Abstract
Background: Extracellular circulating DNA (cfDNA) can be found in small amounts in plasma of healthy individuals. Increased levels of cfDNA have been reported in patients with cancer of breast, cervix, colon, liver and it was shown that cfDNA can originate from both tumour and non-tumour cells.
Objectives: Levels of cfDNA of a large series of children with lymphoma were evaluated and analyzed in relation with clinical characteristics.
Methods: plasma cfDNA levels obtained at diagnosis in 201 paediatric lymphoma patients [43 Hodgkin lymphomas (HL), 45 anaplastic large cell lymphomas (ALCL), 88 Burkitt lymphomas (BL), 17 lymphoblastic (LBL), 8 diffuse large B cell lymphoma (DLBCL)] and 15 healthy individuals were determined using a quantitative PCR assay for POLR2 gene and, in addition, for NPM-ALK fusion gene in ALCL patients. Wilcoxon rank sum test was used to compare plasma levels among different patient subgroups and controls and to analyze relationship between levels of cfDNA and clinical characteristics.
Results: Levels of cfDNA in lymphoma patients were significantly higher compared with controls (p<0.0001). CfDNA was associated with median age (p=0.01) in HL, and with stage in ALCL (p=0.01). In HL patients high cfDNA levels were correlated with poor prognosis (p=0.03). In ALCL we found that most of the cfDNA (77%) was non-tumor DNA.
Conclusion: level of plasma cfDNA might constitute an important non-invasive tool at diagnosis in lymphoma patients' management; in particular in patients with HL, cfDNA seems to be a promising prognostic biomarker.
Keywords: lymphoma, childhood, cfDNA, prognosis, Hodgkin.
INTRODUCTION
Circulating cell-free DNA (cfDNA) represents extracellular DNA occurring in blood. It can be found in small amounts in plasma of healthy individuals. Likely, cfDNA originates from apoptotic cells of hematopoietic origin1 but also living cells may actively release DNA fragments.1,2 Increased levels of cfDNA have been reported in autoimmune diseases, myocardial infarction, pregnancy-associated complications.3,4 Most importantly, cfDNA was isolated in plasma/serum from patients with cancer of the breast, cervix, colon, liver, lung, prostate and thyroid.5 Investigations on characteristics of the DNA found in plasma of cancer patients showed that plasma DNA can originate from both tumor and non-tumor cells. Jahar et al. showed that the fraction of tumor-derived DNA appears to be higher when the total level of plasma DNA is low, supporting the idea that fraction of tumor DNA in plasma may correlate with the state of tumor progression: advanced tumors that infiltrate the surrounding normal tissues shed more DNA including that from adjacent non tumor cells in the blood stream.6,7,8 While cfDNA has been widely described in solid tumors,9,10 there is only few reports available for patients with lymphoproliferative diseases,11,12,13 in particular there is a single report on circulating cfDNA in pediatric lymphoma.14 Machado et al.14 detected total cfDNA and EBV plasma DNA in 30 pediatric B non-Hodgkin Lymphoma concluding that detection of cfDNA is a non-invasive technique that could become a valuable tool for disease detection in pediatric EBV-associated lymphomas and for monitoring treatment response. Indeed, the prognostic role of minimal disseminated disease (MDD), as molecular biomarker at diagnosis, in bone marrow or peripheral blood, was recently demonstrated for some subgroups of pediatric lymphomas.15,16,17 MDD is predictive of high risk of failure in BL15 and pediatric Anaplastic Large Cell Lymphoma (ALCL) MDD-positivity, using NPM-ALK fusion transcript as tumor marker, conferred a relapse risk of approximately 50% in two different studies.18,19 However, for some subgroups of lymphomas, such as Diffuse Large B Cell Lymphoma (DLBCL) or Hodgkin Lymphoma (HL) the absence of a tumor specific molecular marker prevent to assess the impact of MDD on survival.
In the present study, we investigated the presence of circulating cfDNA at diagnosis of 201 Italian pediatric lymphomas and in 15 healthy controls, using an accurate and reproducible real-time quantitative PCR for the POLR2 gene and analyzed its association with clinical characteristics and prognosis. All patients were treated in the Associazione Italiana Emato-Oncologia Pediatrica (Italian Association of Pediatric Hematology-Oncology) (AIEOP) clinical trials. Circulating cfDNA might potentially serve as a prognostic marker, in particular in tumors still lacking specific biological markers able to be used for disease monitoring, such as HL.
MATERIALS AND METHODS
Patients and Controls
We studied plasma samples obtained at initial diagnosis in 201 paediatric patients affected by lymphoma. Samples were collected in our laboratory in the time period 1998-2011 from AIEOP centers and written consent forms were obtained from parents or legal guardians of each patient before enrolment. The study was approved by the ethics committee or by the internal review board of each participating Institution. Patients characteristics are detailed in Table 1. A control group consisting of 15 healthy individuals (7 males, 8 females; median age 12 yrs, range 5-16) was included in order to determine the normal range of circulating cfDNA in plasma.
Table 1.
Main Characteristics of the Study Population.
| Characteristic | Total (n=201) |
ALCL (n=45) |
HL (n=43) |
BL (n=88) |
DLBCL (n=8) |
LBL (n=17) |
|---|---|---|---|---|---|---|
| Gender | ||||||
| Female | 64 | 13 | 25 | 18 | 3 | 5 |
| Male | 137 | 32 | 18 | 70 | 5 | 12 |
| Age (years) | ||||||
| ≤ 10.0 | 98 | 20 | 8 | 56 | 4 | 10 |
| > 10.0 | 103 | 25 | 35 | 32 | 4 | 7 |
| Stage | ||||||
| I+II | 59 | 6 | 18 | 30 | 3 | 2 |
| III+IV | 142 | 39 | 25 | 58 | 5 | 15 |
| LDH UI/L | ||||||
| ≤ 1000 | 97 | 32 | - | 57 | 8 | - |
| >1000 | 37 | 6 | - | 31 | 0 | - |
| B Symptoms | ||||||
| No | 133 | 8* | 23 | 80 | 6 | 16 |
| Yes | 64 | 33 | 20 | 8 | 2 | 1 |
| Bulky disease | ||||||
| No | 113 | 26* | 18* | 53 | 6 | 10 |
| Yes | 84 | 17 | 24 | 35 | 2 | 7 |
| Event (relapse, disease progression) | ||||||
| No | 168 | 31 | 38 | 77 | 8 | 14 |
| Yes | 33 | 14 | 5 | 10 | 0 | 3 |
ALCL, Anaplastic Large Cell Lymphoma; HL, Hodgkin Lymphoma; BL, Burkitt Lymphoma; DLBCL, Diffuse Large B Cell Lymphoma; LBL, Lymphoblastic Lymphoma; LDH, lactate dehydrogenase; *incomplete data available.
DNA extraction and quantification of cell-free DNA in plasma
Peripheral blood samples in sodium citrate (5mL) were shipped to our laboratory and processed within 24 hours from drawing. All blood samples were centrifuged at 820 x g for 10 min. To avoid contamination by blood cells, supernatants were carefully removed, and ri-centrifuged at 2500 x g for 10 min. DNA was isolated from 500 µl plasma samples using QIAmp DNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions, and stored at -20°C until use. Plasma DNA was quantified using a Taq-Man based real-time PCR assay for the POLR2 gene. Amplification was set up in a reaction volume of 25 µl, containing 5 µl of the extracted plasma DNA, TaqMan Universal Master 1X (ABI, Foster City, CA, USA), 300nM of each forward and reverse primer [5'-CCCAGGTGACATGGAATCTTG-3' and 5'-GCAGAGGCACGTTCAGGAA-3'] and 200 nM of TaqMan probe [VIC'- AGCCTTGTGCAGTGGCAGCCACT-TAMRA]. In addition, for ALCL patients, we also performed a SYBER green-based Real-time PCR assay with specific primers for amplification of tumor fusion gene NPM-ALK (NPM 5'-CTCCCCAGGCTTCTTGCAT-3' and ALK 5'-AGTGGATTTGAGGGTGCAGC-3'). Amplification was carried out on ABI Prism 7000 sequence detection system and consisted of 10-min initial activation at 95°C, followed by 40 cycles at 95°C for 15 s and 60° for 1 min. A standard curve was used to determine the starting target quantity, using 5-fold serial dilutions of DNA in deionized water prepared from total DNA extracted from the Karpas-299 cell line. Each PCR run was performed in triplicate and the mean values of results were calculated.
Minimal Disseminated Disease
MDD was analyzed in ALCL and BL cases as previously reported.15,19 Briefly, for ALCL patients with NPM-ALK positive tumour biopsy, total RNA obtained from bone marrow (BM) cells at diagnosis, was isolated using Trizol reagent (Invitrogen, Carlsbad, CA) in accordance with the manufacturer's instructions. An amount of 1 µg of total RNA was reverse transcribed. The 5' and 3' primers specific for the chimeric transcript NPM-ALK amplification were TCCCTTGGGGGCTTTGAAATAACACC (5'NPM) and CGAGGTGCGGAGCTTGCTCAGC (3'ALK). Each reaction mixture contained 10X buffer, 1.5mM MgCl2, 1.6mM dNTPs, 400 nM of each primer, 0.2 IU of Taq polymerase and 5% of the RT product in a final 20 µl reaction volume. PCR reaction consisted of initial denaturation at 94°C for 2 min, followed by 40 cycles of 94°C for 15 s, 68°C for 15 s, 72°C for 30 s and a final extension at 72°C for 10 min. PCR products were analyzed by 3% agarose gel electrophoresis and visualized under UV illumination after ethidium bromide staining. Ladder 50 (Invitrogen, Milan, Italy) was used as a molecular weight marker.
For BL cases, we investigated the presence of t(8;14) by LD-PCR in the tumor and, if positive, also in the corresponding BM. We used one primer for the cMYC gene (5' ACAGTCCTGGATGATGATGTTTTTGATGAAGGTCT-3') combined, alternatively, with one of four primers for the IgH locus: three primers for the constant regions (Cγ/03: 5'-TGCTGCTGATGTCAGAGTTGTTCTTGTATTTCCAG-3';Cµ/02: 5'- AGGGCACGGTCACCACGCTGCTGAGGGAGTAGAGT-3'; Cα/01: 5'-TCGTGTAGTGCTTCACGTGGCATGTCACGGACTTG-3') and one for the joining region (JH:5'- ACCTGAGGAGACGGTGACCAGGGT-3'). Quality of genomic DNA was assessed in each patient by using a combination of primers of the Human tPA Control Primer set (Roche Diagnostics, Milan, Italy). Each reaction mixture (50 µL) contained 250 ng of DNA, 60 pmol of each primer, and amounts of deoxynucleotide triphosphates, buffer III, oligonucleotides, and a combination of Taq and Pwo polymerases (Expand Long template PCR System, Roche Diagnostics), as indicated by the manufacturer. PCR was performed in an ABI Thermal Cycler 9700 (Applied Biosystems, Foster City,CA),using cycle characteristics suggested by the manufacturer and previously described.15 PCR products were analyzed by 0.8% agarose gel electrophoresis and were visualized under ultraviolet illumination after ethidium bromide staining.
Statistical analysis
As the values of circulating cfDNA do not follow a normal distribution, Wilcoxon rank sum test was used to compare plasma levels between different patient subgroups and controls. This test was also used to compare the mean values of cfDNA and clinical characteristics. A two-sided p≤0.05 was considered to be statistically significant. Survival analysis was performed according to Kaplan-Meier method.20 Event free survival (EFS) was calculated from the date of diagnosis to the date of the first event (tumor progression, relapse and death for any cause) or to the date of last follow-up The statistical calculation was carried out using SAS statistical program (SAS-PC, version 9.3; SAS Institute Inc., Cary, NC, USA).
RESULTS
Cell-free circulating DNA concentration in plasma
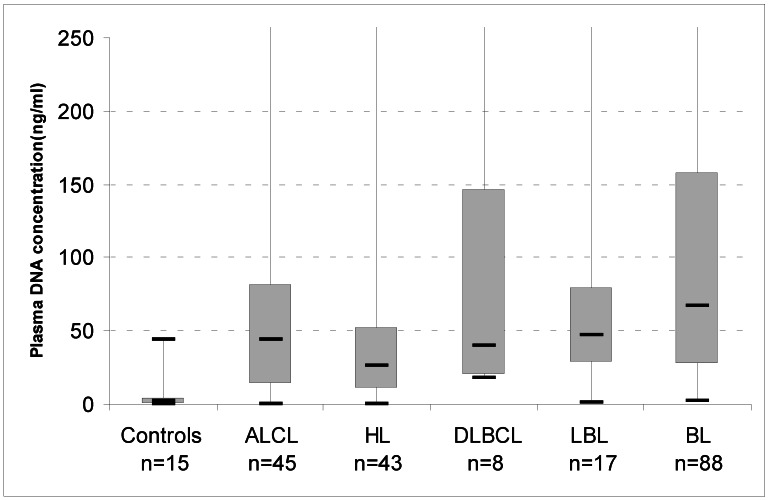
We determined the concentration of DNA in plasma samples of 201 paediatric lymphoma patients using a quantitative PCR assay for the POLR2 gene. Median cfDNA concentration was 1.6 ng/mL (± 11.6ng/mL) in 15 paediatric healthy individuals. Levels of cfDNA in lymphoma patients [43 cases of HL, 45 of ALCL, 88 BL, 17 lymphoblastic (LBL), 8 DLBCL] were significantly higher than in controls (median 46 ng/mL, p<0.0001) (Figure 1).
Figure 1.
Box-plots of cell-free DNA plasma concentrations in healthy controls and patients with lymphoma at diagnosis. The upper border of the box indicates the upper quartile (75th percentile) while the lower border indicates the lower quartile (25th percentile), and the horizontal line in the box the median. ALCL, Anaplastic Large Cell Lymphoma; HL, Hodgkin Lymphoma; BL, Burkitt Lymphoma; DLBCL, Diffuse Large B Cell Lymphoma; LBL, Lymphoblastic Lymphoma. Maximun values: ALCL, 2034 ng/mL; HL, 1650 ng/mL: DLBCL, 700ng/mL; LBL, 360ng/mL;BL, 3660 ng/mL.
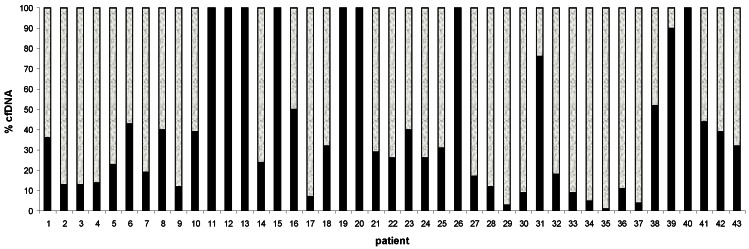
To establish the amount of plasma DNA originating from tumour cells, a quantitative PCR assay for NPM-ALK fusion gene in ALCL cases was performed. We examined 43 out of 45 ALCL plasma DNA samples and found that fraction of tumour DNA varies from 1% to 100% (Figure 2). Interestingly, samples with a high percentage of tumour-specific DNA tend to belong to a group with low concentration of cfDNA: #12, 20 ng/mL; #40, 6 ng/mL; in contrast, samples with low percentage of tumor-specific DNA have a high concentration of total cfDNA ( #35, 80ng/mL;# 36,280 ng/mL; #37, 1760 ng/mL) (Figure 2).
Figure 2.
Distribution of cfDNA in 43 ALCL plasma patients using 2 probes:POLR2 probe for tumor-derived and non-tumor derived cfDNA (white color); NPM-ALK probe specific only for tumor-derived cfDNA (black color). cfDNA=cell free DNA.
Minimal Disseminated Disease
The ALCL - BM samples obtained at diagnosis were MDD-positive by RT-PCR in 53% of the patients (24/45) studied. Whereas in BL patients, MDD was positive by LD-PCR in 37% of the patients (14/38) analyzed.
Correlation of plasma cfDNA to patient characteristics
Plasma DNA concentrations in the 3 major diagnostic subtypes (ALCL, BL and HL) were analyzed together with the main presenting clinical features (Table 2). No statistically significant relationship was found between lymphoma histology and baseline cfDNA levels although the patients with higher values have BL. In HL patients, age >10 was associated with increased levels of cfDNA with a p value of 0.01 (age≤10, cfDNA range 0-58 ng/mL; age>10, cfDNA range 10-1650 ng/mL); whereas in ALCL, advanced stages of disease (stage III/IV) were associated with elevated cfDNA. (p= 0.01)
Table 2.
Associations between patient characteristics and plasma cell-free DNA median levels.
| Characteristic | BL (n=88) | ALCL (n=45) | HL (n=43) | |
|---|---|---|---|---|
| Gender | ||||
| Female | 280 | 110 | 133 | |
| Male | 192 | 136 | 42 | |
| P | 0.92 | 0.41 | 0.13 | |
| Age (years) | ||||
| ≤ 10.0 | 220 | 196 | 14 | |
| > 10.0 | 194 | 75 | 114 | |
| P | 0.6 | 0.72 | 0.01 | |
| Stage | ||||
| I+II | 290 | 14 | 51 | |
| III+IV | 170 | 147 | 128 | |
| P | 0.29 | 0.01 | 0.24 | |
| B-Symptoms | ||||
| No | 226 | 43 | 65 | |
| Yes | 60 | 145 | 131 | |
| P | 0.14 | 0.48 | 0.81 | |
| LDH | ||||
| ≤ 1000 | 226 | 71 | - | |
| > 1000 | 183 | 403 | - | |
| P | 0.39 | 0.20 | ||
| Bulky disease | ||||
| No | 258 | 76 | 122 | |
| Yes | 140 | 207 | 80 | |
| P | 0.11 | 0.88 | 1.0 | |
| MDD | ||||
| No | 24 | 21 | - | |
| Yes | 14 | 24 | - | |
| 0.41 | 0.39 | |||
P-values of Wilcoxon rank tests are given. ALCL, Anaplastic Large Cell Lymphoma; HL, Hodgkin Lymphoma; BL, Burkitt Lymphoma; DLBCL, Diffuse Large B Cell Lymphoma; LBL, Lymphoblastic Lymphoma; LDH, lactate dehydrogenase; MDD= Minimal Disseminated Disease.
We did not find any correlation between cfDNA and other clinical characteristics, such as B-symptoms, LDH levels and bulky disease. In particular, we didn't find any correlation between MDD and cfDNA (Table 2).
The role of the cfDNA at diagnosis as a prognostic marker was separately analyzed for the 3 lymphoma subtypes. The follow-up was available for a total of 176 lymphoma patients who completed chemotherapy treatment. The median time of follow-up was 3 years (range 0,13 yrs-9,6 yrs). Based on median value of cfDNA, the 3 years EFS in HL series was statistically significant with a EFS of 92% (±6%) for patients with cfDNA≤ 46ng/mL and 60% (±18%) for patients with cfDNA> 46ng/mL (p=0.03) (Figure 3). In the two groups the median time of EFS was 1.8 years (range 0.3-6.9 years) and 1,0 year (range 0.4-6.6 years) respectively.
Figure 3.
Event Free Survival (EFS) in 43 HL patients according to the DNA plasma concentration (median value cfDNA= 46ng/mL) at diagnosis. EFS was 92% (±6) for patients with cfDNA≤ 46ng/mL and 60% (±18%) for patients with cfDNA> 46ng/mL (p=0.03).
DISCUSSION
We investigated the plasma of paediatric lymphoma patients and found detectable concentrations of cfDNA in the different subtypes and in a series of healthy controls. Our study demonstrates that lymphoma patients have significantly higher levels of cfDNA at diagnosis, compared to normal plasma, that could be accurately quantified by real-time PCR. Recently, many studies proposed quantification of cfDNA for cancer screening;21 cfDNA has been suggested as a cancer biomarker in adult solid cancer such as lung, breast, prostate cancer; in adult lymphoma patients, Hohaus et al. demonstrated that cfDNA in HL, DLBCL, and mantle cell NHL was significantly higher than in controls.13 Increased levels of cfDNA were associated with advanced stage disease, presence of B-symptoms, elevated lactate dehydrogenase levels and age >60 years.13 Our report is the first study investigating cfDNA in an exclusive cohort of paediatric patients. We found that the range of cfDNA in the circulation of lymphoma patients varies widely, from levels similar to controls (10-30 ng/mL) to levels that exceed values of 500 ng DNA/mL plasma. In particular, we observed higher DNA levels in BL patients (Figure 1).
ALCL patients
Plasma DNA can originate from both tumor and non-tumor cells. In ALCL patients, we evaluated the amount of the two different DNAs based of NPM-ALK and POLR2 quantification. NPM-ALK fusion gene is a specific tumour DNA marker in ALCL, so an estimation of the fraction of tumour specific DNA in the samples could be evaluated. We found that in ALCL patients most of the cfDNA (in 79% of the cases) was non-tumor DNA (Figure 2). Some samples of plasma DNA consisted exclusively of tumor cell DNA (#11,12,13,19,20,26,40, Figure 2); however, in these cases, the absolute amounts of tumor-derived DNA was generally low: <60ng/mL, with a range of 6 to 60 ng/mL. In all the other ALCL samples, the range was 26 to 1650 ng/mL. We found that stage I-II ALCL had a mean value of 14 ng/mL compared to 157 ng/mL of stage III-IV (p= 0.01), (Table 2) and that fraction of tumor-derived DNA appeared to be higher when the total level of plasma DNA was low. Thus, we hypothesize that the fraction of cfDNA and tumor extension are correlated with small tumors shedding small amount of DNA and larger tumors -that infiltrate the surrounding normal tissues- shedding more DNA, including DNA from adjacent non tumor cells. These data are in agreement with results obtained in adult lymphoma patients.13
Association analysis was also performed with other clinical characteristics for ALCL, BL, and HL patients, whereas our DLBCL and T-LBL patient groups were too small for this analysis (Table 2).
HL patients
In HL patients we observed an association between age and cfDNA level (mean cfDNA= 14ng/mL for cases ≤10 years and mean cfDNA= 114ng/mL for cases >10 years, p= 0.014). In adult HL cases, Hohaus et al13 found association between DNA concentrations and age and elevated LDH, suggesting that circulating DNA may reflect an active proliferating disease. In addition, in our pediatric HL patients, we found that plasma DNA level seems to be a prognostic predictor (Figure 3). This is in line with HL adult data, where the 2-year probability of FFTF was 62% for patients with elevated plasma DNA, while patients with normal DNA levels had an 88% 2-year probability of FFTF (p=0.03).13
HL lacks of a biological tumor marker, such MDD in ALCL and BL, that constitutes a prognostic tool not only at diagnosis but also during follow-up. This likely depends on the absence of known characteristic genetic aberrations in HL, such as the NPM-ALK fusion transcript in ALCL. The possibility of using a biological markers not related to specific genetic aberrations, such as cfDNA, could give the opportunity to follow HL patients in the same way as ALCL and BL patients.
CfDNA was discovered over 60 years ago, but research on cfDNA has lagged considerably due to the lack of robust and sensitive assays. Efficient isolation procedures of cfDNA and specific florescent dyes developed during the last two decades have fostered research in this area. Quantitative PCR assay is easily reproducible, relatively inexpensive, and highly sensitive and allows accurate target quantification.
To our knowledge, this is the first report on cfDNA study in paediatric lymphoma. However, further studies are needed prospectively to validate this biomarker.
Acknowledgments
The study was supported by Fondazione Citta` Della Speranza, Associazione Italiana contro le Leucemie (AIL) and by Camera di Commercio di Venezia. We thank the Colleagues from various AIEOP centers for contributing biological samples and patient clinical information.
References
- 1.Stroun M, Lyautey J, Lederrey C. et al. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001;313:139–42. doi: 10.1016/s0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 2.Stroun M, Lyautey J, Lederrey C. et al. Alu repeat sequences are present in increased proportions compared to a unique gene in plasma/serum DNA: evidence for a preferential release from viable cells ? Ann N Y Acad Sci. 2001;945:258–64. doi: 10.1111/j.1749-6632.2001.tb03894.x. [DOI] [PubMed] [Google Scholar]
- 3.Gerovassili A, Garner C, Nicolaides KH. et al. Free fetal DNA in maternal circulation: a potential prognostic marker for chromosomal abnormalities? Prenat Diagn. 2007;27(2):104–10. doi: 10.1002/pd.1607. [DOI] [PubMed] [Google Scholar]
- 4.Bulicheva N, Fidelina O, Mkrtumova N. et al. Effect of cell-free DNA of patients with cardiomyopathy and rDNA on the frequency of contraction of electrically paced neonatal rat ventricular myocytes in culture. Ann N Y Acad Sci. 2008;1137:273–7. doi: 10.1196/annals.1448.023. [DOI] [PubMed] [Google Scholar]
- 5.Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer--a survey. Biochim Biophys Acta. 2007;1775:181–232. doi: 10.1016/j.bbcan.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Jahr S, Hentze H, Englisch S. et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–65. [PubMed] [Google Scholar]
- 7.García-Olmo DC, Samos J, Picazo MG. et al. Release of cell-free DNA into the bloodstream leads to high levels of non-tumor plasma DNA during tumor progression in rats. Cancer Lett. 2008;272:133– 40. doi: 10.1016/j.canlet.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 8.García-Olmo DC, Ruiz-Piqueras R, García-Olmo D. et al. Circulating nucleic acids in plasma and serum (CNAPS) and its relation to stem cells and cancer metastasis: state of the issue. Histol Histopathol. 2004;19:575–83. doi: 10.14670/HH-19.575. [DOI] [PubMed] [Google Scholar]
- 9.Paci M, Maramotti S, Bellesia E. et al. Circulating plasma DNA as diagnostic biomarker in non-small cell lung cancer. Lung Cancer. 2009;64:92–7. doi: 10.1016/j.lungcan.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 10.Nawroz-Danish H, Eisenberger CF, Yoo GH. et al. Microsatellite analysis of serum DNA in patients with head and neck cancer. Int J Cancer. 2004;111:96–100. doi: 10.1002/ijc.20240. [DOI] [PubMed] [Google Scholar]
- 11.Frickhofen N, Müller E, Sandherr M. et al. Rearranged Ig heavy chain DNA is detectable in cell-free blood samples of patients with B-cell neoplasia. Blood. 1997;90:4953–60. [PubMed] [Google Scholar]
- 12.Deligezer U, Yaman F, Erten N. et al. Frequent copresence of methylated DNA and fragmented nucleosomal DNA in plasma of lymphoma patients. Clin Chim Acta. 2003;335:89–94. doi: 10.1016/s0009-8981(03)00279-1. [DOI] [PubMed] [Google Scholar]
- 13.Hohaus S, Giachelia M, Massini G. et al. Cell-free circulating DNA in Hodgkin's and non-Hodgkin's lymphomas. Ann Oncol. 2009;20:1408–13. doi: 10.1093/annonc/mdp006. [DOI] [PubMed] [Google Scholar]
- 14.Machado AS, Da Silva R, Magalhães MC. et al. Circulating cell-free and Epstein-Barr virus DNA in pediatric B-non-Hodgkin lymphomas. Leuk Lymphoma. 2010;51:1020– 7. doi: 10.3109/10428191003746331. [DOI] [PubMed] [Google Scholar]
- 15.Mussolin L, Pillon M, d'Amore ES. et al. Minimal disseminated disease in high-risk Burkitt's lymphoma identifies patients with different prognosis. J Clin Oncol. 2011;29:1779–84. doi: 10.1200/JCO.2010.32.8161. [DOI] [PubMed] [Google Scholar]
- 16.Mussolin L, Bonvini P, Ait-Tahar K. et al. Kinetics of humoral response to ALK and its relationship with minimal residual disease in pediatric ALCL. Leukemia. 2009;23:400– 2. doi: 10.1038/leu.2008.184. [DOI] [PubMed] [Google Scholar]
- 17.Ait-Tahar K, Damm-Welk C, Burkhardt B. et al. Correlation of the autoantibody response to the ALK oncoantigen in pediatric anaplastic lymphoma kinase-positive anaplastic large cell lymphoma with tumor dissemination and relapse risk. Blood. 2010;115:3314–9. doi: 10.1182/blood-2009-11-251892. [DOI] [PubMed] [Google Scholar]
- 18.Damm-Welk C, Busch K, Burkhardt B. et al. Prognostic significance of circulating tumor cells in bone marrow or peripheral blood as detected by qualitative and quantitative PCR in pediatric PM-ALK-positive anaplastic large-cell lymphoma. Blood. 2007;110:670–7. doi: 10.1182/blood-2007-02-066852. [DOI] [PubMed] [Google Scholar]
- 19.Mussolin L, Pillon M, d'Amore ES. et al. Prevalence and clinical implications of bone marrow involvement in pediatric anaplastic large cell lymphoma. Leukemia. 2005;19:1643–7. doi: 10.1038/sj.leu.2403888. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan E. Non-parametric estimation from incomplete observations. Am Stat Assos. 1958;53:457–481. [Google Scholar]
- 21.Sozzi G, Conte D, Leon M. et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol. 2003;21:3902–8. doi: 10.1200/JCO.2003.02.006. [DOI] [PubMed] [Google Scholar]