from the telomere to the 5S rDNA locus, compared with the proximal region between the 5S rDNA locus and the centromere in the 2H (t-test; P=0.0001) and 3H (t-test; P=0.0025) bivalents. This is consistent with the expected preponderance of COs in distal regions. The observed numbers of 2H and 3H bivalents with 0–5 foci were compared using a χ2 goodness-of-fit test to the numbers expected on the basis of a Poisson distribution (Table 1).The differences for both 2H and 3H were not significant, although the P-values were not high (2H=0.1602; 3H=0.0508), indicating either that class I COs are more randomly distributed in these bivalents or, more probably, that insufficient numbers of bivalents were sampled, or that some foci were not detected.
Distribution of HvMLH3 foci in bivalents 2H and 3H
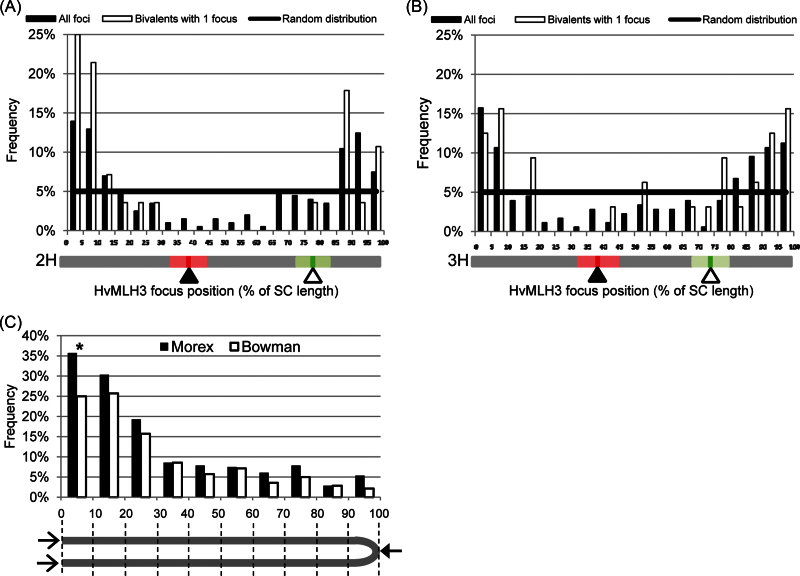
Sequential immunolocalization of ZYP1 and HvMLH3 proteins and FISH with rDNA and centromere probes enabled the construction of high resolution recombination maps based upon a sample of 93 2H and 3H bivalents. Figure 3A and 3B clearly shows a preponderance of HvMLH3 foci in the two distal regions of both bivalents, with a dearth of foci in proximal regions. The regions occupying the distal 20% of each end of 2H (40% of total bivalent length) have 146 foci (73% of total), and the equivalent regions of 3H have 132 foci (74% of total). This contrasts with the central 40% of the 2H and 3H bivalents where only 35 foci (17% and 20% of the respective totals) were recorded. If a subset of 2H and 3H bivalents with one focus only is taken, the skewed distribution of foci is similar but exaggerated (Fig. 3A, B); 25 foci (89%) occupy the distal 20% of 2H, and 24 (75%) occupy the same region of 3H. This suggests that the constraints placed upon the positioning of a single focus in bivalents are no different from those upon multiple foci.
Fig. 3.
Frequencies of HvMLH3 foci along the SC. (A and B) Frequencies of all and single HvMLH3 foci along the length of the 2H (A) and 3H (B) bivalents. The diagram under each graph shows the position of the centromere (filled triangle) and 5S rDNA locus (open triangle), with standard deviations of the means indicated by the boxes. (C) Comparison of focus frequencies along all the bivalents of Morex and Bowman. Because centromeres have not been detected, the frequencies of foci have been calculated on the basis of combined 5% length intervals from both ends of the bivalent (open arrows) to the centre (filled arrow), as indicated in the diagram below the figure. Differences in HvMLH3 frequencies at each interval were tested using Fisher’s exact test (*P-value ≤0.05; **P-values ≤0.01). (This figure is available in colour at JXB online.)
In order to compare the relative distributions of foci in both arms, the numbers of foci in each 10% length interval from the distal end of each arm were plotted together. The distributions of foci in the short arms of 2H and 3H (Supplementary Fig. S4B at JXB online) and long arms of 2H and 3H (Supplementary Fig. S4C) were similar, with significant differences only in two intervals. Supplementary Fig. S4D shows that there is no significant difference in the distribution pattern of foci in the two arms of bivalent 2H, reflecting symmetry in the process governing recombination in both arms. The patterns are comparable in bivalent 3H, but two intervals show a significant difference in the numbers of foci (Supplementary Fig. S4E).
The numbers of MLH3 foci on chromosomes 2H and 3H in relation to the rRNA loci were compared with the genetic map lengths in the standard Steptoe×Morex map on which the rDNA has been mapped as restriction fragment length polymorphisms (RFLPs; Kleinhofs et al., 1993). The recombination events within this segregating population of 150 doubled haploid lines have been the subject of careful study using both genic single nucleotide polymorphisms (SNPs; Close et al., 2009) and other transcript-derived markers (Luo et al., 2007; Potokina et al., 2008). The comparison of the MLH3 foci counts with the recombination observed genetically indicates that the HvMLH3 CO counts are lower (recombination frequency of 1.08 compared with 1.499 Morgans for 2H and 0.96 and 1.535 for 3H). These differences are especially pronounced on the part of the long arms distal to the 5S rDNA loci (being respectively 0.38:0.672 for 2H and 0.36:0.881 for 3H) compared with the proximal regions (0.18:0.206 for 2H and 0.215:0.17 for 3H) (Fig. 4)
Fig. 4.
Genetic versus physical map. Comparison of the physical locations of the centromere and 5S rDNA locus of chromosomes 2H and 3H of Morex with the maps calculated from counts of HvMLH3 foci of Morex and from the genetic map from a Steptoe×Morex mapping family. The numbers in parentheses represent the proportion of the total length of the bivalent.
The distributions of foci along the lengths of all the pooled bivalents of Morex and Bowman are not significantly different, with the exception of the first 10% length interval (Fig. 3C). This implies that the mechanism governing class I CO distribution is similar in the two cultivars, even though the CO frequencies are quite different.
Interference is weak and varies along the bivalent
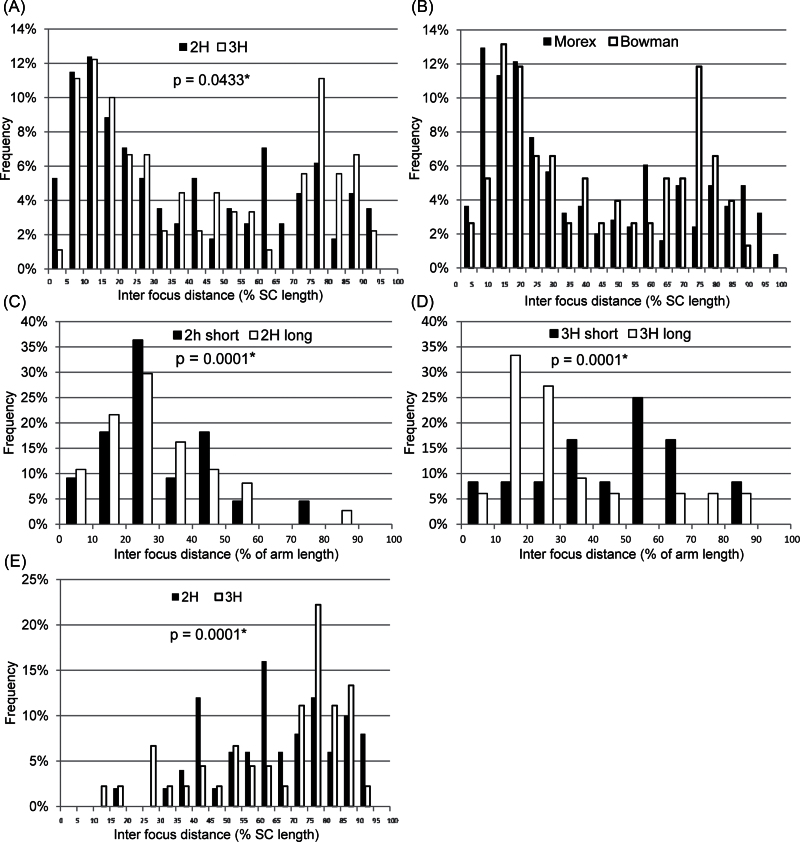
In order to gauge the magnitude of interference along the 2H and 3H bivalents, the distance between each adjacent pair of HvMLH3 foci was calculated as a proportion of total SC length (Fig. 5A). The mean interfocus distance for bivalent 2H is 38.5% (n=113; SD=28.21; range=3.3–93.4%) and for bivalent 3H is 42.6% (n=90; SD=29.6; range=3.1–94.6%). A total of 38% and 34% of foci are <20% apart on bivalents 2H and 3H, respectively, indicating that a relatively high proportion of foci are in close proximity. There is also a preponderance of interfocus distances >70%, reflecting the dearth of foci in proximal and interstitial regions. A similar frequency distribution of interfocus distances was obtained for a pool of all bivalents of the complement (Fig. 5B), suggesting that the forces governing interference act uniformly across the genome. Interference was quantified using the gamma model (unit=nu) of the CODA software developed by Gauthier et al. (2011); a value of nu=1 equates to no interference, nu >1 equates to positive interference, and nu <1 equates to negative interference. The results showed that there is relatively weak positive interference in barley (see Discussion) and similar interference values for bivalents 2H, 3H, and all the bivalents pooled together (Table 2).
Fig. 5.
Comparing interfocus distances. (A and B) Frequencies of interfocus distances for bivalents 2H and 3H (A) and for all bivalents (B). (C and D) Comparison of the frequencies of interfocus distances for the short and long arm of 2H (C) and 3H (D) bivalents. (E) Comparison of the frequencies of interfocus distances spanning the centromere. Frequencies were compared statistically using a χ2 goodness-of-fit test (*P-value ≤0.05).
Table 2.
Interference values for Morex.
| 2H | 3H | All | |
|---|---|---|---|
| Whole chromosome | 1.44 | 1.58 | 1.58 |
| Short arm | 1. 91 | 2.44 | – |
| Long arm | 1.16 | 1.39 | – |
| Across the centromere | 5.86 | 6.42 |
Calculated using the gamma model (unit=nu) using the CODA software (Gauthier et al., 2011).
In order to ascertain if interference is different between the long and short arms of bivalents, foci of the long and short arms of chromosomes 2H and 3H were segregated from the data set, and interfocus distances were calculated (Fig. 5C, D). The mean interfocus distance of the short arm of 2H is 29.5% (n=22; SD=16.7; range; 8.1–75.6%) and that for the long arm is 29.2% (n=37; SD=17.5; range 6.1–87.9%). The frequency distributions of interfocus distances of the two arms are significantly different (P=0.0001), and the interference value for the short arm is greater than that for the long arm (Table 2). The mean interfocus distance of the short arm of 3H is 44.35% (n=12; SD=22.1; range 8.1–82.1%) and that of the long arm is 32.6% (n=33; SD=22.3; range 9.2–80.6%). The frequency distributions of interfocus distances of the two arms are significantly different (P=0.0001), and the interference value for the short arm is again greater than that for the long arm (Table 2).
In order to determine if the centromere exerts a disproportionate effect upon interference, the distances between the two foci flanking the centromere were segregated from the data set (Fig. 5E). The mean interfocus distance for 2H is 65.3% (n=50; SD=17.9; range 20–93.4%) and 65.7% (n=45; SD=21.6; range 11.2–91.4%) for the 3H bivalent, which are considerably higher than the mean interfocus distance for the two bivalents. This is reflected in the relatively high interference values (Table 2), and confirms that COs are far less probable in pericentromeric regions.
Discussion
Isolation and utility of HvMLH3 protein
This study explores the utility of a new cytological approach to probe the CO landscape of barley, without recourse to complex genetic linkage analysis. It was predicated upon the successful isolation and exploitation of HvMLH3 protein which is involved in late recombination events. Barley HvMLH3 protein is encoded by a single gene, as in most other plants species, and is highly expressed in meiotic spikes. However, unlike Arabidopsis where expression is restricted to buds, in barley HvMLH3 is also expressed at lower levels in other tissues (Jackson et al., 2006). The alignment of plant MLH3 protein sequences revealed the three main domains characteristic of the MutL family, including the DQHA(X)2E(X)4E metal-binding site and the MutL-C domain at the C-terminus, that are essential for the catalytic endonuclease activity of the protein (Kosinski et al., 2008; Nishant et al., 2008; Pillon et al., 2010). The antibody raised against this protein clearly and reliably marks mature recombination events, and provides an effective tool for assaying CO frequency and distribution in this crop.
Estimates of CO frequencies are not consistent
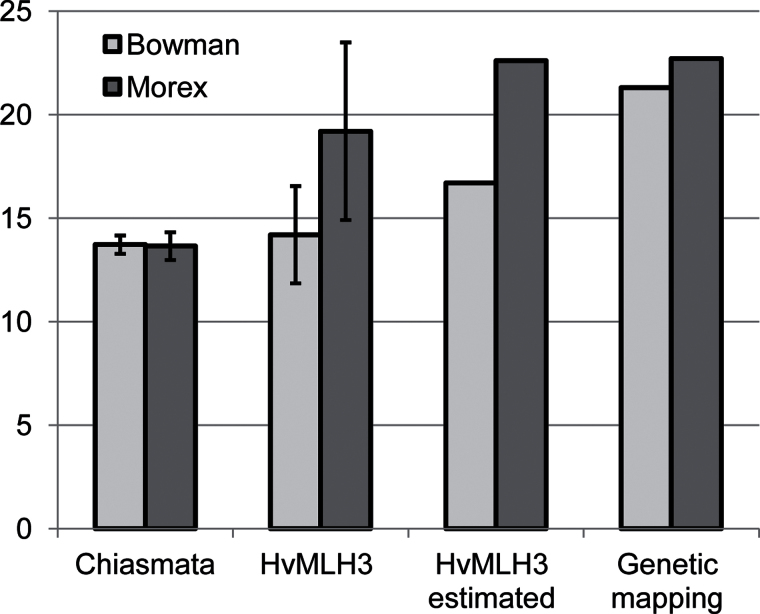
For ease of comparison, the relative frequencies of crossover events in Bowman and Morex are summarized in Fig. 6. The chiasma frequency scored from male meiosis at metaphase I in the two-row cultivar Bowman is 13.72, which is not significantly (t-test; P=0.6983) different from the chiasma frequencies of the six-row cultivar Morex (13.65) and the published values of the two-row cultivar Sultan (14.05; n=20) and six-row breeding line H350-1554 (13.81; n=280) (Bennett et al., 1973). The mean frequency of HvMLH3 foci in Morex (19.2 per nucleus) is considerably higher than the mean chiasma frequency. In contrast, the chiasma frequency of Bowman corresponds closely to its mean frequency of HvMLH3 foci (14.2 per nucleus), indicating that chiasma scores equate with class I CO frequencies in this cultivar (see below). This indicates variation in class I CO frequencies, and may reflect important and potentially useful differences in the mechanism controlling recombination in barley.
Fig. 6.
Relative frequencies of crossover (CO) events in the two barley cultivars. The mean chiasma frequencies were scored from cytological squashes of metaphase I, and the mean frequencies of HvMLH3 foci were obtained from optical sections through pachytene nuclei. CO frequencies were derived from the mean frequencies of HvMLH3 foci by assuming 15% extra class II COs (HvMLH3 estimated), and were derived from genetic linkage analysis (Genetic mapping).
The difference in CO frequency estimated by counting HvMLH3 foci and chiasmata in Morex may be attributed to the difficulty in resolving closely adjacent chiasmata from squashed metaphase I preparations using conventional light microscopy, which would inevitably underestimate the true CO frequencies. Indeed, this study detected over a third of all HvMLH3 foci within 20% of total SC length of each other. Given that the mean SC length of 2H is 105 μm, and that this chromosome is only 10.2 μm long on average at mitotic metaphase (Phillips et al., 2010), the inference is that two CO events could be separated by about only 2 μm at metaphase I, which is too close to resolve as two chiasmata. The same argument can be applied to question the accuracy of the chiasma counts of Bowman. The number of HvMLH3 foci per bivalent does not correlate with the pattern of chiasmata at metaphase I; the former predicts that 34% of bivalents should have more than two chiasmata, whereas only ring bivalents with two chiasmata were observed.
The composite total genetic map length of barley is 1136 cM (Li et al., 2010) which is equivalent to 22.7 COs per nucleus. This is higher than the CO frequency estimated in this study, presumably because the genetic map includes both CO classes, whereas HvMLH3 only detects class I CO events. In mutant S. cerevisiae and Arabidopsis lacking core components of the ZMM protein complex, class I CO formation is inhibited. However, 15% of COs still form, indicating that they must have arisen via an alternative class II pathway (Börner et al., 2004; Higgins et al., 2004). The presence of two CO pathways has yet to be confirmed experimentally in barley. However, if it is assumed that 15% of COs are formed by a class II pathway, the predicted mean number of COs per cell would be 22.6 for Morex. This figure is remarkably close to the mean CO frequency estimated from the genetic map length of barley. The predicted mean CO frequency for Bowman would be 16.7, if the same assumptions are made. This is considerably less than the CO frequency of 21.3 estimated from the genetic map length of a two-row barley (albeit not Bowman) mapping population (1065.5 cM) (Muñoz-Amatriaín et al., 2011). These observations point to the presence of a second CO pathway in Bowman too. Twenty-one percent of bivalents contain only one HvMLH3 focus, whereas only 3.5% of bivalents at metaphase I have one chiasma. This implies that some bivalents contain additional COs that are not detected by the antibody. There are two possible explanations to reconcile the differences between the chiasma scores, the HvMLH3 data, and the genetic mapping data of Bowman. The observations could indicate that Bowman has an atypically low level of recombination compared with other two-rowed cultivars, or that the proportion of class II COs in this cultivar is considerably higher than 15%. The similar patterns of foci in the two cultivars may indicate that the skewed distribution of class I COs may be the norm for meiosis in barley.
The skewed distribution of HvMLH3 foci
Genetic mapping gives a precise estimate of CO frequency in barley, but to date it cannot reliably describe the distribution of CO events along the length of each of the bivalents due to the poor integration of genetic and physical maps and scarcity of polymorphic markers around the centromeres (Künzel et al., 2000). Previous efforts to integrate the genetic markers with physical chromosome maps have identified an uneven distribution of recombination in barley (Pedersen et al., 1995; Künzel et al., 2000). Künzel et al. (2000) used translocation lines to determine the physical location of the RFLP markers used in their study. They proposed that if COs are evenly distributed throughout the genome, their average incidence would be 4.3Mb cM–1, given a haploid size of the barley genome of 5350Mb (Bennett and Smith, 1976), and a genetic length of 1241.2 cM (Künzel et al., 2000). Taking chromosome 2H as an example, they showed that 56% of the short arm and 37% of the long arm had highly suppressed levels of recombination, with pericentromeric regions having a recombination rate of ≤298Mb cM–1. This contrasts with five hotspots they defined as having recombination rates ranging from 0.2 to 0.8Mb cM–1, spanning 24% of the chromosome length and having 81% of the markers (Künzel et al., 2000). The uneven pattern of recombination mirrors the cytological findings of this study—the highest levels of class I recombination occur in distal regions.
The comparison of the counts of MLH3 foci with the genetically determined recombination patterns in the Steptoe×Morex mapping population (Close et al., 2009) indicated that class I COs show a pronounced skewed distribution in keeping with the distal localization of chiasmata. However, the inference is that the distribution of class II COs is even more skewed and not random in terms of the physical or genetic map. Whilst class II COs do not interfere, these observations suggest that they are subject to the same distal localization as their class I counterparts.
The skewed distribution of class I CO events reported in this study may not be exclusive to barley, as the same general pattern of homologous recombination has been noted in wheat (Saintenac et al., 2009), maize (Anderson et al., 2003), ryegrass (King et al., 2007), and, to a lesser extent, in Brachypodium distachyon (Huo et al., 2011). It is likely that the mechanism driving this distinctive pattern of COs is common to other members of the Poaceae. In barley and wheat (Maestra et al., 2002; Corredor and Naranjo, 2007; Phillips et al., 2012), it has been shown that synapsis is initiated in subtelomeric regions when the telomeres are clustered in the bouquet. Phillips et al. (2012) also showed that the AtASY1 protein [orthologous to HOP1 in S. cerevisiae (Kironmai et al., 1998)] is also preferentially loaded to telomeric regions. The asymmetry in the initial loading of these proteins and the subsequent preferential synapsis from the telomeres is reflected in the asymmetry of earlier events of recombination, such as double-strand break (DSB) formation, and may also reflect asymmetry of DNA replication (Higgins et al., 2012). As the decision to repair a DSB via a CO or non-CO pathway is thought to be established prior to SC formation or single end invasion (de Boer et al., 2007), it is feasible, given the skewed loading of the key proteins, that there may be a preference for repair via the CO pathway at the distal regions in barley. Additionally, the non-homologous pairing of centromeres in both wheat and barley at early stages of meiosis may constitute another mechanism to discourage repair via homologous recombination around the centromeres (Maestra et al., 2002 Phillips et al., 2012). There may, of course, be other factors yet to be determined in barley which affect CO distribution. For example, the as1 mutant of tomato has reduced levels of cohesin proteins in the axial elements and lateral elements of the SC, and has longer elements, altered numbers of MLH1 foci, and reduced interfocus distances (Qiao et al., 2012).
Interference of class I COs
Morgan (1919; cited by Whitehouse, 1965) called interference the fifth law of heredity. Interference has become a defining feature of class I COs marked by the MLH1/MLH3 heterodimer. Previous studies have demonstrated that these foci show positive interference, resulting in their spacing along bivalents in a non-random manner (Lhuissier et al., 2007; de Boer et al., 2009).
This study has shown that HvMLH3 foci in barley display weak positive interference. The values for interference (gamma model; measured in nu) were always greater than nu=1, indicating that the foci positively interfere with each other. The interference values for the bivalent arms varied from nu=1.16 to nu=2.44, with the short arms consistently having the lower interference values. The interference values in other organisms are larger than those of barley; Lhuissier et al. (2007) obtained an interference value of nu=7.9 and nu=6.9 for chromosomes 1 and 2, respectively, of tomato, and de Boer et al. (2009) recorded interference values of nu=7.5 and nu=10.1 for chromosomes 1 and 2, respectively, in mouse. Additionally, Giraut et al. (2011) recorded interference values in the range of nu=2 to nu=5 in a genetic mapping study of Arabidopsis, and concluded that there was no difference in interference between the five chromosomes of the complement once interval size was taken into account. The results obtained in this study are not consistent with these other studies, indicating that the factors that define interference in barley must therefore differ from the perceived norm.
Colombo and Jones (1997) stated clearly that interference acts across the centromere and that the action of interference is not changed by the presence of an intervening centromere. Their findings have since been supported by numerous studies (Broman and Weber, 2000; Esch, 2005; Drouaud et al., 2006), and Lhuissier et al. (2007) concluded that in tomato the presence of an MLH1 focus on one arm influenced the position of an MLH1 focus on the other. This observation is consistent with barley, where interference is greatest for pairs of HvMLH3 foci flanking the centromere (Table 2). The dearth of homologous recombination in pericentromeric regions could to a certain extent be the consequence of the superimposition of the mechanism responsible for distal localization, which could give the impression of strong interference across the centromere. In conclusion, there seems to be a two-tier commitment to homologous recombination and establishment of CO interference in barley, where distal regions have a higher propensity to recombine via the class I pathway and display weaker interference, and interstitial and proximal regions are less recombinogenic and have strong interference.
Neither intact SCs nor intact axial elements are required for the establishment of interference (de Boer et al., 2007), indicating that it is established prior to axis formation. The proposed stress model of interference (Kleckner et al., 2004) suggests that the expansion of the chromatin during early leptotene acting against a rigid chromatin-related structure creates mechanical stress. The tension in such regions is relieved by the formation of a single CO, eliminating the requirement for more COs in its vicinity and establishing interference. Differential chromatin compaction has been observed during early leptotene in barley (Phillips et al., 2012), with lower density chromatin in a polarized region of the nucleus containing the telomeres. This region may experience the mechanical stress that promotes CO formation, and the polar loading of ASY1 and ZYP1 proteins (Phillips et al., 2012) and the distal localization of HvMLH3 may merely reflect this early stress release created by the initial wave of DSB formation occurring near the telomere bouquet (Higgins et al., 2012). The skewed distribution of COs could also be reinforced by the condensed nature of the pericentromeric chromatin which may not provide the mechanical stress necessary for the promotion of COs. Some histone modifications (H3K9me3, H3K4me3, and H4K16ac) are distally localized during early meiosis (Higgins et al., 2012), but the significance of these signals is not known in barley.
HvMLH3 distribution in relation to gene density
The production of the first scaffold of the barley genome arranged 21,766 genes in putative linear order along each of the seven chromosomes (Mayer et al., 2011). As previously stated, 3125 genes fell within the genetic centromeres defined as regions around the centromeres that have little or no homologous recombination, making the ordering of the genes or markers in this region difficult if not impossible. Künzel et al. (2000) recognized that the genetic centromeres occupy large proportions of the chromosome arms, which led Mayer et al. (2001) to conclude that the genes are distributed throughout the physical length of the chromosome. The physical sequencing of the wheat 3B chromosome also revealed the scattering of genes along the entire length of the chromosome, with subtelomeric regions having twice the density of genes (Choulet et al., 2010). The precise mapping of all DSB events in S. cerevisiae found that the Spo11 protein was an opportunistic cutter and that hotspots represent windows of opportunity (Pan et al., 2011). A key determinant of these windows is chromatin structure with regions of low nucleosome occupancy, such as promoters, allowing the Spo11 protein and its accessories free access to the DNA strand. The distribution of genes and therefore promoters in barley may indicate that Spo11 may have the opportunity to form DSBs throughout the genome. Indeed, HvMLH3 foci were found, to differing degrees, along the entire length of the bivalents, giving credence to this possibility. The lack of a DSB map for any plant species precludes accurate definition of these sites. Studies of S. cerevisiae revealed that not all DSB hotspots are CO hotspots and that additional levels of control operate that ultimately decide whether DSBs are processed down a CO or a non-CO pathway.
Conclusions
The physical mapping of HvMLH3 foci has provided unprecedented access to the recombination landscape of barley. It has allowed the frequencies and distributions of class I COs and interference to be quantified for the first time, and inferences to be drawn from these in the context of published chiasma counts and genetic mapping data. There is compelling evidence that there are at least two CO pathways in barley. The mapping of HvMLH3 foci will provide a rapid and robust method to assay the effectiveness of future interventions to manipulate recombination in this crop.
Supplementary data
Supplementary data are available at JXB online
Figure S1. Immunolocalization of HvMLH3 pre-immune serum.
Figure S2. Alignment of plant MLH3 protein sequences. Only the Pfam domains HATPase_c_3, DNA_mis_repair, and the MutL_C domain are shown.
Figure S3. Graphs showing the ranked SC lengths in ascending order of the 93 2H and 93 3H bivalents.
Figure S4. Plot of the number of HvMLH3 foci against SC length in Morex and Bowman; the comparison of the frequencies of foci along the short arms and long arms of bivalents 2H and 3H.
Table S1. Primers used in the study.
Table S2. Measurements relating to the 2H and 3H bivalents.
Acknowledgements
We thank Chris Franklin and Sue Armstrong for the supply of AtZYP1 antibodies, and Linda Milne for conducting the RNA-seq assembly. We acknowledge with gratitude the award of an IBERS PhD studentship to JW. This work was supported by the Biotechnology and Biological Sciences Research Council (grant ref. BB/F018754/1).
References
- Akhunov ED, Goodyear AW, Geng S, et al. 2003. The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms. Genome Research 13, 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LK, Doyle GG, Brigham B, Carter J, Hooker KD, Lai A, Rice M, Stack SM. 2003. High-resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics 165, 849–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LK, Reeves A, Webb LM, Ashley T. 1999. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151, 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LK, Stack SM. 2005. Recombination nodules in plants. Cytogenetics and Genome Research 109, 198–204. [DOI] [PubMed] [Google Scholar]
- Baker SM, Plug AW, Prolla TA, et al. 1996. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nature Genetics 13, 336–342. [DOI] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Finch RA, Smith JB, Rao MK. 1973. Time and duration of female meiosis in wheat, rye and barley. Proceedings of the Royal Society B: Biological Sciences 183, 301–319. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Smith JB. 1976. Nuclear-DNA amounts in Angiosperms. Philosophical Transactions of the Royal Society B: Biological Sciences 274, 227–274. [DOI] [PubMed] [Google Scholar]
- Berchowitz LE, Copenhaver GP. 2009. Visual markers for detecting gene conversion directly in the gametes of Arabidopsis thaliana . Methods in Molecular Biology 557, 99–114. [DOI] [PubMed] [Google Scholar]
- Börner GV, Kleckner N, Hunter N. 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117, 29–45. [DOI] [PubMed] [Google Scholar]
- Broman KW, Weber JL. 2000. Characterization of human crossover interference. American Journal of Human Genetics 66, 1911–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L, Grandont L, Vrielynck N, le Guin S, Mercier R, Grelon M. 2010. An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenetics and Genome Research 129, 143–153. [DOI] [PubMed] [Google Scholar]
- Choulet F, Wicker T, Rustenholz C, et al. 2010. Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. The Plant Cell 22, 1686–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close TJ, Bhat PR, Lonardi S, et al. 2009. Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 10, 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo PC, Jones GH. 1997. Chiasma interference is blind to centromeres. Heredity 79, 214–227. [DOI] [PubMed] [Google Scholar]
- Corredor E, Naranjo T. 2007. Effect of colchicine and telocentric chromosome conformation on centromere and telomere dynamics at meiotic prophase I in wheat–rye additions. Chromosome Research 15, 231–245. [DOI] [PubMed] [Google Scholar]
- de Boer E, Dietrich AJ, Hoog C, Stam P, Heyting C. 2007. Meiotic interference among MLH1 foci requires neither an intact axial element structure nor full synapsis. Journal of Cell Science 120, 731–736. [DOI] [PubMed] [Google Scholar]
- de Boer E, Lhuissier FG, Heyting C. 2009. Cytological analysis of interference in mouse meiosis. Methods in Molecular Biolology 558, 355–382. [DOI] [PubMed] [Google Scholar]
- de Massy B. 2003. Distribution of meiotic recombination sites. Trends in Genetics 19, 514–522. [DOI] [PubMed] [Google Scholar]
- Drouaud J, Camilleri C, Bourguignon PY, et al. 2006. Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination ‘hot spots’. Genome Research 16, 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erayman M, Sandhu D, Sidhu D, Dilbirligi M, Baenziger PS, Gill KS. 2004. Demarcating the gene-rich regions of the wheat genome. Nucleic Acids Research 32, 3546–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch E. 2005. Estimation of gametic frequencies from F2 populations using the EM algorithm and its application in the analysis of crossover interference in rice. Theoretical and Applied Genetics 111, 100–109. [DOI] [PubMed] [Google Scholar]
- Falque M, Anderson LK, Stack SM, Gauthier F, Martin OC. 2009. Two types of meiotic crossovers coexist in maize. The Plant Cell 21, 3915–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier F, Martin OC, Falque M. 2011. CODA (crossover distribution analyzer): quantitative characterization of crossover position patterns along chromosomes. BMC Bioinformatics 12, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach WL, Dyer TA. 1980. Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Research 11, 4851–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mezard C. 2011. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genetics 7, e1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systems Biology 52, 696–704. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. 1931. The cytological basis of genetical interference. Cytologia 3, 54–65. [Google Scholar]
- Higgins JD, Armstrong SJ, Franklin FCH, Jones GH. 2004. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis . Genes and Development 18, 2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Perry RM, Barakate A, Ramsay L, Waugh R, Halpin C, Armstrong SJ, Franklin- FCH. 2012. Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. The Plant Cell 24, 4096–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FCH. 2005. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes and Development 19, 2488–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo NX, Garvin DF, You FM, McMahon S, Luo MC, Gu YQ, Lazo GR, Vogel JP. 2011. Comparison of a high-density genetic linkage map to genome features in the model grass Brachypodium distachyon . Theoretical and Applied Genetics 123, 455–464. [DOI] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. 2007. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8, 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Barley Genome Sequencing Consortium. 2012. A physical, genetic and functional sequence assembly of the barley genome. Nature 491, 711–716. [DOI] [PubMed] [Google Scholar]
- Jackson N, Sanchez-Moran E, Buckling E, Armstrong SJ, Jones GH, Franklin FCH. 2006. Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis . EMBO Journal 25, 1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen-Seaman MI, Furey TS, Payseur BA, Lu Y, Roskin KM, Chen CF, Thomas MA, Haussler D, Jacob HJ. 2004. Comparative recombination rates in the rat, mouse, and human genomes. Genome Resarch 14, 528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones GH. 1984. The control of chiasma distribution. Symposium of the Society of Experimental Biology 38, 293–320. [PubMed] [Google Scholar]
- Kakeda K, Jordan ND, Conner A, Ride JP, Franklin-Tong VE, Franklin FCH. 1998. Identification of residues in a hydrophilic loop of the Papaver rhoeas S protein that play a crucial role in recognition of incompatible pollen. The Plant Cell 10, 1723–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J, Armstead IP, Donnison SI, Roberts LA, Harper JA, Skot K, Elborough K, King IP. 2007. Comparative analyses between Lolium/Festuca introgression lines and rice reveal the major fraction of functionally annotated gene models is located in recombination-poor/very recombination-poor regions of the genome. Genetics 177, 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kironmai KM, Muniyappa K, Friedman DB, Hollingsworth NM, Byers B. 1998. DNA-binding activities of Hop1 protein, a synaptonemal complex component from Saccharomyces cerevisiae . Molecular Cell Biology 18, 1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino H, Hasegawa M. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. Journal of Molecular Evolution 29, 170–179. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Storlazzi A, Zickler D. 2003. Coordinate variation in meiotic pachytene SC length and total crossover/chiasma frequency under conditions of constant DNA length. Trends in Genetics 19, 623–628. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, Henle J, Hutchinson J. 2004. A mechanical basis for chromosome function. Proceedings of the National Academy of Sciences, USA 101, 12592–12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinhofs A, Kilian A, Maroof MAS, et al. 1993. A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theoretical and Applied Genetics 86, 705–712. [DOI] [PubMed] [Google Scholar]
- Kosinski J, Plotz G, Guarne A, Bujnicki JM, Friedhoff P. 2008. The PMS2 subunit of human MutLalpha contains a metal ion binding domain of the iron-dependent repressor protein family. Journal of Molecular Biology 382, 610–627. [DOI] [PubMed] [Google Scholar]
- Künzel G, Korzun L, Meister A. 2000. Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 154, 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EK, Cibrian-Jaramillo A, Kolokotronis SO, et al. 2011. A functional phylogenomic view of the seed plants. PLoS Genetics 7, e1002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhuissier FG, Offenberg HH, Wittich PE, Vischer NO, Heyting C. 2007. The mismatch repair protein MLH1 marks a subset of strongly interfering crossovers in tomato. The Plant Cell 19, 862–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HB, Kilian A, Zhou MX, Wenzl P, Huttner E, Mendham N, McIntyre L, Vaillancourt RE. 2010. Construction of a high-density composite map and comparative mapping of segregation distortion regions in barley. Molecular Genetics and Genomics 284, 319–331. [DOI] [PubMed] [Google Scholar]
- Lukaszewski AJ, Curtis CA. 1993. Physical distribution of recombination in B-genome chromosomes of tetraploid wheat. Theoretical and Applied Genetics 86, 121–127. [DOI] [PubMed] [Google Scholar]
- Luo ZW, Potokina E, Druka A, Wise R, Waugh R, Kearsey MJ. 2007. SFP genotyping from affymetrix arrays is robust but largely detects cis-acting expression regulators. Genetics 176, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestra B, de Jong JH, Shepherd K, Naranjo T. 2002. Chromosome arrangement and behaviour of two rye homologous telosomes at the onset of meiosis in disomic wheat-5RL addition lines with and without the Ph1 locus. Chromosome Research 10, 655–667. [DOI] [PubMed] [Google Scholar]
- Mayer KF, Martis M, Hedley PE, et al. 2011. Unlocking the barley genome by chromosomal and comparative genomics. The Plant Cell 23, 1249–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 25, 126–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Kolas NK, Tarsounas M, Marcon E, Cohen PE, Spyropoulos B. 2002. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA–DNA interactions without reciprocal recombination. Journal of Cell Science 115, 1611–1622. [DOI] [PubMed] [Google Scholar]
- Muller HJ. 1916. The mechanism of crossing-over. American Naturalist 50, 193–221. [Google Scholar]
- Muñoz-Amatriaín M, Moscou M, J., Bhat P, et al. 2011. An improved consensus linkage map of barley based on flow-sorted chromosomes and single nucleotide polymorphism markers. Plant Genome 4, 238–249. [Google Scholar]
- Nasuda S, Hudakova S, Schubert I, Houben A, Endo TR. 2005. Stable barley chromosomes without centromeric repeats. Proceedings of the National Academy of Sciences, USA 102, 9842–9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishant KT, Plys AJ, Alani E. 2008. A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae . Genetics 179, 747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeso D, Dawson DS. 2010. Temporal characterization of homology-independent centromere coupling in meiotic prophase. PLoS One 5, e10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Sasaki M, Kniewel R, et al. 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144, 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen C, Giese H, LindeLaursen I. 1995. Towards an integration of the physical and the genetic chromosome maps of barley by in situ hybridization. Hereditas 123, 77–88. [Google Scholar]
- Phillips D, Nibau C, Ramsay L, Waugh R, Jenkins G. 2010. Development of a molecular cytogenetic recombination assay for barley. Cytogenetics and Genome Research 129, 154–161. [DOI] [PubMed] [Google Scholar]
- Phillips D, Nibau C, Wnetrzak J, Jenkins G. 2012. High resolution analysis of meiotic chromosome structure and behaviour in barley (Hordeum vulgare L.). PLoS One 7, e39539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon MC, Lorenowicz JJ, Uckelmann M, et al. 2010. Structure of the endonuclease domain of MutL: unlicensed to cut. Molecular Cell 39, 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potokina E, Druka A, Luo Z, Wise R, Waugh R, Kearsey M. 2008. Gene expression quantitative trait locus analysis of 16 000 barley genes reveals a complex pattern of genome-wide transcriptional regulation. The Plant Journal 53, 90–101. [DOI] [PubMed] [Google Scholar]
- Qiao H, Offenberg HH, Anderson LK. 2012. Altered distribution of MLH1 foci is associated with changes in cohesins and chromosome axis compaction in an asynaptic mutant of tomato. Chromosoma 121, 291–305. [DOI] [PubMed] [Google Scholar]
- Saintenac C, Falque M, Martin OC, Paux E, Feuillet C, Sourdille P. 2009. Detailed recombination studies along chromosome 3B provide new insights on crossover distribution in wheat (Triticum aestivum L.). Genetics 181, 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman JD, Herickhoff LA, Stack SM. 1992. Silver staining two types of meiotic nodules. Genome Research 35, 907–915. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Oliver-Bonet M, Liehr T, Starke H, Turek P, Ko E, Rademaker A, Martin RH. 2006. Variation in MLH1 distribution in recombination maps for individual chromosomes from human males. Human Molecular Genetics 15, 2376–2391. [DOI] [PubMed] [Google Scholar]
- Sym M, Engebrecht JA, Roeder GS. 1993. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72, 365–378. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systems Biology 56, 564–577. [DOI] [PubMed] [Google Scholar]
- Van Bel M, Proost S, Wischnitzki E, Movahedi S, Scheerlinck C, Van de Peer Y, Vandepoele K. 2012. Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiology 158, 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TF, Kleckner N, Hunter N. 1999. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proceedings of the National Academy of Sciences, USA 96, 13914–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse HLK. 1965. Towards an understanding of the mechanism of heredity. London: Edward Arnold. [Google Scholar]
- Wijeratne AJ, Ma H. 2007. Genetic analyses of meiotic recombination in Arabidopsis. Journal of Integral Plant Biology 49, 1199–1207. [Google Scholar]
- Youds JL, Boulton SJ. 2011. The choice in meiosis—defining the factors that influence crossover or non-crossover formation. Journal of Cell Science 124, 501–513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.