Abstract
Plants have duplicate versions of the oxidative pentose phosphate pathway (oxPPP) enzymes with a subset localized to the chloroplast. The chloroplast oxPPP provides NADPH and pentose sugars for multiple metabolic pathways. This study identified two loss-of-function alleles of the Zea mays (maize) chloroplast-localized oxPPP enzyme 6-phosphogluconate dehydrogenase (6PGDH). These mutations caused a rough endosperm seed phenotype with reduced embryo oil and endosperm starch. Genetic translocation experiments showed that pgd3 has separate, essential roles in both endosperm and embryo development. Endosperm metabolite profiling experiments indicated that pgd3 shifts redox-related metabolites and increases reducing sugars similar to starch-biosynthetis mutants. Heavy isotope-labelling experiments indicates that carbon flux into starch is altered in pgd3 mutants. Labelling experiments with a loss of cytosolic 6PGDH did not affect flux into starch. These results support the known role for plastid-localized oxPPP in oil synthesis and argue that amyloplast-localized oxPPP reactions are integral to endosperm starch accumulation in maize kernels.
Key words: Defective kernel, endosperm, maize, pentose phosphate pathway, PGD3, starch.
Introduction
The pentose phosphate pathway (PPP) is central to plant metabolism (Kruger and von Schaewen, 2003). The oxidative section of the PPP (oxPPP) converts glucose-6-P into ribulose-5-P. Glucose-6-P dehydrogenase (G6PDH, EC 1.1.1.49) and 6-phosphogluconate dehydrogenase (6PGDH, EC 1.1.1.44), catalyse the irreversible oxidation of glucose-6-P, generating two molecules of NADPH and one molecule of CO2. The product of the first oxidative reaction, 6-phosphoglucono-δ-lactone, can hydrolyse to 6-phosphogluconate spontaneously or by the catalysis of 6-phosphogluconolactonase (6PGL) (Miclet et al., 2001). The non-oxidative part of the PPP (noxPPP) regenerates glucose-6-P from ribulose-5-P through reversible reactions catalysed by ribulose-5-P epimerase, ribose-5-P isomerase, transaldolase, and transketolase. The noxPPP delivers carbon intermediates for the biosynthesis of amino acids, nucleic acids, lignin, and polyphenols.
The oxPPP is an important source of reducing power in multiple cellular compartments. Non-photosynthetic cells generate NADPH and NADH through the oxPPP, glycolysis, malic enzyme, and the tricarboxylic acid cycle whereas photosynthetic cells generate reducing equivalents primarily from the light reactions. Although multiple transporters move metabolic intermediates between membrane-bound subcellular compartments (Linka and Weber, 2010), direct transport mechanisms for NADH and NADPH are not known. Therefore, subcellular compartments produce reductants independently and plant oxPPP operates at least in the plastid and cytosol based on subcellular localization of enzymes (Kruger and von Schaewen, 2003). In pea and Arabidopsis, G6PDH, 6PGL, and 6PGDH were localized to peroxisomes (Corpas et al., 1998; Reumann et al., 2007; Xiong et al., 2009; Meyer et al., 2011), but not all of these enzymes have peroxisomal targeting signals. By contrast, noxPPP enzymes are always found in the plastid, while localization to other cellular compartments is dependent on plant species and tissue (Debnam and Emes, 1999; Kopriva et al., 2000).
G6PDH is considered the rate-limiting and flux-controlling enzyme of the oxPPP due to feedback inhibition by NADPH in spinach chloroplasts (Lendzian and Bassham, 1975). G6PDH is associated with nitrogen assimilation, oxidative stress, and defence responses (Bowsher et al., 1992; Debnam et al., 2004; Scharte et al., 2009). Tobacco antisense lines of a plastidic G6PDH isoform and Arabidopsis G6PDH mutants disrupting one of two plastidic or two cytosolic isozymes show no obvious phenotype (Debnam et al., 2004; Wakao et al., 2008). By contrast, loss of chloroplast-localized 6PGL in Arabidopsis are seed lethal (Xiong et al., 2009).
The second dehydrogenase, 6PGDH, is not thought to have regulatory properties similar to G6PDH and is not considered a rate-limiting step. All higher plants so far examined have distinct cytosolic and plastidic 6PGDH isoforms (Tanksley and Kuehn, 1985; Bailey-Serres and Nguyen, 1992; Bailey-Serres et al., 1992; Signorini et al., 1995; Krepinsky et al., 2001). The maize genome encodes three copies of 6PGDH. Double mutants of the maize cytosolic isozymes, PGD1 and PGD2, have no obvious phenotype (Bailey-Serres et al., 1992). The third 6PGDH isozyme, PGD3, is predicted to be plastid localized, based on the available data in the literature and protein targeting prediction algorithms (Averill et al., 1998; Settles et al., 2007). However, multiple sources of inference are needed to hypothesize that PGD3 is plastid localized, with no one experiment being conclusive. The current study group previously identified a transposon insertion mutant disrupting enzyme activity of PGD3 (Settles et al., 2007). The pgd3-umu1 knock-out allele co-segregates with a rough endosperm (rgh) kernel phenotype generating a plausible hypothesis that PGD3 has a role in seed development. In addition, a small fraction of the rgh kernels, which was pgd3 loss-of-function due to genetic linkage, could be rescued by tissue culture allowing the conclusion that loss of PGD3 is not completely lethal to maize plants.
This paper proves that loss of PGD3 causes severely reduced grain-fill phenotypes. A combination of isozyme activity, protein subcellular localization, and translocation genetic data indicate that plastidic 6PGDH activity is associated with sink tissues in maize and has distinct functions in the endosperm versus the embryo. Histology, metabolite profiling, and stable isotope-labelling experiments suggest an active involvement of plastidic 6PGDH in starch biosynthesis. Combined, these results suggest the oxPPP has a more important role for starch synthesis in the cereal endosperm than recent models indicate (Hannah and James, 2008; Comparot-Moss and Denyer, 2009).
Materials and methods
Chemicals and plant materials
Chemicals were from Sigma-Aldrich (St Louis, MO, USA) or VWR (Radnor, PA, USA). [U-13C6]glucose was from Isotec (Miamisburg, OH, USA). UniformMu (McCarty et al., 2005) alleles of pgd3-umu1 and pgd3-umu2 were maintained as heterozygotes. Homozygous pgd1;pgd2 double mutants were from Julia Bailey-Serres (University of California, Riverside). The TB-4Sa, B-A translocation stock was maintained as a transposon-marked hyperploid (Birchler and Alfenito, 1993) and crossed onto pgd3/+, bt2/bt2, and dek11/+ plants. Mature non-concordant kernels were selected visually. Maize plants were either grown in field conditions in Citra, FL or in greenhouse conditions in Gainesville, FL.
Screen for pgd3-umu2
Genomic DNA of 72 rgh isolates was pooled into a 6×12 grid. The pgd3-umu2 insertion was amplified and sequenced using Mutator (Mu) and Pgd3 specific primers as described (Settles et al., 2007). Genetic complementation was tested by crossing genotyped heterozygous plants.
Morphology
Mature kernels and sagittal hand sections, cut with a utility knife, were imaged on a flatbed scanner. Normal and pgd3 kernels from segregating ears were harvested for microscopic analysis at 16 days after pollination (DAP). Toluidine blue-stained 500nm sections were prepared as described (Kang et al., 2009).
Biochemical analyses
6PGDH activity was assayed on a total protein basis by native PAGE as described (Settles et al., 2007). Tissues were harvested from seedling roots and shoots 7 days after planting, tassels and immature ears just before flowering, and adult leaves after flowering. Protein was extracted from 0.15g tissue using a 1:1 (ml:g) ratio of extraction buffer to tissue freshweight for kernels and a 2:1 (ml:g) ratio for all other tissues. Plastids were purified and subfractionated as described (Cline et al., 1993; Mori et al., 1999).
For metabolomics, three ears per genotype were harvested at 20 DAP, and metabolite analysis was performed on 3–7 replicates per ear. Kernels were frozen in liquid nitrogen and dissected, and lyophilized tissues were ground using a MiniBeadBeater-96 (BioSpec Products, OK). Analysis of metabolites using GC-MS was completed on 100mg endosperm as described (Witt et al., 2012). Ribitol was the quantitative internal standard. Normalized relative ratios of each metabolite were calculated based on sample dryweight and percentage of solubles (normalized relative ratio = response ratio × 100/sample dryweight/% solubles). Remaining endosperm tissue after extraction was analysed for starch and protein as described (Spielbauer et al. 2009). Principal component analysis was performed with XLSTAT.
NADP+ and NADPH were determined with a modified cycling assay (Kolbe et al., 2005). For this, 400mg 21-DAP kernels were ground in a mortar for 2min with 1.5ml boiling 0.1M NaOH or HCl for NADPH or NADP+, respectively. Extracts were transferred to ice, neutralized with 1.5ml 0.1M HCl or NaOH, and adjusted to 10ml with 0.1M Tricine-OH (pH 8.0). A 1ml aliquot was centrifuged at 15,000 g for 10min at 4 °C. The supernatant was diluted 1:5 in 0.1M Tricine, and 150 µl was mixed with 900 µl 0.1M Tricine and 200 µl of assay mix (10mM EDTA pH 8.0, 4.2mM phenazine ethosulfate, 1.05mM MTT, 12.5mM glucose-6-P). For the cycling reaction, 10 µl of G6PDH (Sigma G5885, 35U ml–1 and 0.1M Tricine) was added, incubated for 30min at 37 °C, and absorbance was measured at 570nm.
13C labelling experiments
W22, pgd3-umu1/+, and homozygous pgd1;pgd2 were grown in field conditions. Homozygous pgd3-umu1 plants were recovered via embryo culture as described (Settles et al., 2007). Ears were self-pollinated, harvested at 8 DAP and ear block tissue cultures were initiated in a modified MS media as described (Ettenhuber et al., 2005), except that 2,4-dichlorphenoxyacetic acid was omitted from all media. Kernel blocks were transferred onto fresh culture media containing a 1:30 mixture of [U-13C6]glucose and naturally labelled glucose from 11–18 DAP. Three biological replicates were performed for each genotype with one replicate deriving from one plant for W22 and homozygous pgd3 or pgd1;pgd2. The 25% frequency of mutant kernels on pgd3-umu1/+ blocks required 3–5 plant pools per replicate. Starch was purified from endosperm tissue, and the isotopologue distribution from starch-derived glucose moieties was determined by isotope ratio mass spectrometry and 1H- and 13C-NMR spectroscopy as described (Spielbauer et al., 2006). 1H and 13C NMR spectra were recorded at 500.13 MHz and 125.76 MHz, respectively, using a Bruker DRX500 spectrometer at 25 °C. The experimental setup, signal assignments, coupling constants, isotope shifts, and the notation of isotopologues of the biolabelled α- and β-glucose were as described (Eisenreich et al., 2004). The analysis of 13C abundance and isotopologue composition was as described (Spielbauer et al., 2006). Percentage 13C-fractions (f i) were calculated from the excess enrichment (e i) of each isotopologue, the total excess amount (t) of 13C incorporated into starch during the labelling period, and the number (n) of 13C atoms of a certain isotopologue by applying f i = (n × e i)/(t × 6).
Results
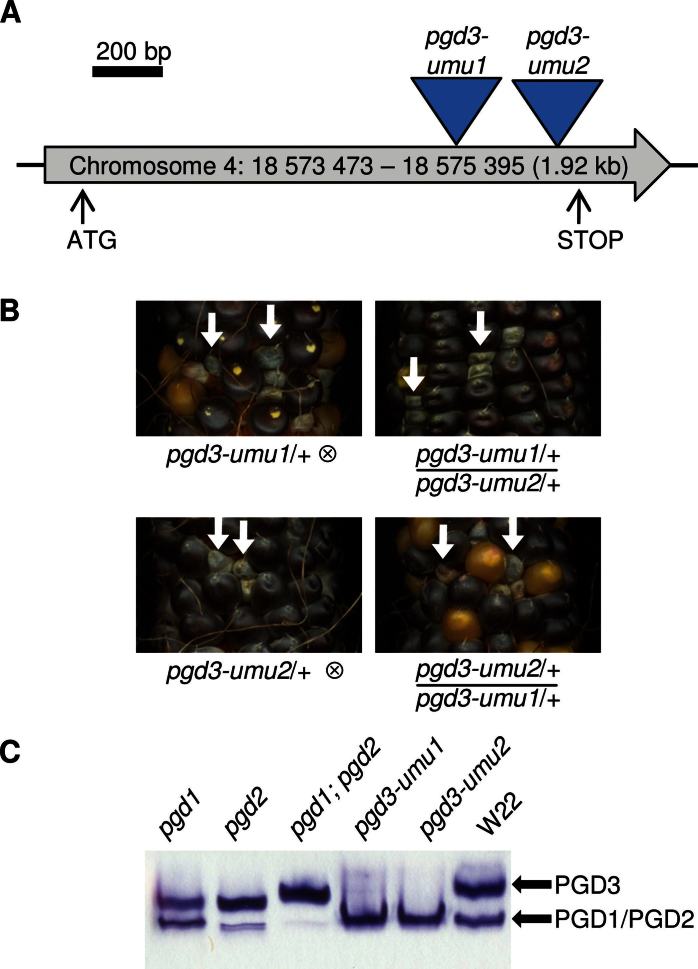
The pgd3-umu1 allele was identified within UniformMu Mu transposon flanking sequence tags and perfectly co-segregates with an rgh phenotype at a map resolution <0.31 cM (Settles et al., 2007). A PCR screen of 72 independent rgh isolates also from UniformMu found an additional pgd3 allele with an insertion in the 3’ end of the Pgd3 ORF (Fig. 1A). This pgd3-umu2 allele co-segregated with an rgh phenotype at <1 cM map resolution (0 recombinants/100 meiotic products) and failed to complement pgd3-umu1 in reciprocal crosses between heterozygotes (Fig. 1B). To determine the effect of pgd3-umu2 on enzyme activity, 16-DAP seeds were assayed for 6PGDH activity (Fig. 1C). The native PAGE assay detects two mobility classes of 6PGDH (Settles et al., 2007). Single mutants of pgd1 or pgd2 reduced the activity of the fast migrating band and pgd1;pgd2 double mutants eliminated this band. Both pgd3 alleles eliminated the slower migrating, PGD3 band. The enzyme, seed, and molecular lesions of pgd3-umu1 and pgd3-umu2 indicated that the alleles were equivalent null mutations. Thus, two independent alleles of pgd3 conferred the same seed phenotype.
Fig. 1.
Cloning of Pgd3. (A) Schematic of the Pgd3 locus; two Mu insertions disrupt the predicted ORF of 6PGDH. (B) Complementation test of pgd3-umu1/+ with pgd3-umu2/+; self-pollinated ears and reciprocal crosses segregate for the pgd3 mutation (arrows). (C) 6PGDH native PAGE activity assays for 16-DAP kernels mutated for 6PGDH isoforms.
Similar to pgd3-umu1 (Settles et al., 2007), pgd3-umu2 mutants had severely reduced grain-fill, and sagittal hand-sections of mature kernels showed developmental defects in endosperm and embryo (Supplementary Fig. S1, available at JXB online). The bulk seed composition of mature kernels from segregating ears revealed that pgd3 alleles reduced fat and starch content while increasing fibre (Supplementary Table S1). Dissected pgd3 seeds showed reduced dryweight of the embryo, endosperm, and maternal pericarp by 30-, 8-, and 2-fold, respectively (Supplementary Table S2). The reduction in fat and embryo weight is consistent with the role of oxPPP in lipid biosynthesis (Schwender et al., 2003; Hutchings et al., 2005; Alonso et al., 2010). The pericarp is predominantly cell-wall material, and the apparent increase of fibre in pgd3 mutants is likely to derive from the increased proportion of maternal tissue. The reduction of starch content was still significant when measured directly from endosperm at 20 DAP and maturity, suggesting a starch synthesis defect in pgd3 (Supplementary Table S3).
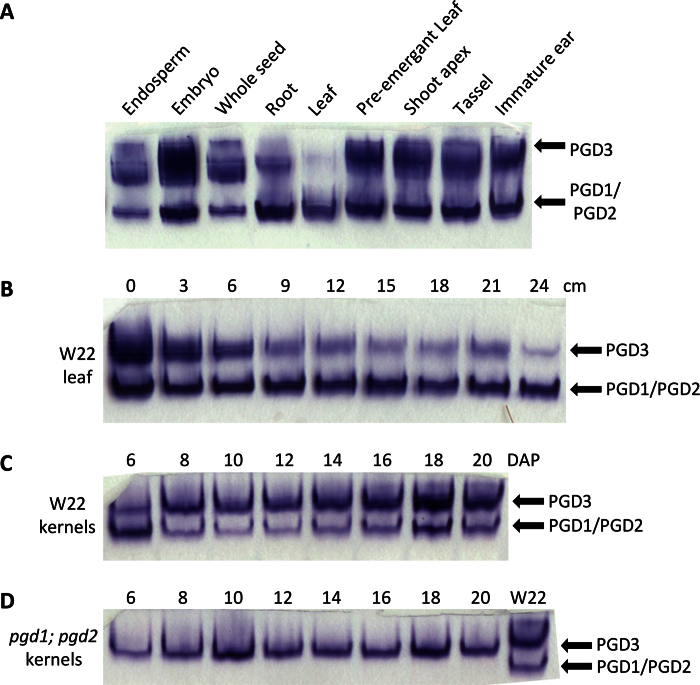
A small fraction of pgd3-umu1 embryos can be rescued with tissue culture (Settles et al., 2007). Although morphologically normal, pgd3 plants were shorter in stature and grew slower with recently emerged leaves being pale green. The seed and plant phenotypes of pgd3 mutants suggested that PGD3 activity is most critical in the developing seed. Potentially, PGD3 activity is only required in a limited number of tissues or plant developmental stages. This study surveyed 6PGDH enzyme activity with wild-type plants to determine tissues in which PGD3 was active (Fig. 2A). PGD1 and PGD2 activity was detected at similar levels in all tissues sampled. By contrast, PGD3 activity showed greater tissue specificity. PGD3 activity was low in fully expanded photosynthetic leaves and was relatively low in roots when compared to PGD1 and PGD2. PGD3 showed a gradient of activity in partially emerged leaves, with the proximal region of the leaf having the greatest activity and the photosynthetic tip having the lowest (Fig. 2B). High levels of PGD3 activity were observed in other sink tissues, such as pre-emergent leaf, shoot apex, tassel, and immature ear, with the most intense activity in 16-DAP embryos (Fig. 2A). PGD3 activity may also show a gradual increase during seed development (Fig. 2C), and the loss of PGD1 and PGD2 did not alter PGD3 activity in developing seeds, indicating independent regulation of 6PGDH isoforms (Fig. 2D).
Fig. 2.
6PGDH native PAGE enzyme activity assays. (A) Wild-type plant tissues from the inbred W22; seed tissues were extracted at 16 DAP and 40 µg total protein was loaded per lane. (B) PGD3 activity is reduced in photosynthetic cells; an emerging leaf was harvested and divided in 3cm increments from the shoot apex (0cm) to the photosynthetic tip (24cm). (C, D) Seed developmental time course from W22 (C) and pgd1;pgd2 double mutant (D) kernels; the W22 lane was extracted at 20 DAP. In B–D, 30 µg total protein was loaded per lane (this figure is available in colour at JXB online).
6PGDH has been co-purified with peroxisomes in pea and Arabidopsis (Corpas et al., 1998; Reumann et al., 2007; Reumann et al., 2009). In maize, the majority of PGD3 enzyme activity is found in the soluble fraction of total protein extracts with approximately one-third of the activity associated with crude membrane fractions (Averill et al., 1998). The current study found PGD1, PGD2, and PGD3 to all be associated with purified chloroplasts and etioplasts, although PGD3 was enriched relative to PGD1 and PGD2 in the plastid stroma fraction (Supplementary Fig. S2C). Protein sequence alignment of higher plant 6PGDH enzymes showed two major clades (Supplementary Fig. S2A). Based on purified spinach enzymes, one clade was predicted to be cytosolic and many enzymes of this clade contain PTS1-type peroxisome targeting signals (Neuberger et al., 2003). The other clade was predicted to be plastid localized (Krepinsky et al., 2001), but subcellular localization prediction algorithms such as TargetP (Emanuelsson et al., 2000), PSORT (Horton et al., 2007), MultiLoc2 (Blum et al., 2009), and Predotar (Small et al., 2004) gave low confidence or conflicting predictions. Only the spinach chloroplast isoform had the characteristic, long N-terminal extension for a chloroplast transit peptide in a multiple sequence alignment (Supplementary Fig. S2B). Predicted chloroplast-localized enzymes from Populus and Arabidopsis lack a transit peptide, while cereal proteins, including PGD3, had short N-terminal extensions that are atypical for transit peptides. To resolve these data, PGD3 subcellular localization was determined by expressing a C-terminal GFP fusion in Arabidopsis protoplasts (Supplementary Fig. S2D). PGD3-GFP signal was found in a subcellular compartment that overlapped completely with chlorophyll autofluorescence, indicating that PGD3 is a plastid-localized enzyme.
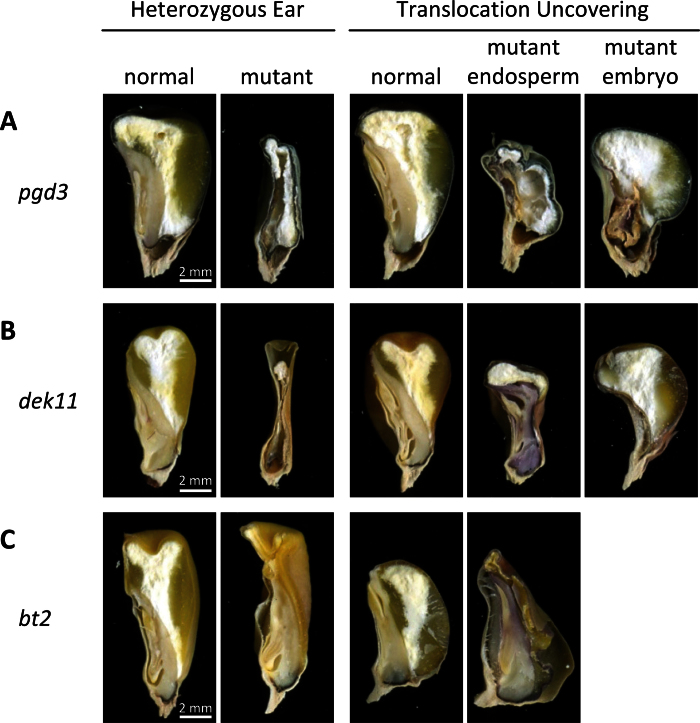
The high level of PGD3 activity in the embryo and the requirement for oxPPP in oil synthesis (Alonso et al., 2010) suggests PGD3 may only be required in the embryo during seed development. This hypothesis predicts that pgd3 mutant embryos would have a non-autonomous effect on endosperm growth and development. The current study generated kernels with non-concordant endosperm and embryo genotypes for the pgd3-umu2 allele by crossing pgd3-umu2/+ with the TB-4Sa B-A translocation stock. B-A translocations undergo non-disjunction in the second mitosis of microsporogenesis resulting in one sperm cell carrying a deletion for the translocated segment and the other being hyperploid (Roman, 1947). Double-fertilization of pgd3 female gametes produced non-concordant endosperm and embryo tissues that ‘uncovered’ either the pgd3 endosperm or embryo phenotype. If pgd3 mutant embryos caused defective endosperm development, a single uncovering class similar to what is observed for defective kernel24 (dek24) would be expected (Chang and Neuffer, 1994). However, two classes of uncovered kernels were observed. A pgd3 embryo had relatively little impact on wild-type endosperm development and growth (Fig. 3A, Supplementary Table S2). In contrast, the pgd3 endosperm disrupted wild-type embryo growth with a reduction in embryo weight (Supplementary Table S2) and development (Fig. 3A). The same TB-4Sa translocation was crossed to two mutant controls. The dek11 locus is a non-viable seed mutant with autonomous defects in the embryo and endosperm (Neuffer and Sheridan, 1980; Fouquet et al., 2011), while brittle2 (bt2) is defective for the small subunit of the rate-limiting enzyme of endosperm starch synthesis, ADP-glucose pyrophosphorylase (AGPase) (Preiss et al., 1990). Fig. 4B shows the autonomous endosperm and embryo phenotypes in non-concordant dek11 kernels, although dek11 endosperm uncovering reduced embryo weight (Supplementary Table S2). The bt2 mutant only showed a phenotype in the endosperm-uncovering kernel class (Fig. 4C) with no effect on embryo development or weight (Supplementary Table S2). The controls showed that pgd3 non-concordant kernel development is not a general phenomenon for seed mutants on chromosome arm 4S or endosperm starch-biosynthetis mutants. The data showed that PGD3 is specifically required within the endosperm and that the endosperm function of PGD3 has an impact on embryo development.
Fig. 3.
Non-concordant seed phenotypes. Sagittal hand sections of mature kernels from self-pollinations and TB-4Sa translocation crosses of pgd3-umu2/+ (A), dek11/+ (B), and bt2/+ (C) plants. Presence of a normal embryo in the mutant endosperm class indicates that a wild-type embryo can develop in conjunction with a mutant endosperm. Bars = 2mm.
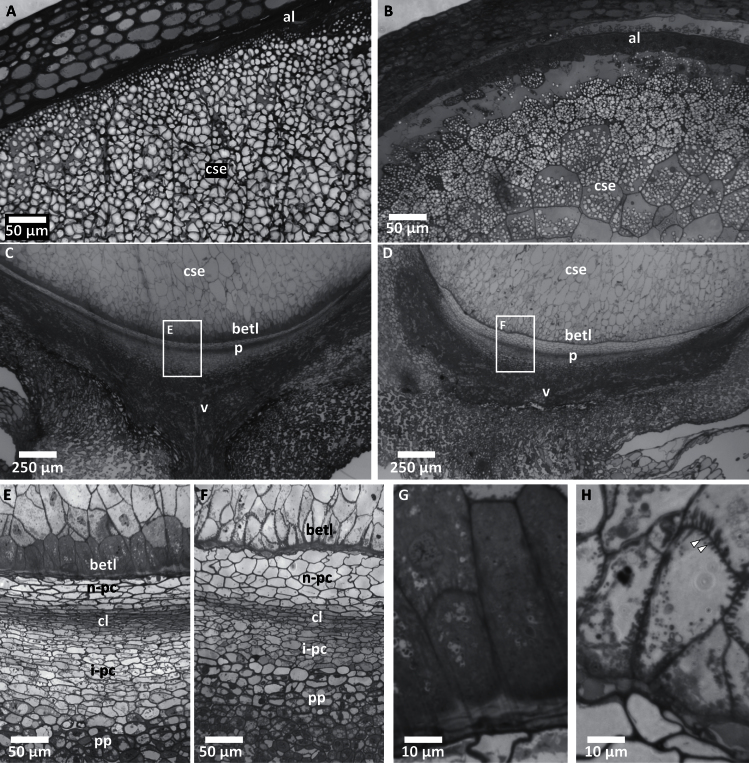
To understand the cellular basis of pgd3 endosperm development defects, this study sectioned 16-DAP kernels and found multiple morphological differences. In the wild type, the starchy endosperm was densely filled with starch granules (Fig. 4A) while pgd3 kernels contained fewer and smaller starch granules (Fig. 4B). Moreover, 20-DAP and mature endosperm starch content was reduced in pgd3-umu1 and pgd3-umu2 (Supplementary Table S3). The basal endosperm transfer cell layer (BETL) showed aberrant morphology in pgd3 mutants. The pgd3 BETL appeared separated from the densely stained closing layer (Fig. 4C, D). At higher magnification, the nucellar pedicel parenchyma of pgd3 mutants was not as compressed as in the wild type, and pgd3 BETL cells were defective (Fig. 4E, F). Wild-type BETL cells had extensive secondary cell-wall ingrowths for efficient uptake of nutrients and were visualized with dark staining throughout the entire cell (Fig. 4G). Short ingrowths were found in pgd3 BETL cells (Fig. 4H). Similar BETL cell morphology was found in 12-DAP wild-type plants at the onset of starch biosynthesis, but the ingrowths were fully developed at 17 DAP at the peak of starch production (Kang et al., 2009). Aberrant BETL morphology similar to pgd3 was found in multiple grain-fill mutants, including defective endosperm17 (de17), minature1 (mn1), baseless1 (bsl1), and rough endosperm3 (rgh3) (Lowe and Nelson, 1946; Brink and Cooper, 1947; Gutierrez-Marcos et al., 2006; Kang et al., 2009; Fouquet et al., 2011). BETL defects are interpreted as an indicator of reduced sink strength (Cheng et al., 1996; Kang et al., 2009).
Fig. 4.
Cellular defects in pgd3-umu1 kernels. Toluidine blue-stained semithin sections of 16-DAP kernels of wild-type sibling (A, C, E, G) and pgd3 (B, D, F, H). (A, B) Apical region of the seed showing central starchy endosperm (cse) and aleurone (al); bars = 50 µm. (C, D) Basal region showing the cse, basal endosperm transfer layer (betl), pedicel (p), and vascular tissue (v); boxes indicate regions magnified in E and F; bars = 250 µm. (E, F) Pedicel region showing the betl, nucellar placento-chalazal layer (n-pc), closing layer (cl), integumental placento-chalazal layer (i-pc), and pedicel parenchyma (pp); bars = 50 µm. (G, H) Basal endosperm transfer layer cells; white arrows indicate short secondary cell-wall ingrowths found in pgd3 mutants; bars = 10 µm.
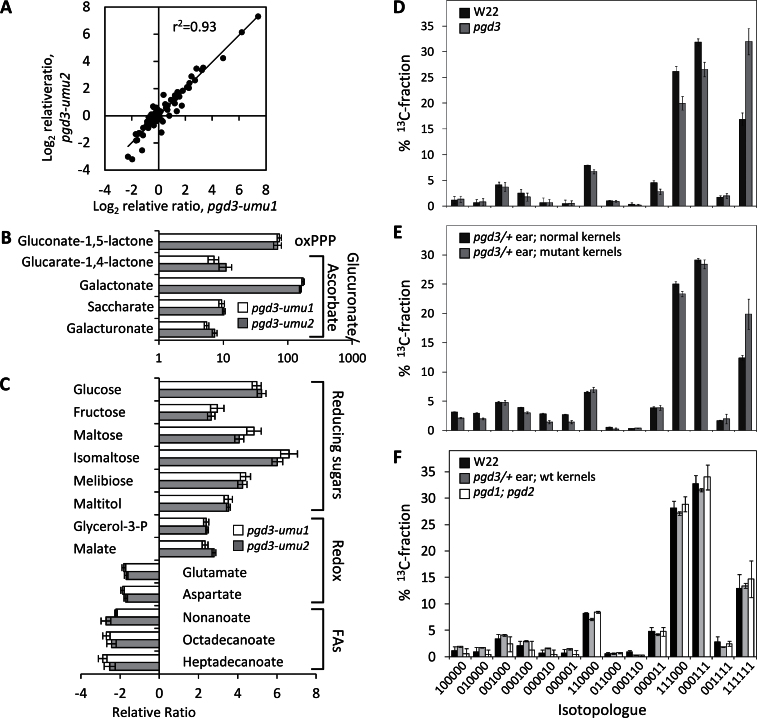
To determine the metabolic consequences of pgd3 defects in the endosperm, this study measured NADP+ and NADPH levels in 21-DAP kernels (Supplementary Table S4) and profiled 74 metabolites in 20-DAP pgd3-umu1, pgd3-umu2, and corresponding normal tissues (Supplementary Table S5). For profiling, GC-MS peak area ratios were normalized to a ribitol internal quantitative standard and the percentage of methanol extractable soluble compounds (Roessner et al., 2000; Witt et al., 2012). Principal components analysis of the data separated pgd3 and normal tissue in PC1, while PC2 separated the genetic pedigrees of the alleles (Supplementary Fig. S3). The relative fold-change in metabolite levels for pgd3:normal endosperm was highly correlated between the alleles, suggesting nearly identical effects of the mutants relative to normal siblings (Fig. 5A). The product of G6PDH, gluconate-1,5-lactone, accumulated significantly in mutants consistent with loss of 6PGDH (Fig. 5B). Total levels of NADP+ and NADPH were reduced, but the ratio of NADP+:NAPDH was unchanged (Supplementary Table S4). Intermediates for glucuronic acid and ascorbic acid pathways were increased, while glycerol-3-phosphate, malate, glutamate, and aspartate showed shifts in relative concentration, suggesting alterations in intracellular redox shuttling mechanisms (Fig. 5B, C). Additional amino acids were changed without a clear trend (Supplementary Table S5). Several fatty acids were reduced in the mutants consistent with the requirement for NAPDH in fatty acid synthesis (Fig. 5C). Finally, pgd3 increased reducing sugars similarly to the increases found in starch-biosynthetis mutants (Creech, 1965; Fig. 5C).
Fig. 5.
Metabolic analysis of pgd3. (A–C) Metabolite profiling in 20-DAP pgd3 endosperm. Response ratios are relative to normal sibling kernels; values close to 1 indicate no change in metabolite concentration for mutants. (A) Scatterplot of normalized relative ratios for the 74 metabolites profiled in both alleles of pgd3. (B, C) Relative ratios for selected metabolites relevant to oxPPP, glucuronic acid, ascorbic acid, redox shuttles, and fatty acids; error bars are standard errors from three biological replicates. (D–F) Altered metabolic flux into starch in pgd3-umu1. Ear block tissues were labelled with [U-13C]glucose and glucose isotopologue abundances were measured from starch by 13C-NMR spectroscopy. Relative abundance of each isotopologue from the total 13C-fractions is plotted. Each isotopologue is identified with 1 indicating 13C and 0 indicating 12C at each C position of the glucose molecule; error bars are standard deviation of three biological replicates. (D) Comparison of W22 and pgd3 homozygous ears. (E) Comparison of pgd3 mutant and normal kernels within segregating ears. (F) Comparison of W22 inbred, normal kernels within pgd3 segregating ears, and pgd1;pgd2 ears.
Heavy isotope-labelling experiments of developing maize kernels have shown the bulk of glucose incorporated into starch derives from products of glycolysis and the PPP (Spielbauer et al., 2006). This system infers relative metabolic pathway usage based on the incorporation of different isotopologues, molecules that differ only in their isotopic composition, into starch (Ettenhuber et al., 2005). However, these experiments cannot identify the subcellular localization of the PPP enzymes that produce substrates for starch synthesis (Masakapalli et al., 2010). In the current study, the reduced starch content of mature and developing endosperm in pgd3 mutants as well as the increase in reducing sugars in 20-DAP endosperm suggested that plastidic 6PGDH activity is needed for starch synthesis. It was then predicted that metabolic flux of glucose into starch would be altered in pgd3 seeds. Steady-state labelling was performed by feeding [U-13C]glucose to cob tissue with attached developing seeds. Fig. 5D–F shows the pattern of glucose isotopologues incorporated into starch in homozygous pgd3-umu1 ears and W22 inbred controls. The most prominent effect was a strong increase in direct incorporation of label into starch relative to fluxes through other pathways (11111 in Fig. 5D–F).
In vitro growth of kernels in sterile culture requires cob tissue (Felker, 1992). Potentially, isotopologue patterns in pgd3 homozygous plants are due to altered cob metabolic activities in ears lacking PGD3 (Alonso et al., 2011). To control for ear effects, the current study labelled pgd3/+ ears segregating for pgd3 mutant kernels. Mutant and normal kernels were visually separated for isotopologue analysis, resulting in the same trend, with pgd3 showing increased direct incorporation (Fig. 5E). The differences observed were not as large as in homozygous pgd3 ears, possibly due to aborted normal kernels being included in pgd3 samples. These results indicate maternal cob tissue has, if any, only minor effects on 13C distribution in the labelling system. It is concluded the altered 13C pattern of pgd3 mutants was due to metabolic reactions in the kernel. Normal kernels from heterozygous ears, W22, and pgd1;pgd2 double mutants all showed similar isotopologue patterns, indicating a specific effect on starch synthesis from loss of plastid-localized oxPPP (Fig. 5F).
Ratios of specific isotopologues can approximate relative flux through carbon metabolic pathways (Spielbauer et al., 2006). The ratio of uniformly labelled glucose {111111} to {111000} (see legend to Fig. 5F) estimates flux partitioning between direct incorporation into starch and hexose cycling, i.e. entry into glycolysis or PPP. This ratio was approximately doubled in pgd3 (Supplementary Table S6). Reforming glucose via triose-hexose exchange by glycolytic reactions should give an equal ratio of {000111} to {111000}, while exchange reactions of transaldolase in the noxPPP will only produce {000111}. Thus, a ratio of {000111} to {111000} above 1 indicates flux through noxPPP (Ettenhuber et al., 2005). Mutant pgd3 kernels showed a statistically significant 5–10% increase in this ratio compared to wild-type and pgd1;pgd2 seeds, suggesting increased flux through noxPPP (Supplementary Table S6). Finally, {110000} is an indicator of oxPPP as it is primarily formed after loss of the C1-atom in the oxPPP and glucose-6-P is regenerated via the noxPPP. Surprisingly, neither pgd1;pgd2 double mutants nor pgd3 changed the ratio of {110000} to {111000}, suggesting that pentoses produced in the pgd3 cytosol can enter the amyloplast to regenerate into glucose. In summary, isotopologue patterns of pgd3 starch suggest that fluxes through glycolysis and PPP are reduced and that flux through noxPPP may be increased.
Discussion
This study shows that loss of PGD3 enzyme activity caused a severe defective kernel and indicates a non-redundant role for Pgd3 with the other 6PGDH loci, Pgd1 and Pgd2. Prior work found PGD3 was enriched in crude membrane fractions, and it was assumed PGD3 is plastid localized based on sequence similarity (Averill et al., 1998; Kruger and von Schaewen, 2003). However, all three 6PGDH isoforms co-purified with plastids, and the PGD3 chloroplast targeting peptide was non-canonical (Supplementary Fig. S2). Thus, this study provides conclusive evidence that PGD3 is plastid localized with a GFP fusion.
The data contrast with prior conclusions that PGD1 and PGD2 enzymes act jointly with PGD3 to satisfy cellular demand for NADPH in all compartments via a proposed shuttling mechanism for reducing equivalents (Averill et al., 1998). Double mutants of pgd1;pgd2 have subtle growth phenotypes and a reduced rate of nitrite uptake in roots (Averill et al., 1998). In light of the severe pgd3 phenotype, the data argue that cytosolic 6PGDH is not essential for maize metabolism. Metabolite analysis of pgd3 endosperm suggests that reductant shuttles are shifted, yet these do not compensate for loss of PGD3 (Fig. 5B, C). These data are consistent with the general model that the cytosol and plastid produce reductant independently.
A simple interpretation of the pgd3 kernel phenotype is an essential role for oil accumulation that impacts kernel development. Almost all maize oil is stored in the embryo (Doehlert and Lambert, 1991), and oxPPP provides the majority of the NADPH needed for oil synthesis (Alonso et al., 2010). However, PGD3 is not required continuously in embryos, because a small percentage of pgd3 embryos can be rescued in culture and develop normally (Settles et al., 2007). Moreover, B-A translocation genetics indicated distinct and critical roles for PGD3 in both the endosperm and embryo (Figs. 2 and 3). PGD3 is specifically required in the endosperm and this endosperm function promotes embryo development.
The essential role for plastid oxPPP in endosperm is not easy to explain. The major storage component of the maize endosperm is starch, and demand for reductant is thought to be low (Alonso et al., 2011). The extensive changes in the pgd3 metabolite profiles suggest a critical role throughout the metabolic network, while the cellular defects, endosperm starch reduction, and heavy isotope-labelling experiments indicate a requirement for PGD3 in starch synthesis. Although pgd3 is more pleiotropic than conventional sweet corn mutants, starch reductions are comparable on a per kernel basis. Mutant pgd3 accumulates 15% the starch of the wild type, while bt1, sh2, and bt2 accumulate 12–21% the starch of wild-type kernels (Spielbauer et al., 2009).
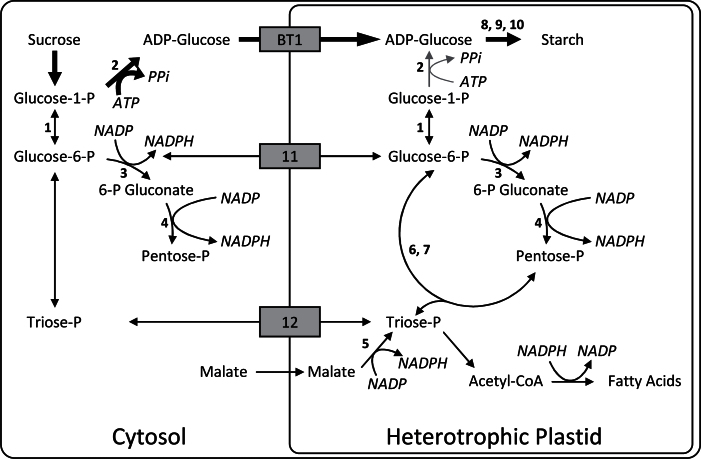
The current model of cereal endosperm starch biosynthesis is known to require ATP and pyrophosphate but not NADPH or NADH (Fig. 6, Comparot-Moss and Denyer, 2009). Sucrose enters the kernel and is hydrolysed by sucrose synthase to yield UDP-glucose and fructose. These hexoses are converted into glucose-1-P and then into ADP-glucose by AGPase, the rate-limiting step of starch biosynthesis. The major AGPase in the cereal endosperm is cytosolic, and ADP-glucose is transported into the plastid by the adenylate translocator for incorporation into starch. Heavy isotope-labelling experiments indicate that much of the glucose incorporated into endosperm starch first passes through the initial reactions of glycolysis and the PPP (Spielbauer et al., 2006). The current study found a significant shift towards direct incorporation of glucose into starch in the pgd3 mutant (Fig. 5D–F). This shift was not due to cob tissue effects, indicating that plastidic oxPPP in the endosperm influences starch synthesis.
Fig. 6.
Starch synthesis and PPP in the maize endosperm. Thick arrows show the conventional biosynthesis of starch: glucose-1-P is converted to ADP-glucose, which is transported into the plastid and polymerized into starch. Some ADP-glucose can be formed in the plastid (grey arrows). oxPPP synthesizes NADPH in the cytosol and plastid. Plastidic NADPH is also synthesized by malic enzyme. NADPH is required for fatty acid synthesis in the plastid, but endosperm amyloplasts primarily accumulate starch. Metabolites are exchanged between cytosol and plastids (grey boxes). 1, phosphoglucomutase; 2, AGPase; 3, G6PDH; 4, 6PGDH; 5, malic enzyme; 6, transaldolase; 7, transketolase; 8, starch synthase; 9, starch branching enzyme; 10, starch debranching enzyme; 11, glucose-6-P/P translocator; 12, triose-P/P translocator and P/phosphoenolpyruvate translocator.
There is no simple explanation for the requirement of plastid-localized 6PGDH in maize endosperm starch biosynthesis. The products of PGD3 are ribulose-5-P and NADPH. Ribulose-5-P can be generated through the reversible reactions of the noxPPP, while NADPH could be generated from NADP-malic enzyme. Malate concentrations are increased up to 2-fold in the pgd3 mutants, while there is an overall 20% reduction of NADP+ and NADPH. These data suggest NADPH may be limiting in pgd3. A role for NADPH in starch biosynthesis is consistent with a metabolic flux model for the maize endosperm in which NADPH is produced well above demand for known endosperm metabolic processes (Alonso et al., 2011).
Redox status in the plastid is potentially important for cereal endosperm starch synthesis. Plastid-produced NADPH is required for the ferredoxin/thioredoxin system identified in wheat amyloplasts (Balmer et al., 2006). Ferredoxin/thioredoxin targets relevant to starch metabolism include α1,4-glucan phosphorylase, starch branching enzyme IIa, and the BT1 transporter (Kirchberger et al., 2007). In contrast to pgd3, bt1 does not change the proportion of glucose that is directly incorporated into starch (Spielbauer et al., 2006), suggesting that BT1 transporter activity is not the primary target of reducing equivalents produced through oxPPP. Posttranslational redox-regulation for the plastid small subunit of AGPase has been established in potato tuber as well as Arabidopsis and pea leaves (Tiessen et al., 2002; Geigenberger et al., 2005; Michalska et al., 2009). There is some debate about the importance of plastid-localized AGPase in cereal endosperm. The majority of AGPase activity has been detected in the cytosol (Denyer et al., 1996). However, residual AGPase activity is found in isolated amyloplasts and in sh2 and bt2 null mutant kernels (Dickinson and Preiss, 1969; Denyer et al., 1996), suggesting that plastid-localized isoforms of APGase are present in maize endosperm. Peptides identical to both SH2 and BT2 subunits co-purify in a complex with plastid-localized starch synthase III supporting a role for plastidic AGPase (Hennen-Bierwagen et al., 2009). In maize endosperm, the genetic identity of the amyloplast-localized small subunit of AGPase has not been resolved. Two candidates are Agpsemzm and Bt2b. Agpsemzm is expressed in the embryo and endosperm (Giroux and Hannah, 1994; Rosti and Denyer, 2007; Cossegal et al., 2008). BT2b is a theoretical alternative product of the Bt2 gene, which is predicted to be targeted to the plastid (Rosti and Denyer, 2007; Cossegal et al., 2008). In contrast to the cytosolic endosperm small subunit of AGPase (BT2) (Hendriks et al., 2003), both plastid-targeted proteins contain the redox-relevant cysteine residue and could be regulatory targets for the reductant produced by PGD3.
Despite the increasing knowledge about redox regulation of starch-biosynthetis enzymes (Geigenberger, 2011), the importance of plastidic oxPPP has been underestimated in the cereal endosperm and has not been integrated in current models (Hannah and James, 2008; Comparot-Moss and Denyer, 2009). The pgd3 mutant provides significant evidence for the requirement of oxPPP in the cereal endosperm amyloplast to enable starch synthesis.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Table S1. Bulk seed composition of selected maize genotypes.
Supplementary Table S2. Dryweights of tissues dissected from mature maize kernels.
Supplementary Table S3. Endosperm composition of the pgd3 mutant and normal kernels from segregating ears.
Supplementary Table S4. NADP+ and NADPH levels in 21 DAP kernels.
Supplementary Table S5. Metabolite ratios in 20 DAP pgd3-umu1 and pgd3-umu2 mutant and normal endosperm.
Supplementary Table S6. Ratios of selected isotopologues from [U-13C6]glucose kernel labelling experiments.
Supplementary Fig. S1. Equivalent phenotypes for pgd3-umu1 and pgd3-umu2 alleles.
Supplementary Fig. S2. PGD3 is a chloroplast-localized 6PGDH.
Supplementary Fig. S3. Principal component analysis of normalized peak area ratios from endosperm metabolic profiles.
Acknowledgements
The authors thank Julia Bailey-Serres, Jim Birchler, Don Auger, and the Maize Genetics Cooperation Stock Center for providing seed stocks, Byung-Ho Kang for technical support in microscopy experiments and Chi-Wah Tseung for technical assistance. This work was supported by the National Science Foundation Plant Genome Research Program (DBI-0606607), the National Institute of Food and Agriculture (2010–04228), the Vasil-Monsanto Endowment, and the Hans-Fischer Gesellschaft.
References
- Alonso AP, Dale VL, Shachar-Hill Y. 2010. Understanding fatty acid synthesis in developing maize embryos using metabolic flux analysis. Metabolic Engineering 12, 488–497. [DOI] [PubMed] [Google Scholar]
- Alonso AP, Val DL, Shachar-Hill Y. 2011. Central metabolic fluxes in the endosperm of developing maize seeds and their implications for metabolic engineering. Metabolic Engineering 13, 96–107. [DOI] [PubMed] [Google Scholar]
- Averill RH, Bailey-Serres J, Kruger NJ. 1998. Co-operation between cytosolic and plastidic oxidative pentose phosphate pathways revealed by 6-phosphogluconate dehydrogenase-deficient genotypes of maize. The Plant Journal 14, 449–457. [Google Scholar]
- Bailey-Serres J, Nguyen MT. 1992. Purification and characterization of cytosolic 6-phosphogluconate dehydrogenase isozymes from maize. Plant Physiology 100, 1580–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Tom J, Freeling M. 1992. Expression and distribution of cytosolic 6-phosphogluconate dehydrogenase isozymes in maize. Biochemical Genetics 30, 233–246. [DOI] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Cai N, Manieri W, Schurmann P, Hurkman WJ, Buchanan BB. 2006. A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proceedings of the National Academy of Sciences, USA 103, 2988–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Alfenito MR. 1993. Marker systems for B-A translocations in maize. Journal of Heredity 84, 135–138. [Google Scholar]
- Blum T, Briesemeister S, Kohlbacher O. 2009. MultiLoc2: integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction. BMC Bioinformatics 10, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowsher CG, Boulton EL, Rose JKC, Nayagam S, Emes MJ. 1992. Reductant for glutamate synthase is generated by the oxidative pentose-phosphate pathway in nonphotosynthetic root plastids. The Plant Journal 2, 893–898. [Google Scholar]
- Brink RA, Cooper DC. 1947. Effect of the De17 allele on development of the maize caryopsis. Genetics 32, 350–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MT, Neuffer MG. 1994. Endosperm–embryo interaction in maize. Maydica 39, 9–18. [Google Scholar]
- Cheng WH, Taliercio EW, Chourey PS. 1996. The Miniature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. The Plant Cell 8, 971–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Henry R, Li C, Yuan J. 1993. Multiple pathways for protein transport into or across the thylakoid membrane. The EMBO Journal 12, 4105–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comparot-Moss S, Denyer K. 2009. The evolution of the starch biosynthetic pathway in cereals and other grasses. Journal of Experimental Botany 60, 2481–2492. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, Sandalio LM, Distefano S, Palma JM, Lupianez JA, Del, Rio LA. 1998. A dehydrogenase-mediated recycling system of NADPH in plant peroxisomes. The Biochemical Journal 330, 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossegal M, Chambrier P, Mbelo S, Balzergue S, Martin-Magniette ML, Moing A, Deborde C, Guyon V, Perez P, Rogowsky P. 2008. Transcriptional and metabolic adjustments in ADP-glucose pyrophosphorylase-deficient bt2 maize kernels. Plant Physiology 146, 1553–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creech RG. 1965. Genetic control of carbohydrate synthesis in maize endosperm. Genetics 52, 1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnam PM, Emes MJ. 1999. Subcellular distribution of enzymes of the oxidative pentose phosphate pathway in root and leaf tissues. Journal of Experimental Botany 50, 1653–1661. [Google Scholar]
- Debnam PM, Fernie AR, Leisse A, Golding A, Bowsher CG, Grimshaw C, Knight JS, Emes MJ. 2004. Altered activity of the P2 isoform of plastidic glucose 6-phosphate dehydrogenase in tobacco (Nicotiana tabacum cv. Samsun) causes changes in carbohydrate metabolism and response to oxidative stress in leaves. The Plant Journal 38, 49–59. [DOI] [PubMed] [Google Scholar]
- Denyer K, Dunlap F, Thorbjornsen T, Keeling P, Smith AM. 1996. The major form of ADP-glucose pyrophosphorylase in maize endosperm is extra-plastidial. Plant Physiology 112, 779–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DB, Preiss J. 1969. Presence of ADP-glucose pyrophosphorylase in shrunken-2 and brittle-2 mutants of maize endosperm. Plant Physiology 44, 1058–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlert DC, Lambert RJ. 1991. Metabolic characteristics associated with starch, protein and oil deposition in developing maize kernels. Crop Science 31, 151–157. [Google Scholar]
- Eisenreich W, Ettenhuber C, Laupitz R, Theus C, Bacher A. 2004. Isotopolog perturbation techniques for metabolic networks: metabolic recycling of nutritional glucose in Drosophila melanogaster . Proceedings of the National Academy of Sciences, USA 101, 6764–6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of Molecular Biology 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Ettenhuber C, Spielbauer G, Margl L, Hannah LC, Gierl A, Bacher A, Genschel U, Eisenreich W. 2005. Changes in flux pattern of the central carbohydrate metabolism during kernel development in maize. Phytochemistry 66, 2632–2642. [DOI] [PubMed] [Google Scholar]
- Felker FC. 1992. Participation of cob tissue in the uptake of medium components by maize kernels cultured in vitro . Journal of Plant Physiology 139, 647–652. [Google Scholar]
- Fouquet R, Martin F, Fajardo DS, Gault CM, Gómez E, Tseung CW, Policht T, Hueros G, Settles AM. 2011. Maize Rough Endosperm3 encodes an RNA splicing factor required for endosperm cell differentiation and has a non-autonomous effect on embryo development. The Plant Cell 23, 4280–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P. 2011. Regulation of starch biosynthesis in response to a fluctuating environment. Plant Physiology 155, 1566–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Kolbe A, Tiessen A. 2005. Redox regulation of carbon storage and partitioning in response to light and sugars. Journal of Experimental Botany 56, 1469–1479. [DOI] [PubMed] [Google Scholar]
- Giroux MJ, Hannah LC. 1994. ADP-glucose pyrophosphorylase in shrunken-2 and brittle-2 mutants of maize. Molecular and General Genetics 243, 400–408. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Costa LM, Evans MMS. 2006. Maternal gametophytic baseless1 is required for development of the central cell and early endosperm patterning in maize (Zea mays). Genetics 174, 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah LC, James M. 2008. The complexities of starch biosynthesis in cereal endosperms. Current Opinion in Biotechnology 19, 160–165. [DOI] [PubMed] [Google Scholar]
- Hendriks JH, Kolbe A, Gibon Y, Stitt M, Geigenberger P. 2003. ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiology 133, 838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennen-Bierwagen TA, Lin Q, Grimaud F, Planchot V, Keeling PL, James MG, Myers AM. 2009. Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts. Plant Physiology 149, 1541–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. 2007. WoLF PSORT: protein localization predictor. Nucleic Acids Research 35, W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings D, Rawsthorne S, Emes MJ. 2005. Fatty acid synthesis and the oxidative pentose phosphate pathway in developing embryos of oilseed rape (Brassica napus L.). Journal of Experimental Botany 56, 577–585. [DOI] [PubMed] [Google Scholar]
- Kang BH, Xiong YQ, Williams DS, Pozueta-Romero D, Chourey PS. 2009. Miniature1-encoded cell wall invertase is essential for assembly and function of wall-in-growth in the maize endosperm transfer cell. Plant Physiology 151, 1366–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchberger S, Leroch M, Huynen MA, Wahl M, Neuhaus HE, Tjaden J. 2007. Molecular and biochemical analysis of the plastidic ADP-glucose transporter (ZmBT1) from Zea mays . Journal of Biological Chemistry 282, 22481–22491. [DOI] [PubMed] [Google Scholar]
- Kolbe A, Tiessen A, Schluepmann H, Paul M, Ulrich S, Geigenberger P. 2005. Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proceedings of the National Academy of Sciences, USA 102, 11118–11123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Koprivova A, Suss KH. 2000. Identification, cloning, and properties of cytosolic D-ribulose-5-phosphate 3-epimerase from higher plants. Journal of Biological Chemistry 275, 1294–1299. [DOI] [PubMed] [Google Scholar]
- Krepinsky K, Plaumann M, Martin W, Schnarrenberger C. 2001. Purification and cloning of chloroplast 6-phosphogluconate dehydrogenase from spinach–Cyanobacterial genes for chloroplast and cytosolic isoenzymes encoded in eukaryotic chromosomes. European Journal of Biochemistry 268, 2678–2686. [DOI] [PubMed] [Google Scholar]
- Kruger NJ, von Schaewen A. 2003. The oxidative pentose phosphate pathway: structure and organisation. Current Opinion in Plant Biology 6, 236–246. [DOI] [PubMed] [Google Scholar]
- Lendzian K, Bassham JA. 1975. Regulation of glucose-6-phosphate dehydrogenase in spinach chloroplasts by ribulose 1,5-diphosphate and NADPH-NADP+ ratios. Biochimica and Biophysica Acta 396, 260–275. [DOI] [PubMed] [Google Scholar]
- Linka N, Weber AP. 2010. Intracellular metabolite transporters in plants. Molecular Plant 3, 21–53. [DOI] [PubMed] [Google Scholar]
- Lowe J, Nelson OE. 1946. Miniature seed – a study in the development of a defective caryopsis in maize. Genetics 31, 525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masakapalli SK, Le Lay P, Huddleston JE, Pollock NL, Kruger NJ, Ratcliffe RG. 2010. Subcellular flux analysis of central metabolism in a heterotrophic Arabidopsis cell suspension using steady-state stable isotope labeling. Plant Physiology 152, 602–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Settles AM, Suzuki M, et al. 2005. Steady-state transposon mutagenesis in inbred maize. The Plant Journal 44, 52–61. [DOI] [PubMed] [Google Scholar]
- Meyer T, Holscher C, Schwoppe C, von Schaewen A. 2011. Alternative targeting of Arabidopsis plastidic glucose-6-phosphate dehydrogenase G6PD1 involves cysteine-dependent interaction with G6PD4 in the cytosol. The Plant Journal 66, 745–758. [DOI] [PubMed] [Google Scholar]
- Michalska J, Zauber H, Buchanan BB, Cejudo FJ, Geigenberger P. 2009. NTRC links built-in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proceedings of the National Academy of Sciences, USA 106, 9908–9913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miclet E, Stoven V, Michels PA, Opperdoes FR, Lallemand JY, Duffieux F. 2001. NMR spectroscopic analysis of the first two steps of the pentose-phosphate pathway elucidates the role of 6-phosphogluconolactonase. Journal of Biological Chemistry 276, 34840–34846. [DOI] [PubMed] [Google Scholar]
- Mori H, Summer EJ, Ma X, Cline K. 1999. Component specificity for the thylakoidal Sec and Delta pH-dependent protein transport pathways. Journal of Cell Biology 146, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger G, Maurer-Stroh S, Eisenhaber B, Hartig A, Eisenhaber F. 2003. Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. Journal of Molecular Biology 328, 581–592. [DOI] [PubMed] [Google Scholar]
- Neuffer MG, Sheridan WF. 1980. Defective kernel mutants of maize I. Genetic and lethality studies. Genetics 95, 929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss J, Danner S, Summers PS, Morell M, Barton CR, Yang L, Nieder M. 1990. Molecular characterization of the Brittle-2 gene effect on maize endosperm ADP glucose pyrophosphorylase subunits. Plant Physiology 92, 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Luder F, Weckwerth W, Jahn O. 2007. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. The Plant Cell 19, 3170–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Quan S, Aung K, et al. 2009. In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiology 150, 125–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. 2000. Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. The Plant Journal 23, 131–42. [DOI] [PubMed] [Google Scholar]
- Roman H. 1947. Mitotic nondisjunction in the case of interchanges involving the B-type chromosome in maize. Genetics 32, 391–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosti S, Denyer K. 2007. Two paralogous genes encoding small subunits of ADP-glucose pyrophosphorylase in maize, Bt2 and L2, replace the single alternatively spliced gene found in other cereal species. Journal of Molecular Evolution 65, 316–327. [DOI] [PubMed] [Google Scholar]
- Scharte J, Schon H, Tjaden Z, Weis E, von Schaewen A. 2009. Isoenzyme replacement of glucose-6-phosphate dehydrogenase in the cytosol improves stress tolerance in plants. Proceedings of the National Academy of Sciences, USA 106, 8061–8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge JB, Shachar-Hill Y. 2003. A flux model of glycolysis and the oxidative pentosephosphate pathway in developing Brassica napus embryos. Journal of Biological Chemistry 278, 29442–29453. [DOI] [PubMed] [Google Scholar]
- Settles AM, Holding DR, Tan BC, et al. 2007. Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics 8, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signorini M, Bregoli AM, Caselli L, Bergamini CM. 1995. Purification and properties of 6-phosphogluconate dehydrogenase from beet leaves. Biochemistry and Molecular Biology International 35, 669–675. [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. 2004. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4, 1581–1590. [DOI] [PubMed] [Google Scholar]
- Spielbauer G, Armstrong P, Baier JW, Allen WB, Richardson K, Shen B, Settles AM. 2009. High-throughput near-infrared reflectance spectroscopy for predicting quantitative and qualitative composition phenotypes of individual maize kernels. Cereal Chemistry 86, 556–564. [Google Scholar]
- Spielbauer G, Margl L, Hannah LC, Romisch W, Ettenhuber C, Bacher A, Gierl A, Eisenreich W, Genschel U. 2006. Robustness of central carbohydrate metabolism in developing maize kernels. Phytochemistry 67, 1460–1475. [DOI] [PubMed] [Google Scholar]
- Tanksley SD, Kuehn GD. 1985. Genetics, subcellular-localization, and molecular characterization of 6-phosphogluconate dehydrogenase isozymes in tomato. Biochemical Genetics 23, 441–454. [DOI] [PubMed] [Google Scholar]
- Tiessen A, Hendriks JH, Stitt M, Branscheid A, Gibon Y, Farre EM, Geigenberger P. 2002. Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: a novel regulatory mechanism linking starch synthesis to the sucrose supply. The Plant Cell 14, 2191–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakao S, Andre C, Benning C. 2008. Functional analyses of cytosolic glucose-6-phosphate dehydrogenases and their contribution to seed oil accumulation in Arabidopsis . Plant Physiology 146, 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt S, Galicia L, Lisec J, Cairns J, Tiessen A, Araus JL, Palacios-Rojas N, Fernie AR. 2012. Metabolic and phenotypic responses of greenhouse-grown maize hybrids to experimentally controlled drought stress. Molecular Plant 5, 401–417. [DOI] [PubMed] [Google Scholar]
- Xiong Y, DeFraia C, Williams D, Zhang X, Mou Z. 2009. Characterization of Arabidopsis 6-phosphogluconolactonase T-DNA insertion mutants reveals an essential role for the oxidative section of the plastidic pentose phosphate pathway in plant growth and development. Plant Cell Physiology 50, 1277–1291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.