Abstract
S-RNase-based gametophytic self-incompatibility evolved once before the split of the Asteridae and Rosidae. In Prunus (tribe Amygdaloideae of Rosaceae), the self-incompatibility S-pollen is a single F-box gene that presents the expected evolutionary signatures. In Malus and Pyrus (subtribe Pyrinae of Rosaceae), however, clusters of F-box genes (called SFBBs) have been described that are expressed in pollen only and are linked to the S-RNase gene. Although polymorphic, SFBB genes present levels of diversity lower than those of the S-RNase gene. They have been suggested as putative S-pollen genes, in a system of non-self recognition by multiple factors. Subsets of allelic products of the different SFBB genes interact with non-self S-RNases, marking them for degradation, and allowing compatible pollinations. This study performed a detailed characterization of SFBB genes in Sorbus aucuparia (Pyrinae) to address three predictions of the non-self recognition by multiple factors model. As predicted, the number of SFBB genes was large to account for the many S-RNase specificities. Secondly, like the S-RNase gene, the SFBB genes were old. Thirdly, amino acids under positive selection—those that could be involved in specificity determination—were identified when intra-haplotype SFBB genes were analysed using codon models. Overall, the findings reported here support the non-self recognition by multiple factors model.
Key words: gametophytic self-incompatibility, molecular evolution, positively selected amino acid sites, SFBB, Sorbus aucuparia, S-RNase.
Introduction
Self-incompatibility (SI) is a genetic barrier to self-fertilization in which the female reproductive cells discriminate between genetic relative and non-relative pollen, and reject the former (De Nettancourt, 1977). In order to maintain functional incompatibility, the S genes, those determining the pistil and pollen specificities, must co-evolve as a genetic unit (for details on co-evolution see, Newbigin et al., 2008). In gametophytic SI (GSI), the pollen is rejected when it expresses a specificity that matches either of those expressed in the style. In this system, because of frequency-dependent selection, many specificities are maintained in natural populations (Wright, 1939).
The self-incompatibility S-pistil gene product in Rosaceae, Rubiaceae, Solanaceae, and Plantaginaceae is an extracellular ribonuclease, called S-RNase (Roalson and McCubbin, 2003; Nowak et al., 2011). Phylogenetic analyses suggest that S-RNase-based GSI has evolved only once, before the split of the Asteridae and Rosidae, about 120 million years ago (MYA) (Igic and Kohn, 2001; Steinbachs and Holsinger, 2002; Vieira et al., 2008a). Because of the single origin of this system, in principle, similarities are expected when comparing the GSI players in these plant families.
In Rosaceae, for the pistil gene, studies at the molecular level have been performed in species of the tribe Amygdaloideae (Prunus) and subtribe Pyrinae (Malus, Pyrus, Sorbus, and Crataegus; see references in Vieira et al., 2010). The pistil gene shows the expected features for a gene determining GSI specificity, namely, high levels of synonymous and non-synonymous divergence, as well as positively selected amino acid sites that account for the many specificities known to be present in natural populations (Vieira et al., 2007).
The pollen component, always an F-box protein, has been identified as one gene in Prunus (called SFB, S-haplotype-specific F-box gene) (Ushijima et al., 2001; Entani et al., 2003; Ushijima et al., 2003; Ikeda et al., 2004; Romero et al., 2004; Sonneveld et al., 2005; Nunes et al., 2006; Vieira et al., 2008b), but multiple genes in Malus, Pyrus, (called SFBBs, S-locus F-box brothers), Petunia (Solanaceae), and Nicotiana (Solanaceae; called SLFs, S-locus F-boxes) (Cheng et al., 2006; Kakui et al., 2007; Sassa et al., 2007; Wheeler and Newbigin, 2007; Kubo et al., 2010; Minamikawa et al., 2010). In Prunus, the non-self S-RNases taken up by the growing pollen tube are postulated to be inactivated by a general inhibitor and only the self S-RNases are protected from inactivation (Luu et al., 2001; Sonneveld et al., 2005). In the multiple S pollen genes system, found in Malus, Pyrus, Petunia, and Nicotiana, within an S haplotype, each SLF/SFBB is predicted to interact with a subset of non-self S-RNases, and mediates their degradation by the ubiquitin–26S proteasome system. Multiple SLF/SFBB genes are thus required for the recognition of the large repertory of non-self S-RNases (Kubo et al., 2010) under this protein degradation model (Hua and Kao, 2006; Hua et al., 2007). Therefore, SLF/SFBB genes could represent a case of paralogous gene expansions that are often the substrate for adaptive change.
The very different mechanisms of S-RNase inhibition in systems with one or multiple S pollen genes imply remarkable differences at the S-locus region. Indeed, in Prunus, as expected, if a single S pollen gene is involved, the S-locus region is smaller than 45kb (Vieira et al., 2008c). The S-RNase gene is flanked by two F-box like genes, namely, SFB and SLFL1 (SLF-like gene 1). Nevertheless, in contrast to the SLFL1 gene, the SFB gene is expressed only in pollen, and its average diversity is similar to that observed for the S-RNase gene (Ushijima et al., 2001; Entani et al., 2003; Ushijima et al., 2003; Vieira et al., 2008a). Despite evidence for specific associations between SLFL1 and S-RNase - SFB genes, as well as the relatively old age of SLFL1 alleles (Vieira et al., 2008d ), amino acid sites showing strong evidence for positive selection have been identified only in the SFB gene (Nunes et al., 2006; Vieira et al., 2008b , d ). Despite the lack of congruent tree topologies for the S-RNase and SFB genes (Nunes et al., 2006; Tsukamoto et al., 2008), the two genes show evidence for a partially co-evolved history (Tsukamoto et al., 2008).
Under the multiple S pollen genes system, SFBB genes are expected to be in linkage with the S-RNase gene and to have pollen expression only. In Malus, the extent of the S-locus region is unknown but is larger than 317kb (corresponding to the BAC contig s analysed by Sassa et al., 2007, and Minamikawa et al., 2010). In this region, two F-box genes, MdSFBB9-alpha and MdSFBB9-beta (located 42kb upstream and 93kb downstream of the S 9 -RNase, respectively), with expression restricted to pollen, and in linkage with the S-RNase gene, were initially reported as S pollen genes (Sassa et al., 2007). The number of these genes in the Malus S-locus region is currently greater than ten (Minamikawa et al., 2010; Sassa et al., 2010). In Pyrus, the size of the S-locus region is also unknown but is larger than 649kb (Okada et al., 2011), and the number of F-box genes with expression restricted to pollen and in linkage with the S-RNase gene is greater than eight (Minamikawa et al., 2010; Sassa et al., 2010; De Franceschi et al., 2011a , b ; Kakui et al., 2011; Okada et al., 2011). The order of the SFBB genes in different S haplotypes is not conserved (Minamikawa et al., 2010; Okada et al., 2011). Therefore, depending on the haplotype analysed in the segregation experiments, an SFBB gene can show linkage with an S-RNase allele but incomplete linkage with another (Pyrus SFBB alpha, and Pyrus SFBB-gamma in De Franceschi et al., 2011a, and SFBB6 in Kakui et al., 2011). It is possible that SFBB alleles not showing linkage to the S-RNase may have no target S-RNase allele and thus are not being constrained by selection (Kakui et al., 2011). These genes have been assigned as not being involved in S pollen specificity (De Franceschi et al., 2012).
Kakui et al. (2007) reported low polymorphism for the PpSFBB-gamma gene compared with the S-RNase gene. This pattern seems to be a common feature of all SFBB genes (Minamikawa et al., 2010; De Franceschi et al., 2011a; Kakui et al., 2011; Okada et al., 2011) and SLFs (Zhou et al., 2003; Wheeler and Newbigin, 2007; Newbigin et al., 2008; Kubo et al., 2010). Identical alleles at one SFBB gene have been reported from two different S haplotypes (Minamikawa et al., 2010). Another pattern is the high divergence between SFBB genes, comparable to the allelic diversity of the S-RNase gene (Minamikawa et al., 2010; De Franceschi et al., 2011a ; Kakui et al., 2011; Okada et al., 2011), but age estimates for these genes have not been obtained. Phylogenetic analyses have shown that diversification of SFBB genes pre-dates speciation of Pyrus and Malus (Minamikawa et al., 2010; Kakui et al., 2011). These data are compatible with the scenario where a large repertoire of non-self S-RNases are targeted and detoxified by multiple SFBB genes, each of which recognizes a subfraction of S-RNases (Kakui et al., 2011). In Petunia, a non-self recognition pollen rejection mechanism has also been proposed (Kubo et al., 2010; Wang and Kao, 2011). Although in the initial protein degradation model the S pollen genes were assumed to inhibit all S-RNases except that of the corresponding S haplotype (Hua and Kao, 2006; Hua et al., 2008), in vivo functional assays and protein-interaction assays revealed that each SLF protein functions as a pollen determinant and recognizes a subset of non-self S-RNases (Kubo et al., 2010; Wang and Kao, 2011).
As expected for the S pollen, evidence for positive selection acting on amino acid sites located in two (V1 and V2) of the four variable regions was found by calculating the ratio of non-synonymous substitutions per non-synonymous site (K a) divided by the ratio of synonymous substitutions per synonymous site (K s), using Malus (two SFBB-alpha and two SFBB-beta sequences) and Pyrus (two SFBB-alpha, two SFBB-beta, and two SFBB-gamma sequences) SFBB genes (Sassa et al., 2007). Nevertheless, positively selected amino acid sites have not been identified, mainly due to the small sample size. Therefore, it is not known how these amino acids vary among SFBB genes. Hence, it has been suggested that all SFBB genes could act together as the pollen determinant (Sassa et al., 2007). Under the non-self recognition by multiple factors, the high intra-haplotypic diversity of SFBB is the result of natural selection favouring diversification of SFBB genes within an S haplotype (Kakui et al., 2011). Under this model, no strong evidence for positive selection is expected when individual SFBB genes are considered, as observed for Pyrus SFBB-gamma (Vieira et al., 2009; De Franceschi et al., 2011a), SFBB-alpha, SFBB-beta, SFBB-delta, and SFBB-epsilon genes (De Franceschi et al., 2011a).
Another line of evidence in support of Japanese pear GSI non-self recognition by a multiple factors rejection mechanism comes from analyses of loss of function of two SFBB genes on SI phenotypes (Kakui et al., 2011). In the mutant haplotype S 4sm (a mutant derived from the S4 haplotype that shows stylar self-compatibility because of deletion of S4-RNase; Okada et al., 2008), deletion of the SFBB1 gene specifically affects recognition of S1-RNase (Okada et al., 2008). Nevertheless, no effect of loss of function of the SFBB1 gene from the S5 haplotype was observed on the SI phenotype in the pistils with S1-, S2-, S3-, S4-, and S9-RNases (Kakui et al., 2011, and references therein). Moreover in the S1 haplotype, the S1 pollen is rejected and the SFBB1 gene from the S1 haplotype is not truncated. Therefore, the S1-RNase is not always targeted by the SFBB1 gene for degradation and other factors are involved in detoxification of the S1-RNase (Kakui et al., 2011).
As genes determining GSI specificity are under frequency-dependent selection, they are maintained for long periods of time (Wright, 1939). In Pyrinae, the oldest S-RNase gene specificity lineage is about 23 million years old (Vieira et al., 2010). Therefore, it is not surprising that most of the S-RNase allele lineages are found in Malus, Pyrus, Sorbus, and Crataegus species (Vieira et al., 2010). In this work, we showed that all SFBB genes described in Malus and Pyrus are also present in Sorbus aucuparia. Phylogenetic inferences of Pyrinae SFBB genes suggested the presence of 16 S. aucuparia SFBB genes. The age of the SFBB duplications also supported the involvement of these genes in GSI. Furthermore, amino acids under positive selection—those that could be involved in specificity determination—were identified when intra-haplotype SFBB genes were analysed.
Material and methods
Plant material and DNA extraction
S. aucuparia is a self-incompatible species that has been characterized at the molecular level for the S-RNase gene (Raspé and Kohn, 2002, 2007). Due to the ecology of this bird-dispersed, insect-pollinated species, little population structure is found (Raspé et al., 2000; Raspé and Kohn, 2007). Leaves were collected from seven individuals from a natural population located in Bragança, Portugal (assigned as B). Furthermore, for the segregation experiments, 74 individuals from the progeny of the cross between individual Belgium5 (S17-RNase/S20-RNase) and Belgium6 (S2-RNase/S10-RNase) (Raspé and Kohn, 2007), assigned as D, were used. Genomic DNA was extracted from leaves of individual plants using the method of Ingram et al. (1997) or the Puregene® DNA Purification System (Gentra Systems, Minneapolis, MN, USA). No specific permits were required for the field collection, as the plant location was not privately owned or protected, and S. aucuparia is not an endangered or protected species.
Amplification of S-RNases
Three primer combinations (SorbusRNaseF and SorbusRNaseR, S-RNaseFT-F and S-RNase(I/T)W-R, and MaCiF1+ and Mac2/3R1+) were used for the amplification of S-RNases (Supplementary Table S1 at JXB online). Genomic DNA from individuals B2, B4, B5, B6, B8, B10, and B13 was used as template. Standard amplification conditions were 35 cycles of denaturation at 94 °C for 30 s, primer annealing according to Supplementary Table S1 for 30 s, and primer extension at 72 ºC for 3min. All amplification products were cloned using a TA Cloning kit (Invitrogen, Carlsbad, CA, USA). For each individual and amplification product, the insert of an average of 20 colonies was cut separately with RsaI, AluI, AvaII, and Sau3AI restriction enzymes. For each individual and restriction pattern, one colony was sequenced. For those sequences that showed similarity to the S-RNase gene when using BLASTn, two more colonies were sequenced in order to obtain a consensus sequence. An ABI PRISM BigDye Cycle Sequencing kit (Perkin Elmer, Foster City, CA, USA) and specific primers, or the primers for the M13 forward and reverse priming sites of the pCR2.1 vector, were used to prepare the sequencing reactions. Sequencing runs were performed by STABVIDA (Lisboa, Portugal).
Amplification of SFBB genes
Primers SFBBgenF and SFBBgenR (Supplementary Table S1) were designed on the basis of the 12 SLF and SFBB sequences of Cheng et al. (2006) and Sassa et al. (2007) (SFBB3α, AB270795; SFBB9α, AB270793; SFBB9β, AB270794; SFBB3β, AB270796; SLF1, DQ422810; SLF2, DQ422811; SFBB4α, AB270797; SFBB5α, AB270800; SFBB5β, AB270801; SFBB4β, AB270798; SFBB4γ, AB270799; SFBB5γ, AB270802). These primers, although designed based on a small set of sequences, are, however, present in 65.5% of Malus and Pyrus SFBB sequences available in GenBank (n=165, see Fig. 1 and legend for accession numbers). Genomic DNA from individuals B5 and B6 was used as template. Standard amplification conditions were 35 cycles of denaturation at 94 °C for 30 s, primer annealing at 48 ºC for 30 s, and primer extension at 72 °C for 2 min. The amplification products were cloned as described above. For each individual and amplification product, the insert of an average of 100 colonies was cut separately with RsaI, AluI, AvaII, and Sau3AI restriction enzymes. Sequencing reactions were performed as described above.
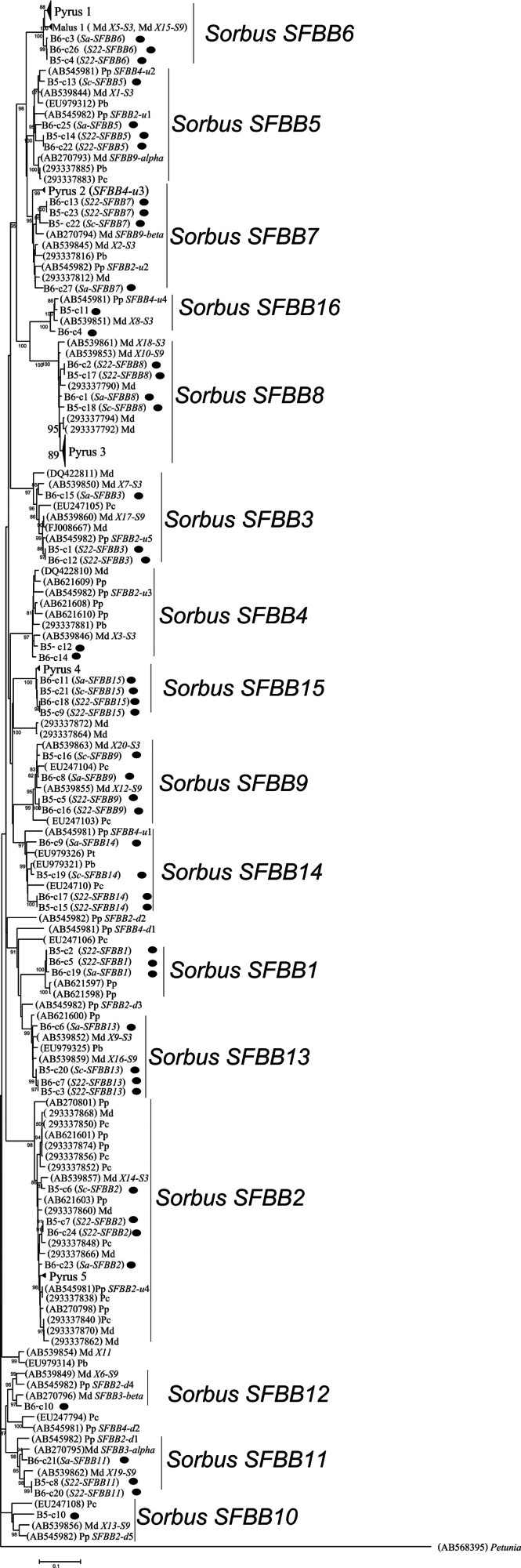
Fig. 1.
Maximum parsimony tree showing the relationship of the Pyrinae SFBB genes. Md, Malus domestica; Pp, Pyrus pyrifolia; Pb, Pyrus bretschneideri; Pu, Pyrus ussuriensis; Ps, Pyrus sinkiangensis; Pc, Pyrus communis; Sa, Sorbus aucuparia (sequences are indicated by a filled circle). Bx-cx represents a Bragança S. aucuparia population (B) and the individual name (x), with cx representing the colony type. Numbers below the branches represent bootstrap values above 60. Genes of known location in Malus are: Md-X1, Md-X2, Md-X3, Md-X5, Md-X7, Md-X8, Md-X9, Md-X14, Md-X18, Md-X20, MdSFBB3α, and MdSFBB3β for the haplotype S3; and Md-X6, Md-X10, Md-X12, Md-X13, Md-X15, Md-X16, Md-X17, Md-X19, MdSFBB9α, and MdSFBB9β for the haplotype S9 (Sassa et al., 2007; Minamikawa et al., 2010). Genes of known location in Pyrus are: PpSFBB4-d1, PpSFBB4-d2, and PpSFBB4-u1–PpSFBB4-u4 for haplotype S4; and PpSFBB2-d1–PpSFBB2-d5, and PpSFBB2-u1–PpSFBB2-u5 for haplotype S5 (Okada et al., 2011). GenBank numbers are: Pyrus 1 sequences: AB621615, 293337907, EU979310, EU979317, AB621617, EU979315, EU979311, EU979313, 293337889, 293337854, AB270800, AB621616, 293337905, 293337893, 293337887, EU979316, 293337891, AB270797, 293337911, 293337895, EU979309, 293337909; Pyrus 2 sequences: 293337814, 293337810, AB545981, 293337808, 293337806, 293337804; Pyrus 3 sequences: 293337784, 293337782, 293337780, 293337786, 293337778, EU422961, EU422960, 293337788, 293337776, EU081892, EU422958, AB270799, 293337802, EU081890, AB297937, EU422956, AB297935, 293337798, EU418249, EU979327, AB297939, AB297934, AB297938, EU081887, AB297933, 293337796, EU418248, AB297940, AB270802, EU081894, 293337800, EU081891, AB297936, EU081893, EU422959, EU422962; Pyrus 4 sequences: EU979319, EU979320, EU979323, 293337878, 293337844, EU979324. Pyrus 5 sequences: AB621602, 293337876, 293337858, 293337846, 293337842; Malus 1 sequences: 293337901, 293337897, AB539848, 293337903, 293337899, AB539858, FJ008668.
S. aucuparia SFBB1–SFBB3, SFBB5–SFBB9, SFBB11, and SFBB13–SFBB15 genes of the S22 and Sa haplotypes
Genomic DNA of the seven B individuals was used to amplify the SFBB1–SFBB3, SFBB5–SFBB9, SFBB11, and SFBB13–SFBB15 genes using specific primers. For all genes except SFBB6, based on the sequences obtained for B5 and B6 individuals, single-nucleotide polymorphisms were used to find restriction fragment length polymorphisms (RFLPs) that allowed the identification of the S22- and Sa- alleles of these 11 SFBB genes (Supplementary Table S2 at JXB online). For the SFBB6 gene, the amplification product of each individual was cloned. For each individual, ten random colonies were sequenced. DNA sequencing was performed as described above. These sequences have been deposited in GenBank (accession numbers KC701664–KC701673).
S. aucuparia SFBB1–SFBB3, SFBB5–SFBB9, SFBB11, and SFBB13–SFBB15 allele sequences in individuals from the progeny of the Belgium5 (S17-RNase/S20-RNase) and Belgium6 (S2-RNase/S10-RNase) cross
For 12 of the 16 SFBB genes studied, we were able to infer the allele that went with both the S22-RNase and the Sa-RNase (see Results). Nevertheless, we wanted to show that these 12 SFBB genes were located in the S-locus region. Therefore, the 74 individuals (assigned as D) from the progeny of the cross between individuals Belgium5 (S17-RNase/S20-RNase) and Belgium6 (S2-RNase/S10-RNase) were genotyped using specific primers (Supplementary Table S1) for the four segregating S-RNase alleles. For each of the 12 SFBB genes, the amplification product obtained using specific primers (Supplementary Table S1) and genomic DNA of individuals D28 (S2-RNase/S20-RNase), D34 (S10-RNase/S17-RNase), D69 (S10-RNase/S20-RNase), and D82 (S2-RNase/S17-RNase) was cloned. These individuals showed the four possible S-RNase combinations, and therefore all segregating SFBB alleles must be present in this sample. For each gene, ten randomly chosen colonies were sequenced to identify the SFBB alleles that were segregating in this cross. For SFBB15, only one allele was identified that was present in all four individuals analysed. Therefore, this gene was not studied further. For the SFBB3, SFBB5–SFBB9, and SFBB14 genes, all four alleles were identified (Supplementary Table S3 at JXB online). For SFBB1 and SFBB11, three alleles were identified, and for SFBB2 and SFBB13, two alleles were identified (Supplementary Table S3). The DNA sequences have been deposited in GenBank (accession numbers KC701674–KC701712). For these genes, in order to determine which individuals had a given SFBB allele, specific primers, as well as RFLPs, were developed (Supplementary Table S3). Because alleles of these genes had low levels of diversity, it was often not possible to develop a diagnostic marker for all observed alleles.
Phylogenetic analyses, summary statistics, and testing for positive selection of the S. aucuparia SFBB genes
The DNA sequences were deposited in GenBank (accession numbers KC701614–KC701663). Translated amino acid sequences were aligned using the accurate CLUSTALW algorithm as implemented in DAMBE (Xia and Xie, 2001). This amino acid alignment was used as a guide to obtain the corresponding nucleotide alignment. Analyses of DNA polymorphisms were performed using DnaSP (version 4.1) (Rozas et al., 2003). Using 216 SFBB sequences, minimum evolutionary trees were built with MEGA5 (Tamura et al., 2011), using CNI (level=1) and complete deletion. For the identification of sites under positive selection, we used ADOPS (Reboiro-Jato et al., 2012) and two datasets of 11 SFBB gene sequences that showed linkage with the S22-RNase and Sa-RNase. We compared the M2–M1 and M8–M7 models. We only considered as positively selected those amino acid sites that showed a probability >90% for both naive empirical Bayes (NEB) and Bayes empirical Bayes (BEB) methods, and that were identified in at least two of the three alignment methods used. Fourteen divergent F-box sequences from Fig. 1 in Vieira et al. (2009) (Medicago truncatula GI61806856; Antirrhinum hispanicum GI38229882; Petunia integrifolia GI162134184; Populus trichocarpa GI158749689, GI167963539, GI159885773, GI116734897, GI159647948, GI116734897; Malus domestica GI90103253; Prunus avium GI33354144; Oryza sativa GI115487495; and Arabidopsis thaliana GI30692063, GI18404533) were also used. Moreover, we used 16 Malus domestica F-box sequences with known location that are not on chromosome 17 (the location of the S-locus region), obtained from BLASTp at the Genome Database for Rosaceae (http://www.rosaceae.org), using Md SFBB3-beta (BAF47180) as the query.
Results
S. aucuparia S-RNase genes
Because of the S-RNase intron size variation, the amplification products of the seven individuals analysed varied from 505 to 1819bp (Table 1). Individuals B2, B5, and B6 had the same S-RNase allele, which was identical in the coding region to the S. aucuparia S22-RNase (EF494760). Individuals B2 and B13 had the S. aucuparia S21-RNase allele (EF494759), whilst individual B10 had the S26-RNase allele (EF494764). Individuals B4, B6, and B8 had one S-RNase allele, called Sa-RNase, which shared 99% amino acid identity with Malus domestica Sf-RNase (D50837) and Pyrus pyrifolia S12-RNase (AB426604). Individuals B5 and B13 presented the same S-RNase allele, called Sc-RNase, which shared 98% amino acid identity with Malus domestica S10-RNase (AF239809). The other allele of individual B8 was S20-RNase (AF504272). For individuals B4 and B10, although three different primer combinations were used and all amplification products obtained were cloned and sequenced, only one S-RNase allele could be characterized.
Table 1.
S-RNase haplotypes in the studied individuals NA, Not applicable.
| Individuals | S-RNases | |
|---|---|---|
| B2 | S21-RNase (523bp) | S22-RNase (505bp) |
| B4 | Sa-RNase (684bp) | NA |
| B5 | S22-RNase (505bp) | Sc-RNase (510bp) |
| B6 | S22-RNase (505bp) | Sa-RNase (684bp) |
| B8 | Sa-RNase (684bp) | S20-RNase (1819bp) |
| B10 | S26-RNase (721bp) | NA |
| B13 | Sc-RNase (510bp) | S21-RNase (523bp) |
| Belgium5a | S17-RNase | S20-RNase |
| Belgium6a | S2-RNase | S10-RNase |
a According to Raspé and Kohn 2007.
SFBB genes in S. aucuparia
For B5 and B6 individuals, both having in common the S22-RNase allele, the 900bp (expected size) amplification product obtained using primers SFBBgenF and SFBBgenR revealed 23 (called B5c1–B5c23) and 27 (B6c1–B6c27) sequences, respectively. Fig, 1 shows the phylogenetic relationship of the 50 S. aucuparia SFBB sequences and the 165 available Malus and Pyrus SFBB–SLF sequences. As S. aucuparia is a diploid species (Castroviejo and Real Jardín Botánico, 1986), the presence of more than two sequences from the same individual means that sequences from different genes are being amplified. Thus, the 27 sequences obtained for B6 individual implied at least 14 genes in S. aucuparia. Nevertheless, using the phylogenetic position of the S. aucuparia sequences and the genes of known location in Malus (Sassa et al., 2007; Minamikawa et al., 2010) and Pyrus (Okada et al., 2011), 16 genes could be considered (Fig. 1). S. aucuparia SFBB genes were present in Malus and Pyrus, thus predating the appearance of these genera (Fig. 1). These 16 SFBB genes in Malus and Pyrus have been shown to be expressed in pollen only (Sassa et al., 2007; Minamikawa et al., 2010; De Franceschi et al., 2011a , b ). Given that the average K s between the Petunia and Sorbus SFBB genes is 1.577 and Solanaceae and Rosaceae have been diverging for 106 million years (Wikstrom et al., 2001), the two most closely related SFBB genes were 8.27 million years old (Table 2).
Table 2.
Synonymous (above the diagonal) and non-synonymous (below the diagonal) divergence levels in S. aucuparia SFBB genes. Synonymous and non-synonymous diversity is presented in the diagonal. The estimated age, in million years, for the SFBB genes split is given in square brackets. –, Only one sequence is available.
| SFBB1 | SFBB2 | SFBB3 | SFBB4 | SFBB5 | SFBB6 | SFBB7 | SFBB8 | SFBB9 | SFBB10 | SFBB11 | SFBB12 | SFBB13 | SFBB14 | SFBB15 | SFBB16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SFBB1 | 0.0036 | 0.2322 | 0.2713 | 0.2495 | 0.2351 | 0.2321 | 0.2480 | 0.3218 | 0.2592 | 0.3199 | 0.2274 | 0.2100 | 0.1231 | 0.2207 | 0.2345 | 0.3140 |
| 0.0071 | [15.61] | [18.24] | [16.77] | [15.80] | [15.60] | [16.67] | [21.63] | [17.42] | [21.50] | [15.28] | [14.12] | [8.27] | [14.83] | [15.76] | [21.11] | |
| SFBB2 | 0.2090 | 0.0452 | 0.2535 | 0.2424 | 0.2427 | 0.2175 | 0.2299 | 0.3167 | 0.2462 | 0.3565 | 0.2485 | 0.2328 | 0.2129 | 0.2142 | 0.2369 | 0.3129 |
| 0.0146 | [17.04] | [16.29] | [16.31] | [14.62] | [15.45] | [21.29 | [16.55] | [23.96] | [16.70] | [15.65] | [14.31] | [14.40] | [15.92] | [21.03] | ||
| SFBB3 | 0.2027 | 0.1916 | 0.0425 | 0.2134 | 0.2111 | 0.2052 | 0.1917 | 0.2752 | 0.2252 | 0.2526 | 0.2213 | 0.1923 | 0.2423 | 0.2095 | 0.1631 | 0.2763 |
| 0.0166 | [14.34] | [14.19] | [13.79] | [12.89] | [18.50] | [15.14] | [16.98] | [14.87] | [12.93] | [16.29] | [14.08] | [10.96] | [18.57] | |||
| SFBB4 | 0.1928 | 0.1720 | 0.1472 | 0.0326 | 0.2355 | 0.2192 | 0.1885 | 0.2702 | 0.2207 | 0.2711 | 0.2183 | 0.1865 | 0.2345 | 0.2185 | 0.1801 | 0.2885 |
| 0.0187 | [15.83] | [14.73] | [12.67] | [18.16] | [14.83] | [18.22] | [14.67] | [12.54] | [15.76] | [14.69] | [12.11] | [19.39] | ||||
| SFBB5 | 0.1884 | 0.1894 | 0.1765 | 0.1659 | 0.0598 | 0.1517 | 0.1382 | 0.2651 | 0.2690 | 0.2770 | 0.2207 | 0.1773 | 0.2214 | 0.2168 | 0.1780 | 0.2585 |
| 0.0244 | [10.20] | [9.29] | [17.82] | [18.08] | [18.62] | [14.83] | [11.92] | [14.88] | [14.57] | [11.96] | [17.38] | |||||
| SFBB6 | 0.2119 | 0.1959 | 0.1800 | 0.1911 | 0.0705 | 0.0124a | 0.1437 | 0.2579 | 0.2689 | 0.3033 | 0.2189 | 0.1801 | 0.2149 | 0.1870 | 0.1762 | 0.2445 |
| 0.0034(N=13) | [9.66] | [17.34] | [18.07] | [20.39] | [14.71] | [12.11] | [14.44] | [12.57] | [11.84] | [16.43] | ||||||
| SFBB7 | 0.1776 | 0.1782 | 0.1619 | 0.1689 | 0.0671 | 0.0843 | 0.0641 | 0.2501 | 0.2276 | 0.2380 | 0.1994 | 0.1533 | 0.2275 | 0.2078 | 0.1631 | 0.2611 |
| 0.0331 | [16.81] | [15.30] | [16.00] | [13.40] | [10.30] | [15.29] | [13.97] | [10.96] | [17.55] | |||||||
| SFBB8 | 0.2611 | 0.2570 | 0.2039 | 0.2002 | 0.1778 | 0.2092 | 0.1796 | 0.0305 | 0.3208 | 0.3279 | 0.2718 | 0.2331 | 0.3016 | 0.2594 | 0.2263 | 0.2459 |
| 0.0114 | [21.56] | [22.04] | [18.27] | [15.67] | [20.27] | [17.44] | 15.21] | [16.53] | ||||||||
| SFBB9 | 0.1709 | 0.1688 | 0.1492 | 0.1215 | 0.1755 | 0.1916 | 0.1664 | 0.2056 | 0.0458 | 0.2781 | 0.2376 | 0.2147 | 0.2256 | 0.2137 | 0.1827 | 0.3174 |
| 0.0110 | [18.69] | [15.97] | [14.43] | [15.16] | [14.36] | [12.28] | [21.33] | |||||||||
| SFBB10 | 0.1848 | 0.1709 | 0.1548 | 0.1455 | 0.1768 | 0.1992 | 0.1721 | 0.2182 | 0.1473 | – | 0.2824 | 0.2417 | 0.2901 | 0.2568 | 0.2376 | 0.3238 |
| [18.98] | [16.25] | [19.50] | [17.26] | [15.97] | [21.76] | |||||||||||
| SFBB11 | 0.1822 | 0.1811 | 0.1677 | 0.1364 | 0.1711 | 0.1918 | 0.1729 | 0.2250 | 0.1378 | 0.1161 | 0.0646 | 0.1658 | 0.1970 | 0.2308 | 0.1699 | 0.2755 |
| 0.0210 | [11.14] | [13.24] | [15.51] | [11.42] | [18.52] | |||||||||||
| SFBB12 | 0.1677 | 0.1712 | 0.1522 | 0.1290 | 0.1572 | 0.1674 | 0.1540 | 0.1970 | 0.1349 | 0.1155 | 0.0844 | – | 0.2046 | 0.2044 | 0.1437 | 0.2756 |
| [13.75] | [13.74] | [9.66] | [18.52] | |||||||||||||
| SFBB13 | 0.1080 | 0.1808 | 0.1715 | 0.1503 | 0.1656 | 0.1871 | 0.1569 | 0.2285 | 0.1344 | 0.1530 | 0.1561 | 0.1375 | 0.0341 | 0.2129 | 0.2036 | 0.3092 |
| 0.0111 | [14.31] | [13.69] | [20.78] | |||||||||||||
| SFBB14 | 0.1844 | 0.1654 | 0.1405 | 0.1315 | 0.1517 | 0.1802 | 0.1671 | 0.2032 | 0.1195 | 0.1534 | 0.1563 | 0.1333 | 0.1508 | 0.0635 | 0.1617 | 0.2556 |
| 0.0247 | [10.87] | [17.18] | ||||||||||||||
| SFBB15 | 0.1797 | 0.1873 | 0.1718 | 0.1446 | 0.1780 | 0.1895 | 0.1687 | 0.2224 | 0.1356 | 0.1793 | 0.1705 | 0.1556 | 0.1616 | 0.1263 | 0.0102 | 0.2256 |
| 0.0028 | [15.16] | |||||||||||||||
| SFBB16 | 0.2227 | 0.2036 | 0.1790 | 0.1966 | 0.1641 | 0.1952 | 0.1698 | 0.1168 | 0.1879 | 0.1764 | 0.1946 | 0.1703 | 0.1977 | 0.1669 | 0.1803 | 0.0654 |
| 0.0136 |
a Diversity levels calculated from a larger data set indicated in brackets.
Although the levels of polymorphism were low (Table 2), for the SFBB1–SFBB3, SFBB5, SFBB7–SFBB9, SFBB11, SFBB13, and SFBB14 genes RFLPs were obtained that allowed identification of the sequences of the S22 and Sa haplotypes (see Materials and method, and Supplementary Table S2). For these genes, the allele assigned as S22 was present in individuals B2, B5, and B6 (all having S22-RNase), but not in individuals B4, B8, B10, and B13 (Table 1). Furthermore, for SFBB1–SFBB3, SFBB5, SFBB7– SFBB9, SFBB11, SFBB13, and SFBB14, the allele assigned as Sa was only present in individuals B4, B6, and B8 that presented Sa-RNase (Table 1). For the SFBB6 gene, the sequences obtained with specific primers revealed that the S22-allele was only present in B2, B5, and B6 individuals, and the Sa-allele was only present in B4, B6, and B8 individuals only.
S. aucuparia SFBB1–SFBB3, SFBB5–SFBB9, SFBB11, SFBB13, and SFBB14 genes are located in the S-locus region
As described in Material and methods, these genes were sequenced using specific primers (Supplementary Table S1) in individuals that had S2-RNase, S10-RNase, S17-RNase, and S20-RNase. These sequences were used to develop specific RFLP markers (Supplementary Table S3) or specific primers for a particular SFBB allele. These markers were used in the segregation analyses of these genes in 74 individuals from the progeny of the cross between individuals Belgium5 (S17-RNase/S20-RNase) and Belgium6 (S2-RNase/S10-RNase). For all 11 genes, we found linkage of a particular SFBB allele with a specific S-RNase allele (Table 3). Therefore, in Sorbus, these genes are located in the S-locus region.
Table 3.
Segregation analyses of S. aucuparia SFBB1–SFBB3, SFBB5–SFBB9, SFBB11, SFBB13, and SFBB14.
| Gene | Allele | S2-RNase (N=39) | S10-RNase (N=35) | S17-RNase (N=34) | S20-RNase (N=40) |
|---|---|---|---|---|---|
| SFBB1 | S2-SFBB1 | 39 | 0 | 0 | 0 |
| SFBB2 | S10-SFBB2 | 0 | 35 | 0 | 0 |
| S20-SFBB2 | 0 | 0 | 0 | 40 | |
| SFBB3 | S17-SFBB3 | 0 | 0 | 34 | 0 |
| S20-SFBB3 | 0 | 0 | 0 | 40 | |
| SFBB5 | S2-SFBB5 | 39 | 0 | 0 | 0 |
| S20-SFBB5 | 0 | 0 | 0 | 40 | |
| SFBB6 | S2-SFBB6 | 39 | 0 | 0 | 0 |
| S17-SFBB6 | 0 | 0 | 34 | 0 | |
| SFBB7 | S10-SFBB7 | 0 | 35 | 0 | 0 |
| S17-SFBB7 | 0 | 0 | 34 | 0 | |
| S20-SFBB7 | 0 | 0 | 0 | 40 | |
| SFBB8 | S2-SFBB8 | 39 | 0 | 0 | 0 |
| S17-SFBB8 | 0 | 0 | 34 | 0 | |
| S20-SFBB8 | 0 | 0 | 0 | 40 | |
| SFBB9 | S2-SFBB9 | 39 | 0 | 0 | 0 |
| S10-SFBB9 | 0 | 35 | 0 | 0 | |
| S17-SFBB9 | 0 | 0 | 34 | 0 | |
| S20-SFBB9 | 0 | 0 | 0 | 40 | |
| SFBB11 | S10-SFBB11 | 0 | 35 | 0 | 0 |
| S17-SFBB11 | 0 | 0 | 34 | 0 | |
| S20-SFBB11 | 0 | 0 | 0 | 40 | |
| SFBB13 | S2-SFBB13 | 39 | 0 | 0 | 0 |
| S10-SFBB13 | 0 | 35 | 0 | 0 | |
| SFBB14 | S2-SFBB14 | 39 | 0 | 0 | 0 |
| S10-SFBB14 | 0 | 35 | 0 | 0 | |
| S17-SFBB17 | 0 | 0 | 34 | 0 | |
| S20-SFBB20 | 0 | 0 | 0 | 40 |
Positively selected amino acid sites in 11 S. aucuparia SFBB genes from the S22 and Sa haplotypes
Adaptive evolution is likely to act on a small subset of amino acids, and thus average substitution rates across the gene may not indicate positive selection (Yang and Bielawski, 2000). Under the non-self recognition by multiple factors system, allelic products of the pollen S gene would be highly homologous with each other and would be expected to target similar fractions of S-RNases because allelic divergence of each type of pollen S is not required and would not be favoured by natural selection (Kakui et al., 2011). Fixation of duplicated genes is an adaptive event, and these duplicated genes can act as a source of protein subfunctionalization by evolution of key positions in the protein (Hurles, 2004). In the case of SFBB genes, protein subfunctionalization would imply a change in the amino acids that determine the specificity recognition of S-RNases. SFBBs would thus be a positively evolving gene family. In fact, the high intra-haplotypic K a values of SFBB (for the S22 haplotype, K a is 0.172, and for the Sa haplotype, K a is 0.161) suggest that natural selection has favoured diversification of SFBB paralogues to target allelic variants of S-RNases in Pyrinae (Kakui et al., 2011). Adaptive evolution of duplicated paralogous gene families using CodeML (Yang, 1997) has been identified at the genome level to identify genes subject to positive selection (Emes and Yang, 2008). The amino acid sites identified as being positively selected with a posterior probability higher than 90% for the 11 SFBB genes of the S22 and Sa haplotypes, using the Yang (1997) method as implemented in ADOPS (Reboiro-Jato et al., 2012), are shown in Fig. 2. There were 12 amino acid sites identified as being positively selected common to the S22 and Sa haplotypes. Amino acid sites at positions 179 and 197 of the Sa haplotype were, however, assigned as positively selected with probabilities higher than 91 and 83% in BEB, and 75 and 71% in NEB, respectively. It should be noted that, within each haplotype, the combination of these amino acid sites was different for every Sorbus SFBB gene analysed, and the minimum number of differences between any two SFBB genes from the same haplotype at these amino acid sites was six (between SFBB9 and SFBB14 at the S22 haplotype; Fig. 2). As expected, in the F-box region, there were no amino acid sites showing evidence for positive selection.
Fig. 2.
Schematic representation of the amino acid sites identified as positively selected (shaded) when using at least two different alignment algorithms and the method of Yang (1997) implemented in ADOPS (Reboiro-Jato et al., 2012) with a probability higher than 90 and 95% (bold) in both NEB (naive empirical Bayes) and BEB (Bayes empirical Bayes) and the 11 S. aucuparia SFBB genes of the S22 and Sa haplotype.
When comparing the amino acid sites identified as positively selected between the two S haplotypes, for the SFBB1, SFBB2, SFBB5, SFBB6, SFBB8, and SFBB13 genes, there are no differences (Supplementary Table S4 at JXB online). For the SFBB9 gene, at these amino acid sites there was one amino acid difference between the two S haplotypes. Two amino acid differences at these amino acid sites were observed in the SFBB11 gene between the two S haplotypes. Four amino acid differences at the amino acid sites identified as positively selected were observed at the SFBB3 and SFBB4 genes between the S22 and Sa haplotypes.
Discussion
In S. aucuparia, there are at least 16 SFBB genes. For these genes, Malus and Pyrus orthologues have been described, for which expression has been shown to be pollen restricted (Sassa et al., 2007; Minamikawa et al., 2010; De Franceschi et al., 2011a , b ). Indeed, all Pyrinae SFBB-like sequences described so far are expressed in pollen only. Although the 11 SFBB genes here studied were in linkage with the S-RNase, not all Malus and Pyrus SFBB genes are in linkage with the S-RNase gene (De Franceschi et al., 2011a ; Kakui et al., 2011), and thus these genes are probably not involved in determining S pollen specificity (see review by Minamikawa et al., 2010; De Franceschi et al., 2011a ; Kakui et al., 2011; Okada et al., 2011).
The characterization of a large number of genes in one species, as performed here, will help to establish gene orthologies in different species using a phylogenetic approach. It should be noted that synteny alone cannot be used to establish orthologies, as gene order is not conserved among S haplotypes (Minamikawa et al., 2010; Okada et al., 2011). Because polymorphism levels at SFBB genes were always below 10% (Table 2, and Table 1 in Kakui et al., 2011), this can also be used as a guide for the presence of multiple genes within species. Indeed, the exception of the less than 10% diversity reported for the Pyrus SFBB1 gene (which includes PpSFBB4-d1 and PpSFBB2-d3 sequences; Kakui et al., 2011) is due to the inclusion of two different genes (Fig. 1). The PpSFBB4-d1 sequence is deleted in the mutant haplotype S 4sm (Okada et al., 2008) and specifically affects recognition of the S1-RNase. The non-functional gene (named SFBB1-like) of the S5 haplotype, which shows no effect in the S5 pollen phenotype when crossed with plants having S1-RNase pistils (Kakui et al., 2011), may thus represent a different gene from PpSFBB4-d1.
As expected for genes determining GSI specificity, the SFBB genes predate the appearance of the Malus, Pyrus, and Sorbus genera. Species from these genera may, however, have diverged in the last 5 million years (under the assumption of a molecular clock for five genes; Table 4). It should be noted that this interpretation is far from being consensual. Indeed, when using information from both the fossil record and molecular data, Campbell et al. (2007) suggested that the genera Malus, Pyrus, and Sorbus, among others, are the result of an ancient, rapid radiation associated with a low mutation rate (as discussed by Vieira et al., 2010). The two most closely related SFBB genes are 8.27 million years old (Table 2). On the other hand, the two most divergent SFBB genes are 23.96 million years old (Table 2). This age is, as expected, similar to the age of the oldest Pyrinae S-RNase specificity lineages (about 23 million years old; Vieira et al., 2010). This implies that the genes identified here do not have a Prunus orthologue (under the assumption of 32 million years for the age of the split between the Amygdaloideae and Pyrinae lineages; Wikstrom et al., 2001).
Table 4.
Average silent site divergence and estimated age in million of years (within brackets) for Pyrinae species.
| Species comparison | Genes | Average | |||||
|---|---|---|---|---|---|---|---|
| trnL–trnF | rbcL | matK1 | 5–8S ribosomal RNA | rpoC1 | |||
| Sorbus | Malus | 0.0050 (1.63) | 0.0 | 0.0120 (4.03) | 0.0643 (9.87) | 0.0201 (8.0) | 5.88 |
| Pyrus | 0.0140 (4.57) | 0.0071 (2.25) | 0.006 (2.01) | 0.0769 (11.81) | 0.0 | 5.16 | |
| Malus | Pyrus | 0.0130 (4.25) | 0.0071 (2.25) | 0.0181 (6.08) | 0.0718 (11.02) | 0.0201 (8. 0) | 6.32 |
| Pyrinae | Prunus | 0.09797 (32a) | 0.1009 (32a) | 0.0953 (32a) | 0.2084 (32a) | 0.0804 (32a) | 32 |
a The split between the Amygdaloideae and Pyrinae lineages has been estimated to be between 29 and 35 million years ago (Wikstrom et al., 2001); thus, we use the average of these values (32).
At the S-RNase gene, 18 S. aucuparia alleles have been characterized (Raspé and Kohn, 2007, and this work), but 40 different specificities have been inferred (Raspé and Kohn, 2007). Furthermore, using a phylogenetic approach, 35 S-RNase specificities have been estimated in Pyrinae (Vieira et al., 2010). Although we cannot be sure that all Sorbus SFBB genes have been characterized, since the approach used here depended on primer specificity, the 16 Sorbus SFBB genes clustered with high support with all SFBB genes described in Malus, and 13 out of 16 with Pyrus sequences obtained from the characterization of BAC libraries using different primers. The three exceptions (Pp SFBB2-d2, PpSFBB2-d3, and PpSFBB4-d1; Fig. 1) may represent Sorbus SFBB genes that have not been characterized, or, genes that recognize S-RNase specificities that have been lost in S. aucuparia. Furthermore, we are assuming that the 16 genes may be involved in pollen GSI specificity, although we have evidence of linkage with the S-RNase for 11 genes only. Nevertheless, the number of S-RNase specificities seems to be higher than the number of SFBB genes. This is expected, as, under the non-self recognition by multiple factors rejection model, each S-RNase can be targeted by multiple SFBB genes (Kubo et al., 2010; Wang and Kao, 2011). Moreover, natural selection favours diversification of SFBB genes within an S haplotype (Kakui et al., 2011). When the 11 Sorbus SFBB genes of the same S haplotype were analysed, amino acids that were associated with recognition of the S-RNase specificities—those under positive selection—were observed using codon models. Because selection acts at the gene level, it was not surprising that the same amino acid sites under positive selection were identified in the two S haplotypes (the 11 SFBBs were the same). The amino acid sites under positive selection could, however, represent different SFBB protein functions. Although there is no functional data for most plant F-box genes, phylogenetic divergent F-box genes, in principle, are associated with different functions. Therefore, to test whether the identified positively selected amino acid sites reflected different functions, we selected 14 F-box sequences from the seven divergent groups presented in Fig. 1 in Vieira et al. (2009), most of which were not expected to be involved in GSI. No amino acid sites under positive selection were identified in the region analysed here. An identical result was obtained when only Malus domestica F-box genes, not located in the S-locus region (chromosome 17), were analysed. Thus, the amino acid sites under positive selection identified in the Sorbus SFBB sequences may represent amino acids involved in specificity determination.
The variability at the amino acid sites under positive selection within SFBB genes may account for the differences observed in SI behaviour when different alleles of the same SFBB gene are analysed. The presence of identical amino acids at the positively selected sites between different S haplotypes, in the same SFBB gene, implies that alleles of these genes are recognizing the same S-RNase specificities and marking them for degradation. This was the case for six of the 11 Sorbus SFBB genes analysed. The S22 and Sa haplotypes were from the same population, and thus they must recognize as non-self the same spectrum of S-RNases, except for Sa- and S22-RNase. In a model of one SFBB gene–one S-RNase specificity, we would expect to find, between the two S haplotypes, differences in one SFBB gene only at the amino acid sites under positive selection. There were, however, five SFBB genes that showed differences at the amino acids under positive selection. This again suggests that one SFBB gene can recognize more than one S-RNase specificity. In Petunia, co-immunoprecipitation experiments and transgenic approaches have shown that the SLF1 gene recognized S17-RNase as non-self in four S haplotypes analysed, but only recognized S9-RNase as non-self in two S haplotypes (Kubo et al., 2010). Thus, one SFBB gene of an S haplotype can interact with two or more S-RNases (Kubo et al., 2010; Wang and Kao, 2011). The same was observed when the Pyrus SFBB1 gene was deleted (see Introduction), although another interpretation for this result may be possible (see above). At present, predictions about recognition of an S-RNase specificity by a particular SFBB gene are very difficult to test as neither transformation nor antisense RNA technology used to induce post-transcriptional gene silencing (Lopez-Gomollon and Dalmay, 2010) are available for these species or for this system. Furthermore, the Pyrinae species studied are shrubs or trees that have a minimum 2–3 years juvenile period following planting (Shulaev et al., 2008). Therefore, at present, only characterization of truncated SFBB genes can be used to confirm these predictions. Nevertheless, as the plants having these truncated SFBB gene sequences show no specific phenotype (the SI phenotype will be different only in the presence of the S-RNase that is recognized by this gene), only by performing detailed studies in several individuals will these mutations be identified. At present, there are only two such mutations in Pyrus (De Franceschi et al., 2011a ; Kakui et al., 2011). The characterization of these genes in a larger number of individuals is thus needed.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. Primers used in this work.
Supplementary Table S2. RFLPs used to identify the SFBB1–SFBB3, SFBB5, SFBB7–SFBB9, SFBB11, SFBB13, and SFBB14 genes of the S22 and Sa haplotypes.
Supplementary Table S3. Patterns used for the identification of alleles from SFBB1–SFBB3, SFBB5–SFBB9, SFBB11, SFBB13, and SFBB14 genes in the segregation experiment.
Supplementary Table S4. Polymorphisms at the amino acid sites under positive selection between the S22 and Sa haplotypes.
Acknowledgements
This work has been funded by the project PTDC/BIA-BEC/100616/2008 comp-01-0124-FEDER-008916, funded by Programa Operacional para a Ciência e Inovação (POCI) 2012, co-funded by Fundo Europeu para o Desenvolvimento Regional (FEDER) funds and Programa Operacional para a Promoção da competividade (COMPETE; FCOMP-01-0124-FEDER-022718; PEst-C/SAU/LA0002/2011). B. A. is the recipient of a PhD grant (SFRH/BD/69207/2010) from FCT.
References
- Campbell CS, Evans RC, Morgan DR, Dickinson TA, Arsenault MP. 2007. Phylogeny of subtribe Pyrinae (formerly the Maloideae, Rosaceae): limited resolution of a complex evolutionary history. Plant Systematics and Evolution 266, 119–145. [Google Scholar]
- Castroviejo S, Jardín Botánico Real. 1986. Flora ibérica: plantas vasculares de la Península Ibérica e Islas Baleares. Madrid: Real Jardín Botánico (Spain), C.S.I.C. [Google Scholar]
- Cheng J, Han Z, Xu X, Li T. 2006. Isolation and identification of the pollen-expressed polymorphic F-box genes linked to the S-locus in apple (Malus×domestica). Sexual Plant Reproduction 19, 175–183. [Google Scholar]
- De Franceschi P, Dondini L, Sanzol J. 2012. Molecular bases and evolutionary dynamics of self-incompatibility in the Pyrinae (Rosaceae). Journal of Experimental Botany 63, 4015–4032. [DOI] [PubMed] [Google Scholar]
- De Franceschi P, Pierantoni L, Dondini L, Grandi M, Sansavini S, Sanzol J. 2011a. Evaluation of candidate F-box genes for the pollen S of gametophytic self-incompatibility in the Pyrinae (Rosaceae) on the basis of their phylogenomic context. Tree Genetics & Genomes 7, 663–683. [Google Scholar]
- De Franceschi P, Pierantoni L, Dondini L, Grandi M, Sanzol J, Sansavini S. 2011b. Cloning and mapping multiple S-locus F-box genes in European pear (Pyrus communis L.). Tree Genetics & Genomes 7, 231–240. [Google Scholar]
- De Nettancourt D. 1977. Incompatibility in angiosperms. Berlin: Springer-Verlag. [Google Scholar]
- Emes RD, Yang Z. 2008. Duplicated paralogous genes subject to positive selection in the genome of Trypanosoma brucei . PLoS One 3, e2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entani T, Iwano M, Shiba H, Che FS, Isogai A, Takayama S. 2003. Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen-expressed F-box gene with allelic diversity. Genes to Cells 8, 203–213. [DOI] [PubMed] [Google Scholar]
- Hua Z, Kao TH. 2006. Identification and characterization of components of a putative Petunia S-locus F-box-containing E3 ligase complex involved in S-RNase-based self-incompatibility. Plant Cell 18, 2531–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Meng X, Kao TH. 2007. Comparison of Petunia inflata S-locus F-box protein (Pi SLF) with Pi SLF like proteins reveals its unique function in S-RNase based self-incompatibility. Plant Cell 19, 3593–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua ZH, Fields A, Kao TH. 2008. Biochemical models for S-RNase-based self-incompatibility. Molecular Plant 1, 575–585. [DOI] [PubMed] [Google Scholar]
- Hurles M. 2004. Gene duplication: the genomic trade in spare parts. PLoS Biology 2, E206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igic B, Kohn JR. 2001. Evolutionary relationships among self-incompatibility RNases. Proceedings of the National Academy of Sciences, USA 98, 13167–13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Igic B, Ushijima K, et al. 2004. Primary structural features of the S haplotype-specific F-box protein, SFB, in Prunus . Sexual Plant Reproduction 16, 235–243. [Google Scholar]
- Ingram GC, Doyle S, Carpenter R, Schultz EA, Simon R, Coen ES. 1997. Dual role for fimbriata in regulating floral homeotic genes and cell division in Antirrhinum . EMBO Journal 16, 6521–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakui H, Kato M, Ushijima K, Kitaguchi M, Kato S, Sassa H. 2011. Sequence divergence and loss-of-function phenotypes of S locus F-box brothers genes are consistent with non-self recognition by multiple pollen determinants in self-incompatibility of Japanese pear (Pyrus pyrifolia). The Plant Journal 68, 1028–1038. [DOI] [PubMed] [Google Scholar]
- Kakui H, Tsuzuki T, Koba T, Sassa H. 2007. Polymorphism of SFBB-γ and its use for S genotyping in Japanese pear (Pyrus pyrifolia). Plant Cell Reproduction 26, 1619–1625. [DOI] [PubMed] [Google Scholar]
- Kubo K, Entani T, Takara A, et al. 2010. Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science 330, 796–799. [DOI] [PubMed] [Google Scholar]
- Lopez-Gomollon S, Dalmay T. 2010. Recent patents in RNA silencing in plants: constructs, methods and applications in plant biotechnology. Recent Patents on DNA Gene Sequences 4, 155–166. [DOI] [PubMed] [Google Scholar]
- Luu DT, Qin X, Laublin G, Yang Q, Morse D, Cappadocia M. 2001. Rejection of S-heteroallelic pollen by a dual-specific S-RNase in Solanum chacoense predicts a multimeric SI pollen component. Genetics 159, 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamikawa M, Kakui H, Wang S, Kotoda N, Kikuchi S, Koba T, Sassa H. 2010. Apple S locus region represents a large cluster of related, polymorphic and pollen-specific F-box genes. Plant Molecular Biology 74, 143–154. [DOI] [PubMed] [Google Scholar]
- Newbigin E, Paape T, Kohn JR. 2008. RNase-based self-incompatibility: puzzled by pollen S. Plant Cell 20, 2286–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MD, Davis AP, Anthony F, Yoder AD. 2011. Expression and trans-specific polymorphism of self-incompatibility RNases in Coffea (Rubiaceae). PLoS One 6, e21019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes MD, Santos RA, Ferreira SM, Vieira J, Vieira CP. 2006. Variability patterns and positively selected sites at the gametophytic self-incompatibility pollen SFB gene in a wild self-incompatible Prunus spinosa (Rosaceae) population. New Phytologist 172, 577–587. [DOI] [PubMed] [Google Scholar]
- Okada K, Tonaka N, Moriya Y, Norioka N, Sawamura Y, Matsumoto T, Nakanishi T, Takasaki-Yasuda T. 2008. Deletion of a 236kb region around S 4 -RNase in a stylar-part mutant S 4 sm-haplotype of Japanese pear. Plant Molecular Biology 66, 389–400. [DOI] [PubMed] [Google Scholar]
- Okada K, Tonaka N, Taguchi T, Ichikawa T, Sawamura Y, Nakanishi T, Takasaki-Yasuda T. 2011. Related polymorphic F-box protein genes between haplotypes clustering in the BAC contig sequences around the S-RNase of Japanese pear. Journal of Experimental Botany 62, 1887–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspé O, Kohn JR. 2002. S-allele diversity in Sorbus aucuparia and Crataegus monogyna (Rosaceae: Maloideae). Heredity 88, 458–465. [DOI] [PubMed] [Google Scholar]
- Raspé O, Kohn JR. 2007. Population structure at the S-locus of Sorbus aucuparia L. (Rosaceae: Maloideae). Molecular Ecology 16, 1315–1325. [DOI] [PubMed] [Google Scholar]
- Raspé O, Saumitou-Laprade P, Cuguen J, Jacquemart AL. 2000. Chloroplast DNA haplotype variation and population differentiation in Sorbus aucuparia L. (Rosaceae: Maloideae). Molecular Ecology 9, 1113–1122. [DOI] [PubMed] [Google Scholar]
- Reboiro-Jato D, Reboiro-Jato M, Fdez-Riverola F, Vieira CP, Fonseca NA, Vieira J. 2012. ADOPS—automatic detection of positively selected sites. Journal of Integrative Bioinformatics 9, 200 [DOI] [PubMed] [Google Scholar]
- Roalson EH, McCubbin AG. 2003. S-RNases and sexual incompatibility: structure, functions, and evolutionary perspectives. Molecular Phylogenetics and Evolution 29, 490–506. [DOI] [PubMed] [Google Scholar]
- Romero C, Vilanova S, Burgos L, Martinez-Calvo J, Vicente M, Llacer G, Badenes ML. 2004. Analysis of the S-locus structure in Prunus armeniaca L. Identification of S-haplotype specific S-RNase and F-box genes. Plant Molecular Biology 56, 145–157. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497. [DOI] [PubMed] [Google Scholar]
- Sassa H, Kakui H, Minamikawa M. 2010. Pollen-expressed F-box gene family and mechanism of S-RNase-based gametophytic self-incompatibility (GSI) in Rosaceae. Sexual Plant Reproduction 23, 39–43. [DOI] [PubMed] [Google Scholar]
- Sassa H, Kakui H, Miyamoto M, Suzuki Y, Hanada T, Ushijima K, Kusaba M, Hirano H, Koba T. 2007. S locus F-box brothers: multiple and pollen-specific F-box genes with S haplotype-specific polymorphisms in apple and Japanese pear. Genetics 175, 1869–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, Korban SS, Sosinski B, et al. 2008. Multiple models for Rosaceae genomics. Plant Physiology 147, 985–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneveld T, Tobutt KR, Vaughan SP, Robbins TP. 2005. Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. Plant Cell 17, 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbachs JE, Holsinger KE. 2002. S-RNase-mediated gametophytic self-incompatibility is ancestral in eudicots. Molecular Biology and Evolution 19, 825–829. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Potter D, Tao R, Vieira CP, Vieira J, Iezzoni AF. 2008. Genetic and molecular characterization of three novel S-haplotypes in sour cherry (Prunus cerasus L.). Journal of Experimental Botany 59, 3169–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H. 2003. Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima K, Sassa H, Tamura M, Kusaba M, Tao R, Gradziel TM, Dandekar AM, Hirano H. 2001. Characterization of the S-locus region of almond (Prunus dulcis): analysis of a somaclonal mutant and a cosmid contig for an S haplotype. Genetics 158, 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J, Ferreira PG, Aguiar B, Fonseca NA, Vieira CP. 2010. Evolutionary patterns at the RNase based gametophytic self-incompatibility system in two divergent Rosaceae groups (Maloideae and Prunus). BMC Evolutionary Biology 10, 200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J, Fonseca NA, Vieira CP. 2008a. An S-RNase-based gametophytic self-incompatibility system evolved only once in eudicots. Journal of Molecular Evolution 67, 179–190. [DOI] [PubMed] [Google Scholar]
- Vieira J, Fonseca NA, Vieira CP. 2009. RNase-based gametophytic self-incompatibility evolution: questioning the hypothesis of multiple independent recruitments of the S-pollen gene. Journal of Molecular Evolution 69, 32–41. [DOI] [PubMed] [Google Scholar]
- Vieira J, Morales-Hojas R, Santos RA, Vieira CP. 2007. Different positively selected sites at the gametophytic self-incompatibility pistil S-RNase gene in the Solanaceae and Rosaceae (Prunus, Pyrus, and Malus). Journal of Molecular Evolution 65, 175–185. [DOI] [PubMed] [Google Scholar]
- Vieira J, Santos RA, Ferreira SM, Vieira CP. 2008b. Inferences on the number and frequency of S-pollen gene (SFB) specificities in the polyploid Prunus spinosa . Heredity 101, 351–358. [DOI] [PubMed] [Google Scholar]
- Vieira J, Santos RA, Habu T, Tao R, Vieira CP. 2008c. The Prunus self-incompatibility locus (S locus) is seldom rearranged. Journal of Heredity 99, 657–660. [DOI] [PubMed] [Google Scholar]
- Vieira J, Teles E, Santos RA, Vieira CP. 2008d. Recombination at Prunus S-locus region SLFL1 gene. Genetics 180, 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Kao T-H. 2011. Self-incompatibility in Petunia: a self/nonself-recognition mechanism employing S-locus F-box proteins and S-RNase to prevent inbreeding. Wiley Interdisciplinary Reviews: Developmental Biology 1, 267–275. [DOI] [PubMed] [Google Scholar]
- Wheeler D, Newbigin E. 2007. Expression of 10 S-class SLF-like genes in Nicotiana alata pollen and its implications for understanding the pollen factor of the S locus. Genetics 177, 2171–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikstrom N, Savolainen V, Chase MW. 2001. Evolution of the angiosperms: calibrating the family tree. Proceedings of the Royal Society of London B—Biological Sciences 268, 2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. 1939. The distribution of self-sterility alleles in populations. Genetics 24, 538–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Xie Z. 2001. DAMBE: software package for data analysis in molecular biology and evolution. Journal of Heredity 92, 371–373. [DOI] [PubMed] [Google Scholar]
- Yang Z, Bielawski JP. 2000. Statistical methods for detecting molecular adaptation. Trends in Ecology & Evolution 15, 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Computer Applications in the Biosciences 13, 555–556. [DOI] [PubMed] [Google Scholar]
- Zhou JL, Wang F, Ma WS, Zhang YS, Han B, Xue YB. 2003. Structural and transcriptional analysis of S-locus F-box genes in Antirrhinum . Sexual Plant Reproduction 16, 165–177. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.