Abstract
Streptomycetes comprise very important industrial bacteria, producing two-thirds of all clinically relevant secondary metabolites. They are mycelial microorganisms with complex developmental cycles that include programmed cell death (PCD) and sporulation. Industrial fermentations are usually performed in liquid cultures (large bioreactors), conditions in which Streptomyces strains generally do not sporulate, and it was traditionally assumed that there was no differentiation. In this work, we review the current knowledge on Streptomyces pre-sporulation stages of Streptomyces differentiation.
Keywords: antibiotic, differentiation, programmed cell death, secondary metabolism, sporulation, Streptomyces
Introduction
Streptomycetes are gram-positive, mycelium-forming, soil microorganisms that play important roles in mineralization processes in nature. They have great socio-economic relevance, as they produce several clinically relevant secondary metabolites (antibiotics, antitumorals, immunosuppressants, etc.) (Hopwood, 2007). Streptomycetes have complex developmental cycles that resemble filamentous fungi, forming hyphae and mycelia. They also have sporulation and programmed cell death (PCD) processes and are considered multicellular prokaryotic models.
Differentiation and development of Streptomyces in solid cultures
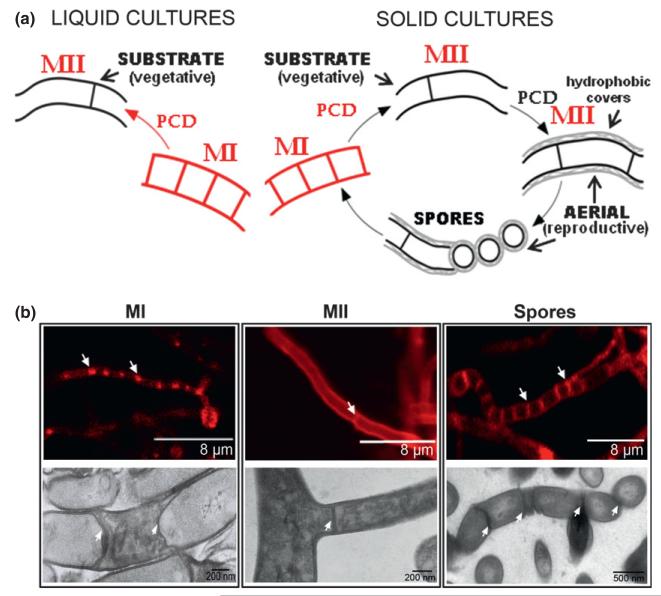
The traditional Streptomyces developmental cycle mainly focused on the sporulation phases occurring in solid cultures. After spore germination, a completely viable vegetative mycelium (substrate) grows on the surface and inside agar until it differentiates to a reproductive (aerial) mycelium that grows into the air, producing spores at the end of the cycle (reviewed in Flärdh & Buttner, 2009). This developmental model has been refined with respect to the stages preceding aerial mycelium formation and sporulation (Fig. 1). A young, compartmentalized mycelium (MI) was reported to die early on, following a highly ordered sequence (Manteca et al., 2005, 2006a). Subsequently, the viable segments of this mycelium differentiate into a multinucleated second mycelium (MII). MII grows inside the culture medium (substrate mycelium) until it starts to express hydrophobic covers and grows into the air (aerial mycelium) and ends by forming spores (Manteca et al., 2007). Prior to sporulation, there is a second round of PCD affecting substrate and aerial mycelium (Wildermuth, 1970; Mendez et al., 1985; Miguelez et al., 1999). MI, MII, and PCD were mainly described in Streptomyces antibioticus ATCC11891 (Miguelez et al., 1999; Manteca et al., 2005) or Streptomyces coelicolor M145 (Manteca et al., 2007). However, the existence of these developmental stages can be considered general to the Streptomyces genus, as they were observed in all the streptomycetes analysed: Streptomyces griseus IFO 13350, Streptomyces avermitillis MA-4680, Streptomyces cinereoruber ATCC19740, as well as hundreds of unclassified streptomycetes, examined during the screening experiments aimed at discovering novel secondary metabolites (Yagüe P, Genilloud O, Manteca A, unpublished results).
Fig. 1.
Streptomyces developmental cycle and mycelium differentiation. (a) Streptomyces developmental cycle in liquid (left) and solid (right) cultures. Newly described structures and the proposed nomenclature (Manteca et al., 2005) are indicated in red: MI, first compartmentalized mycelium; MII, second multinucleated mycelium. Classical nomenclature (substrate and aerial mycelium) and hydrophobic layers are also indicated. PCD, programmed cell death. (b) Different types of mycelia observed under the confocal and electron microscopes. Upper panels, confocal images. Left, MI young compartmentalized hypha (notice the original spore in the right side) stained with membrane stain FM 4-64; mycelium is fully compartmentalized and compartments are separated by membranes (arrows). Centre, MII multinucleated hypha stained with the cell wall stain WGA. Right, sporulated hypha stained with WGA; notice thick cell wall septa separating spores. Some of the septa are indicated by arrows. Figure adapted from Manteca et al. (2006a, 2010b).
Spore germination
Spore germination constitutes the first step of Streptomyces development (Fig. 2). However, the mechanisms activating germination remain somewhat vague. Spore germination comprises a sucession of distinctive steps, which were organized nicely by Hardisson et al. (1978) into three stages: darkening, swelling, and germ tube emergence. Darkening only required exogenous divalent cations (Ca2+, Mg2+ or Fe2+) and spore energy reserves. Calcium was reported to accumulate in the spore covers and be released during germination (Eaton & Ensign, 1980; Salas et al., 1983). Wang et al. (2008) demonstrated that calcium regulation could be mediated, at least in part, by cabC, a gene encoding an EF-hand calcium-binding protein. Trehalose was demonstrated to be consumed during the early stages of germination (Hey-Ferguson et al., 1973; McBride & Ensign, 1987). The second stage, swelling, needed an exogenous carbon source, and the last stage, germ tube emergence, required additional carbon and nitrogen sources.
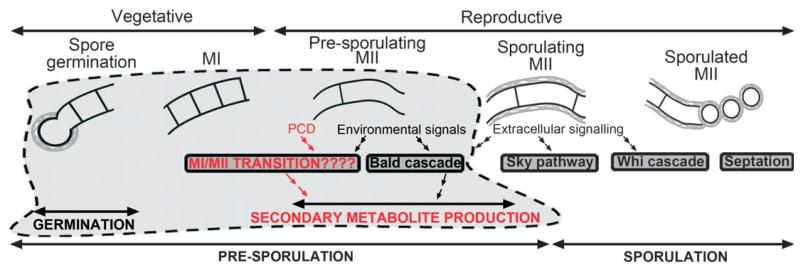
Fig. 2.
Biochemical pathways regulating Streptomyces differentiation. Pathways involved in hydrophobic covers formation (‘bald’, ‘sky’) and sporulation (‘whi’, ‘septation’) are illustrated. New developmental stages (MI/MII; Manteca et al., 2005) and presporulation pathways (‘MI/MII transition’) switching on secondary metabolite production are indicated in red.
Spore germination is highly regulated and can be externally modified. Hirsch & Ensign (1976) reported that the latency preceding germination of S. viridochromogenes spores was eliminated by gentle heat shock, a method that is routinely used to synchronize spore germination in Streptomyces (Kieser et al., 2000). Guijarro et al. (1983) revealed the existence of a protein fraction that rapidly degrades during germination and that might be regulating this process. Mikulík et al. (1984) demonstrated that RNA and protein synthesis began in the first 5 min following spore inoculation, a fact that was later confirmed by Strakova et al. (2013). Ribosomes were described as being complex, with melanine-type pigments forming insoluble aggregates, rendering them inactive in the dormant spores (Mikulík et al., 1984). Haiser et al. (2009) demonstrated the importance of cell wall hydrolases in both spore formation and spore germination. The existence of germination inhibitors excreted by germinating spores was discovered in Streptomyces viridochromogenes by Grund & Ensign (1985) and its chemical nature was subsequently characterized by Petersen et al. (1993). These inhibitors were also identified in S. coelicolor (Song et al., 2006). Cyclic AMP is involved in the regulation of germination (Süsstrunk et al., 1998); this regulation is mediated, at least partially, by the cyclic AMP receptor protein (Crp) (Derouaux et al., 2004; Piette et al., 2005). NepA has been described as a structural cell wall protein involved in maintaining spore dormancy in S. coelicolor (de Jong et al., 2009). Noens et al. (2007) identified SsgA as a protein marking cell wall sites where germination takes place.
Overall, important information has already been obtained concerning Streptomyces germination. However, there is still much to discover to fully understand the biochemical pathways regulating this important process.
Primary compartmentalized mycelium (MI)
MI is completely compartmentalized and is different from substrate and aerial mycelia, which are multinucleated (Manteca et al., 2005). Compartmentalization of this mycelium was studied by fluorescence microscopy, using membrane (FM 4-64, Cell Mask) and cell wall (WGA, vancomycin) fluorescent stains, as well as electron microscopy (Manteca et al., 2005; Manteca & Sánchez, 2009) (Fig. 1). MI septa membranes did not generally display thick cell walls; moreover, they were curved, probably due to the osmotic cellular pressure which could not be supported by their thin cell walls (Manteca et al., 2005; Manteca & Sánchez, 2009) (Fig. 1b). The function of MI thin septa remains unknown. They may facilitate intercellular communication inside Streptomyces hyphae.
Jakimowicz & van Wezel (2012) described the existence of two different septa in Streptomyces: substrate and aerial mycelia septa and spore septa. The formation of the two types of septa is regulated by different mechanisms (Willemse et al., 2011). MI septa would constitute a third type of thin septa that is structurally different from substrate/aerial and sporulation septa. This would make Streptomyces a very unusual organism, with three distinct septa associated with different developmental stages. Mechanisms regulating MI septa formation have yet to be discovered. FtsZ is one of the key proteins involved in cell division in bacteria. FtsZ was proven to participate in the formation of substrate/aerial/sporulation septa and its mutation gives rise to a non-sporulating syncytial mycelium having no septa (McCormick et al., 1994). Surprisingly, FtsZ mutant tolerated strong mechanical breakage (McCormick et al., 1994); the reason for this resistance remains unknown. One possibility could be the existence of some kind of septa in the FtsZ mutant similar to those with thin cell walls present in MI.
Secondary multinucleated mycelium (MII)
MI mycelial segments which remained viable after the first round of PCD started to grow as multinucleated hyphae (MII), whereas dead segments were progressively dismantled (Manteca et al., 2006b) (Fig. 1). Cellular debris generated by MI dead cells was present in the extracellular medium (cytosolic proteins; diaminopimelic acid, D-alanine, and other amino acids originating from cell wall degradation; nucleolytic activities; DNA or RNA fragments, etc.) (Manteca et al., 2006a). They were also observed under the electron microscope: García (1995) reported that substrate mycelium in S. antibioticus was ‘embedded among an intercellular material’ and Manteca et al. (2005) described the complete disorganization of MI dying cells. MII growth is completely viable on the surface and inside agar (substrate mycelium) until it undergoes a new round of PCD (Wildermuth, 1970; Mendez et al., 1985; Miguelez et al., 1999; Manteca et al., 2006a). The remaining viable MII hyphae start to form hydrophobic covers (chaplin-rodlin layer) (reviewed in Claessen et al., 2006) and grow into the air (aerial mycelium). Substrate mycelium was considered the vegetative mycelium, whereas aerial mycelium hyphae were considered specialized hyphae destined to sporulate (Chater, 1984). Aerial mycelium would re-use nutrients released by the substrate mycelium during the second round of PCD (a kind of cannibalism) (Mendez et al., 1985) and antibiotics would be produced by the substrate and/or aerial mycelium to prevent competition with other microorganisms during sporulation.
Substrate and aerial mycelia (MII) are multinucleated (reviewed in Jakimowicz & van Wezel, 2012). The existence of these multinucleated hyphae is very unusual and its biological relevance has yet to be revealed. Other well characterized filamentous bacteria, such as Cyanobacteria, are not multinucleated (reviewed in Singh & Montgomery, 2011). The obvious advantage to being multinucleated would be to facilitate the distribution of nutrients and biochemical signals, but with a very important risk in nature, as any damage would spread to the whole colony. Other mycelial microorganisms, such as fungi, also have multinucleated mycelia at temporary specific stages which are usually related with reproduction (reviewed in Glass & Kaneko, 2003). As discussed below, when Streptomyces development was analysed in soils, MI was the predominant mycelium and MII was a transitory phase preceding sporulation, which suggests that MI may be the true vegetative mycelium in nature.
The transition from substrate to aerial mycelium was extensively studied (Fig. 2). Streptomyces coelicolor mutant strains defective in different stages of hydrophobic cover formation (aerial mycelium) were used for the genetic and biochemical analysis of Streptomyces differentiation. Bald mutants (defective in aerial growth) regulate the ‘sky-pathway’, which activates the expression of genes related with hydrophobic cover formation (Rdls Chps, SapB) (Fig. 2). Elegant revisions of the state of the art of these developmental pathways have already been published (Claessen et al., 2006; McCormick & Flärdh, 2012).
The mechanisms regulating the absence of septa in MII or their presence in MI remain unknown (Fig. 2). Some authors have described the potential of substrate hyphae to septate and form spores prior to aerial mycelium differentiation, a feature known as ‘ectopic sporulation’ or ‘de-programmed sporulation’. Kelemen et al. (1995) described ectopic sporulation in a mutant strain lacking a DNA fragment near glkA in S. coelicolor. Kwak & Kendrick (1996) and Ohnishi et al. (2002) have described the same process in class III bald and NP4 mutants of S. griseus. Sporulation was also reported in substrate mycelium of wild Streptomyces carpinensis strain (Miguelez et al., 1997). Ohnishi et al. (2002) postulated the existence of unknown, specific mechanisms inhibiting septa formation in substrate hyphae. Manteca et al. (2010a, b) analysed differences between MI and MII proteomes, identifying several putative regulatory proteins differentially expressed in both types of mycelia. These experiments were recently extended to MI and MII transcriptomes (Yagüe et al., 2013). Further work will be necessary to characterize the biochemical pathways controlling the transition from MI to MII (Fig. 2).
Compartmentalization of tip ends of aerial mycelium and sporulation
The last stage of Streptomyces development in solid cultures corresponds to hypha septation and spore formation (Fig. 1). Streptomyces whi mutants defective in different stages of sporulation were used for the genetic and biochemical analyses of these developmental stages (Fig. 2). Sporulation is beyond the scope of this review. Revisions of the state of the art of genes and proteins regulating sporulation already exist (Claessen et al., 2006; Flärdh & Buttner, 2009; Jakimowicz & van Wezel, 2012; McCormick & Flärdh, 2012).
Differentiation and development of Streptomyces in liquid cultures
Most Streptomyces species do not sporulate in liquid cultures and it was widely accepted that no morphological differentiation took place in these conditions. Secondary metabolites would be produced by the substrate mycelium at the stationary phase after a transient growth arrest (Granozzi et al., 1990; Neumann et al., 1996; Novotna et al., 2003; Zhou et al., 2005; Chouayekh et al., 2007). Despite that, sporulation was reported in liquid cultures for several streptomycetes, such as Streptomyces venezuelae (Glazebrook et al., 1990), S. griseus (Kendrick & Ensign, 1983), Streptomyces chrysomallus (Kuimova & Soina, 1981), S. antibioticus ETHZ7451 (Novella et al., 1992), Streptomyces albidoflavus SMF301 (Rho & Lee, 1994), or Streptomyces brasiliensis (Rueda et al., 2001). Sporulation was also seen to be activated in several Streptomyces species under nutritional downshifts, including the model strain S. coelicolor (Koepsel & Ensign, 1984; Daza et al., 1989), and was also observed in several streptomycetes liquid cultures during the screening experiments aimed at discovering novel secondary metabolites (Yagüe P, Genilloud O, Manteca A, unpublished results).
New aspects regarding Streptomyces development (MI, MII, PCD) in solid cultures were extended to liquid cultivation (Manteca et al., 2008) (Fig. 1a). Similar to solid cultures, there was a young, compartmentalized mycelium (MI) that differentiated to a multinucleated mycelium (MII). The MII emergence was preceded by a transient growth arrest, which was the consequence of MI PCD. The only mycelial phases present in liquid were MI and MII without hydrophobic layers (Fig. 1). It was demonstrated that MII is the antibiotic-producing mycelium. This was the first time that antibiotic production was associated with differentiation in liquid cultures (Manteca et al., 2008). The lifespan of MI in liquid cultures was longer than in solid media (around 17 h in solid vs. 48 h in liquid) (Manteca et al., 2007, 2008). MI compartmentalization correlated well with the traditionally accepted existence of a specific phase at the beginning of the development – ‘the middle of the exponential phase’ in which protoplasts could be formed in Streptomyces liquid cultures (Okanishi et al., 1974). Protoplast formation by MII multinucleated mycelium was almost non-existent, a feature that can, in fact, be used to fractionate MI and MII mycelia (Manteca et al., 2010a).
Proteomic (Manteca et al., 2010b) and transcriptomic (Yagüe et al., 2013) analyses demonstrated that differentiation in liquid was much more similar to solid cultures than might be expected within the context of the classical Streptomyces developmental model. Proteins and transcripts involved in primary metabolism were up-regulated in MI, whereas proteins and genes involved in secondary metabolite biosynthesis were up-regulated in MII. The most remarkable differences between MII from solid and liquid cultures involved proteins regulating the hydrophobic cover formation and sporulation (Manteca et al., 2008, 2010b). Differentiation of MII after mycelia growth arrest is not enough to guarantee secondary metabolite production, as it can also be regulated by environmental signals, including components of the culture medium, such as nitrogen (Aharonowitz, 1980), carbon (Sánchez et al., 2010) and phosphate (Chouayekh & Virolle, 2002; Martín, 2004).
Streptomyces development in conditions resembling nature (soils)
The significance of the first compartmentalized mycelium was obscured by its short lifespan in usual laboratory culture conditions (Manteca et al., 2005, 2008). This might be attributable to the relatively high cell densities attained in laboratory culture conditions, which provoked massive cell death, differentiation, and sporulation. Natural growth conditions imply discontinuous growth and limited colony development (Williams, 1985). When Streptomyces development was analysed in conditions resembling nature (soils inoculated with poor spore inocula), a new developmental cycle emerged in which MI was the predominant mycelium (Manteca & Sánchez, 2009) (Fig. 3). Spore germination was a very slow, non-synchronous process that commenced at about 7 days and lasted for at least 21 days. The mycelium did not clump into dense pellets and remained in the MI compartmentalized mycelium phase for a long time. Even after 1 month of incubation, PCD, MII or sporulation were not detected. It is clear that in nature, cell death and sporulation must take place at the end of the long vegetative phase (Wellington et al., 1990; Anukool et al., 2004) when the nutrient imbalance gives rise to bacterial differentiation. As already commented above, the absence of compartmentalization in the vegetative Streptomyces mycelium (substrate) was unique in filamentous bacteria and difficult to understand due to the fragility of a multinucleated mycelium in nature. If we consider development in conditions resembling nature, compartmentalized MI would in fact be the dominant stage and multinucleated MII would be a transient antibiotic-producing structure, facilitating nucleic acid division and preceding sporulation (Fig. 3).
Fig. 3.
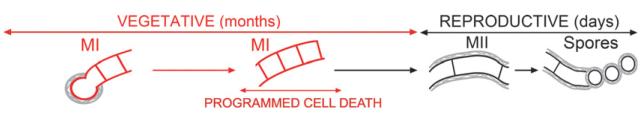
Proposed developmental model for Streptomyces growing in natural soils. Mycelial structures (MI, first mycelium; MII, second mycelium), vegetative and reproductive phases, and PCD are indicated. The vegetative phase is the predominant one. See text for details.
Streptomyces programmed cell death
Bacterial PCD can be defined as any type of genetically controlled cell dismantling involving the activation of specific cell death transducers, regulators, and effectors (Engelberg-Kulka et al., 2006). PCD was described in bacteria from different taxa, such as Bacillus and Escherichia coli (Engelberg-Kulka et al., 2006), Anabaena (Ning et al., 2002), Caulobacter (Hochman, 1997; Bos et al., 2012), Streptococcus (Guiral et al., 2005), Staphylococcus (Chatterjee et al., 2010), and Myxobacteria (Søgaard-Andersen & Yang, 2008). With few exceptions, such as the toxin-antitoxin modules from E. coli, the competence-sporulation processes from Bacillus subtilis (both reviewed in Engelberg-Kulka et al., 2006) and the competence processes of Streptococcus pneumonia (Guiral et al., 2005), the biochemical pathways controlling bacterial PCD, as well as the biological role of this process, are poorly understood (reviewed in Engelberg-Kulka et al., 2006). These three, well characterized bacterial PCDs are regulated in distinct ways, and there is no general biochemical model applicable to all bacterial PCD.
Miguelez et al. (1999) and Manteca et al. (2006a) demonstrated that Streptomyces death phenomena associated with development present the characteristics of programmed cell death. Biochemical parameters, such as the degradation of the cell wall and membrane, DNA/RNA degradation, corroborated the existence of a highly regulated, active cellular suicide that entails the activation of specific degradative enzymes (Manteca et al., 2006a). Among these enzymes there was a precursor of sequence non-specific nucleases involved in massive chromosomal degradation (Nicieza et al., 1999) and the sequence-specific nuclease (endoG) that produced chromosomal bands analogous to those that appear in the programmed cell death of eukaryotic cells (apoptosis) (Cal et al., 1996; Samejima & Earnshaw, 2005). A proteomic analysis revealed that PCD in S. coelicolor was accompanied by the appearance of enzymes involved in the degradation of cellular macromolecules, regulatory proteins, and stress-induced proteins (Manteca et al., 2006b). Sevillano et al. (2012), identified the first functional toxin-antitoxin system in Streptomyces that could be related to PCD. Bacteria having complex life cycles (streptomycetes, cyanobacteria, etc.) harbour several eukaryotic signalling domains and are considered to be the evolutive origin of these domains (Zhang, 1996; Aravind et al., 1999; Koonin & Aravind, 2002; Petrickova & Petricek, 2003). In all, 244 genes (3% of all Streptomyces ORFs) harbour these kinds of domains (Table 1). Further work will be necessary to characterize the biochemical regulation of Streptomyces PCD and to determine whether the genes described above, including those encoding for proteins harbouring eukaryotic type signalling domains, are involved in its regulation.
Table 1.
Genes harbouring eukaryotic type signalling domains in the Streptomyces coelicolor genome according to the Conserved Domain Database
| Eukaryotic type domain | Genes | ||||||
|---|---|---|---|---|---|---|---|
| Htra (cd00987) | SCO2171 | SCO3977 | SCO5149 | SCO6074 | |||
| TIR (cl02060) | SCO0305 | SCO2602 | SCO2680 | SCO5642 | SCO5953 | ||
| AAA ATPases (cd00009) | SCO0025 | SCO1506 | SCO2094 | SCO3373 | SCO4067 | SCO5587 | SCO6529 |
| SCO1024 | SCO1518 | SCO2449 | SCO3404 | SCO4263 | SCO6134 | SCO6623 | |
| SCO1306 | SCO1648 | SCO2617 | SCO3661 | SCO5270 | SCO6394 | SCO7523 | |
| SCO1434 | SCO1726 | SCO3018 | SCO3879 | SCO5285 | SCO6408 | SCO7582 | |
| Ser/Thr kinases (smart00220) | SCO0239 | SCO2244 | SCO3277 | SCO3860 | SCO4507 | SCO4820 | |
| SCO1278 | SCO2450 | SCO3344 | SCO4192 | SCO4775 | SCO4911 | ||
| SCO1468 | SCO2666 | SCO3360 | SCO4377 | SCO4776 | SCO5192 | SCO6951 | |
| SCO1549 | SCO2973 | SCO3621 | SCO4423 | SCO4777 | SCO6077 | SCO7240 | |
| SCO1551 | SCO2974 | SCO3820 | SCO4481 | SCO4778 | SCO6085 | SCO7251 | |
| SCO1724 | SCO3102 | SCO3821 | SCO4487 | SCO4779 | SCO6626 | SCO7291 | |
| SCO2110 | SCO3234 | SCO3848 | SCO4488 | SCO4817 | SCO6681 | ||
| AP-ATPase (cl09099) | SCO0002 | SCO1300 | SCO2677 | SCO3706 | SCO5183 | SCO5920 | |
| SCO0006 | SCO1331 | SCO2681 | SCO3824 | SCO5184 | SCO5923 | SCO6719 | |
| SCO0132 | SCO1433 | SCO2737 | SCO3876 | SCO5188 | SCO5973 | SCO6720 | |
| SCO0163 | SCO1504 | SCO2763 | SCO3886 | SCO5275 | SCO6010 | SCO6742 | |
| SCO0166 | SCO1621 | SCO2767 | SCO3934 | SCO5277 | SCO6047 | SCO6814 | |
| SCO0255 | SCO1671 | SCO2952 | SCO3947 | SCO5280 | SCO6193 | SCO6981 | |
| SCO0322 | SCO1707 | SCO2969 | SCO3958 | SCO5339 | SCO6259 | SCO7008 | |
| SCO0491 | SCO1719 | SCO2975 | SCO4075 | SCO5383 | SCO6295 | SCO7051 | |
| SCO0493 | SCO1742 | SCO3005 | SCO4116 | SCO5387 | SCO6366 | SCO7173 | |
| SCO0504 | SCO1780 | SCO3217 | SCO4259 | SCO5439 | SCO6426 | SCO7689 | |
| SCO0700 | SCO1798 | SCO3235 | SCO4316 | SCO5448 | SCO6512 | SCO7690 | |
| SCO0723 | SCO1840 | SCO3257 | SCO4359 | SCO5449 | SCO6517 | SCO7841 | |
| SCO0742 | SCO1848 | SCO3261 | SCO4405 | SCO5451 | SCO6633 | SCO7845 | |
| SCO0755 | SCO1850 | SCO3351 | SCO4508 | SCO5580 | SCO6635 | SCP1.110 | |
| SCO0756 | SCO1852 | SCO3369 | SCO4585 | SCO5603 | SCO6677 | SCP1.136 | |
| SCO0824 | SCO1966 | SCO3370 | SCO4620 | SCO5633 | SCO6683 | SCP1.169 | |
| SCO1144 | SCO2000 | SCO3372 | SCO4685 | SCO5648 | SCO6684 | SCP1.205c | |
| SCO1147 | SCO2257 | SCO3418 | SCO4797 | SCO5668 | SCO6719 | SCP1.216Ac | |
| SCO1148 | SCO2259 | SCO3453 | SCO4803 | SCO5734 | SCO6720 | SCP1.290c | |
| SCO1152 | SCO2324 | SCO3526 | SCO4909 | SCO5750 | SCO6742 | SCP1.63 | |
| SCO1183 | SCO2463 | SCO3541 | SCO4963 | SCO5802 | SCO6814 | SCP1.90c | |
| SCO1232 | SCO2523 | SCO3550 | SCO5028 | SCO5818 | SCO6849 | SCP2.05c | |
| SCO1246 | SCO2532 | SCO3556 | SCO5166 | SCO5835 | SCO6865 | ||
| IL1 like (cl01077) | SCO2021 | ||||||
| DEADc (cd00046) | SCO3732 | SCO4096 | |||||
| Caspase (cl00042) + Ser/Thr kinases | SCO6861 | ||||||
| AP-ATPase + TIR (cl02060) | SCO4632 | SCO5629 |
HtrA shock-induced-envelope-associated serine proteases (HtrA), Toll/IL-1 Receptor (TIR), AAA ATPases, Ser/Thr kinases, apoptotic ATPases (AP-ATPases), interleukin (IL)-1 receptor-associated kinase (IL1-like), DEAD-like helicases (DEADc), cysteine-aspartic proteases (caspases). Conserved domain database accession numbers are indicated in brackets.
The biological function of Streptomyces PCD remains somewhat unclear. It was reported to be involved in the generation of nutrients to be consumed by the aerial/sporulating mycelium, a kind of cannibalism (Mendez et al., 1985; Miguelez et al., 1999). If we consider that the best known bacterial PCDs, those occurring in Streptococcus and Bacillus, are involved in competence (taking fragmented DNA by transformation) (Guiral et al., 2005; Engelberg-Kulka et al., 2006), and that in the case of Bacillus, this process precedes sporulation and antibiotic production, an analogous process might also be happening in Streptomyces: appropriate DNA fragments would be produced by specific nuclease activities (Cal et al., 1996) and the lysis of MI mycelium (Manteca et al., 2006a) and incorporated by the multinucleated MII followed by recombination and the formation of a huge battery of variable spores. Several authors have hypothesized about the existence of horizontal gene transmission (HGT) phenomena in Streptomyces and other actinomycetes (Wiener et al., 1998; Ueda et al., 1999; Egan et al., 2001; Metsä-Ketelä et al., 2002; García-Vallve et al., 2003; Kawase et al., 2004; Nishio et al., 2004; Doroghazi & Buckley, 2010) but the mechanisms generating this HGT remains poorly understood. Conjugative plasmids (reviewed in Thoma & Muth, 2012) and transduction (Burke et al., 2001) may contribute in some way to this HGT, but competence/transformation may also occur. Streptomyces PCD precedes MII differentiation and sporulation and it was postulated that components released during the degradation of these dying cells could be producing diffusible signals in the form of amino acids/peptides (Sánchez & Braña, 1996) or N-acetylglucosamine (Rigali et al., 2006), thereby inducing differentiation. Further work will need to delve into the biological significance of Streptomyces PCD.
Conclusions and future perspectives
Streptomyces growth in nature differs substantially from that observed in ordinary laboratory cultures, a fact that must be borne in mind when development is analysed. MI is the vegetative mycelium and predominates in nature. Under stress conditions (nutrient/oxygen limitation etc.) it suffers a PCD and differentiates to a multinucleated mycelium (MII) that forms spore chains at the end of the cycle. Multinucleated MII would facilitate rapid growth and nucleoid division prior to sporulation. MII produces antibiotics that are decisive in helping the bacterium compete with other microorganisms.
Streptomyces research has classically focused on the aerial mycelium formation and sporulation phases taking place in solid cultures. By contrast, pre-sporulation stages, including differentiation in liquid cultures, have been largely ignored. The new insights regarding presporulation stages of Streptomyces in combination with future work aimed at understanding the biochemical regulation of these processes will be key to comprehending and optimizing hyphae differentiation in industrial fermentations, as well as improving the screening for new secondary metabolites from natural Streptomyces strains.
Acknowledgements
This research was funded by an ERC Starting Grant (Strp-differentiation 280304) and by grant BIO2010-16303 from the Subdirección General de Proyectos de Investigación, (DGI), Ministry of Science and Innovation (MICINN). We also thank Priscilla A. Chase for proofreading the text.
References
- Aharonowitz Y. Nitrogen metabolite regulation of antibiotic biosynthesis. Annu Rev Microbiol. 1980;34:209–233. doi: 10.1146/annurev.mi.34.100180.001233. [DOI] [PubMed] [Google Scholar]
- Anukool U, Gaze WH, Wellington EM. In situ monitoring of streptothricin production by Streptomyces rochei F20 in soil and rhizosphere. Appl Environ Microbiol. 2004;70:5222–52228. doi: 10.1128/AEM.70.9.5222-5228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Dixit VM, Koonin EV. The domains of death: evolution of the apoptosis machinery. Trends Biochem Sci. 1999;24:47–53. doi: 10.1016/s0968-0004(98)01341-3. [DOI] [PubMed] [Google Scholar]
- Bos J, Yakhnina AA, Gitai Z. BapE DNA endonuclease induces an apoptotic-like response to DNA damage in Caulobacter. P Natl Acad Sci USA. 2012;109:18096–18101. doi: 10.1073/pnas.1213332109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J, Schneider D, Westpheling J. Generalized transduction in Streptomyces coelicolor. P Natl Acad Sci USA. 2001;98:6289–6294. doi: 10.1073/pnas.101589398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cal S, Connolly BA, Nicieza RG, Sánchez J. Interaction of the periplasmic dG-selective Streptomyces antibioticus nuclease with oligonucleotide substrates. Biochemistry. 1996;35:10828–10836. doi: 10.1021/bi960616u. [DOI] [PubMed] [Google Scholar]
- Chater KF. Morphological and physiological differentiation in Streptomyces. In: Losick R, Shapiro L, editors. Microbial Development. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1984. pp. 89–115. [Google Scholar]
- Chatterjee I, Neumayer D, Herrmann M. Senescence of staphylococci: using functional genomics to unravel the roles of ClpC ATPase during late stationary phase. Int J Med Microbiol. 2010;300:130–136. doi: 10.1016/j.ijmm.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Chouayekh H, Virolle MJ. The polyphosphate kinase plays a negative role in the control of antibiotic production in Streptomyces lividans. Mol Microbiol. 2002;43:919–930. doi: 10.1046/j.1365-2958.2002.02557.x. [DOI] [PubMed] [Google Scholar]
- Chouayekh H, Nothaft H, Delaunay S, Linder M, Payrastre B, Seghezzi N, Titgemeyer F, Virolle MJ. Phosphoinositides are involved in control of the glucose-dependent growth resumption that follows the transition phase in Streptomyces lividans. J Bacteriol. 2007;189:741–749. doi: 10.1128/JB.00891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D, de Jong W, Dijkhuizen L, Wösten HA. Regulation of Streptomyces development: reach for the sky! Trends Microbiol. 2006;14:313–319. doi: 10.1016/j.tim.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Daza A, Martín JF, Dominguez A, Gil JA. Sporulation of several species of Streptomyces in submerged cultures after nutritional downshift. J Gen Microbiol. 1989;135:2483–2491. doi: 10.1099/00221287-135-9-2483. [DOI] [PubMed] [Google Scholar]
- Derouaux A, Halici S, Nothaft H, Neutelings T, Moutzourelis G, Dusart J, Titgemeyer F, Rigali S. Deletion of a cyclic AMP receptor protein homologue diminishes germination and affects morphological development of Streptomyces coelicolor. J Bacteriol. 2004;186:1893–1897. doi: 10.1128/JB.186.6.1893-1897.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroghazi JR, Buckley DH. Widespread homologous recombination within and between Streptomyces species. ISME J. 2010;4:1136–1143. doi: 10.1038/ismej.2010.45. [DOI] [PubMed] [Google Scholar]
- Eaton D, Ensign JC. Streptomyces viridochromogenes spore germination initiated by calcium ions. J Bacteriol. 1980;143:377–382. doi: 10.1128/jb.143.1.377-382.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan S, Wiener P, Kallifidas D, Wellington EM. Phylogeny of Streptomyces species and evidence for horizontal transfer of entire and partial antibiotic gene clusters. Antonie Van Leeuwenhoek. 2001;79:127–133. doi: 10.1023/a:1010296220929. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2:e135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flärdh K, Buttner MJ. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol. 2009;7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- García M. A membrane-like structure envelopes substrate mycelium during colony development in Streptomyces. FEMS Microbiol Lett. 1995;131:107–111. doi: 10.1016/0378-1097(95)00246-2. [DOI] [PubMed] [Google Scholar]
- García-Vallve S, Guzman E, Montero MA, Romeu A. HGT-DB: a database of putative horizontally transferred genes in prokaryotic complete genomes. Nucleic Acids Res. 2003;31:187–189. doi: 10.1093/nar/gkg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass NL, Kaneko I. Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot Cell. 2003;2:1–8. doi: 10.1128/EC.2.1.1-8.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook MA, Doull JL, Stuttard C, Vining LC. Sporulation of Streptomyces venezuelae in submerged cultures. J Gen Microbiol. 1990;136:581–588. doi: 10.1099/00221287-136-3-581. [DOI] [PubMed] [Google Scholar]
- Granozzi C, Billetta R, Passantino R, Sollazzo M, Puglia AM. A breakdown in macromolecular synthesis preceding differentiation in Streptomyces coelicolor A3(2) J Gen Microbiol. 1990;136:713–716. doi: 10.1099/00221287-136-4-713. [DOI] [PubMed] [Google Scholar]
- Grund AD, Ensign JC. Properties of the germination inhibitor of Streptomyces viridochromogenes spores. J Gen Microbiol. 1985;131:833–847. doi: 10.1099/00221287-131-4-833. [DOI] [PubMed] [Google Scholar]
- Guijarro JA, Suarez JE, Salas JA, Hardisson C. Pattern of protein degradation during germination of Streptomyces antibioticus spores. Can J Microbiol. 1983;29:637–643. doi: 10.1139/m83-103. [DOI] [PubMed] [Google Scholar]
- Guiral S, Mitchell TJ, Martin B, Claverys JP. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. P Natl Acad Sci USA. 2005;14:8710–8715. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser HJ, Yousef MR, Elliot MA. Cell wall hydrolases affect germination, vegetative growth, and sporulation in Streptomyces coelicolor. J Bacteriol. 2009;191:6501–6512. doi: 10.1128/JB.00767-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardisson C, Manzanal MB, Salas JA, Suárez JE. Fine structure, physiology and biochemistry of arthrospore germination in Streptomyces antibioticus. J Gen Microbiol. 1978;105:203–214. doi: 10.1099/00221287-105-2-203. [DOI] [PubMed] [Google Scholar]
- Hey-Ferguson A, Mitchell M, Elbein AD. Trehalose metabolism in germinating spores of Streptomyces hygroscopicus. J Bacteriol. 1973;116:1084–1085. doi: 10.1128/jb.116.2.1084-1085.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch CF, Ensign JC. Heat activation of Streptomyces viridochromogenes spores. J Bacteriol. 1976;126:24–30. doi: 10.1128/jb.126.1.24-30.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochman A. Programmed cell death in prokaryotes. Crit Rev Microbiol. 1997;23:207–214. doi: 10.3109/10408419709115136. [DOI] [PubMed] [Google Scholar]
- Hopwood DA. Streptomyces in Nature and Medicine: The Antibiotic Makers. Oxford University Press; New York: 2007. [Google Scholar]
- Jakimowicz D, van Wezel GP. Cell division and DNA segregation in Streptomyces: how to build a septum in the middle of nowhere? Mol Microbiol. 2012;85:393–404. doi: 10.1111/j.1365-2958.2012.08107.x. [DOI] [PubMed] [Google Scholar]
- de Jong W, Manteca A, Sánchez J, Bucca G, Smith CP, Dijkhuizen L, Claessen D, Wösten HA. NepA is a structural cell wall protein involved in maintenance of spore dormancy in Streptomyces coelicolor. Mol Microbiol. 2009;71:1591–1603. doi: 10.1111/j.1365-2958.2009.06633.x. [DOI] [PubMed] [Google Scholar]
- Kawase T, Saito A, Sato T, Kanai R, Fujii T, Nikaidou N, Miyashita K, Watanabe T. Distribution and phylogenetic analysis of family 19 chitinases in Actinobacteria. Appl Environ Microbiol. 2004;70:1135–1144. doi: 10.1128/AEM.70.2.1135-1144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen GH, Plaskitt KA, Lewis CG, Findlay KC, Buttner MJ. Deletion of DNA lying close to the glkA locus induces ectopic sporulation in Streptomyces coelicolor A3(2) Mol Microbiol. 1995;17:221–230. doi: 10.1111/j.1365-2958.1995.mmi_17020221.x. [DOI] [PubMed] [Google Scholar]
- Kendrick KE, Ensign JC. Sporulation of Streptomyces griseus in submerged culture. J Bacteriol. 1983;155:357–366. doi: 10.1128/jb.155.1.357-366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. The John Innes Foundation; Norwich: 2000. Growth and preservation of Streptomyces; pp. 49–54. Availble at: http://www.jic.ac.uk/science/molmicro/Strepmanual/Manual.htm. [Google Scholar]
- Koepsel R, Ensign JC. Microcycle sporulation of Streptomyces viridochromogenes. Arch Microbiol. 1984;140:9–14. doi: 10.1007/BF00409764. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 2002;9:394–404. doi: 10.1038/sj.cdd.4400991. [DOI] [PubMed] [Google Scholar]
- Kuimova TF, Soina VS. A submerged sporulation and ultrastructural changes in the mycelium of Streptomyces chrysomallus. Hindustan Antibiot Bull. 1981;23:1–5. [PubMed] [Google Scholar]
- Kwak J, Kendrick KE. Bald mutants of Streptomyces griseus that prematurely undergo key events of sporulation. J Bacteriol. 1996;178:4643–4650. doi: 10.1128/jb.178.15.4643-4650.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manteca A, Sánchez J. Streptomyces development in colonies and soils. Appl Environ Microbiol. 2009;75:2920–2924. doi: 10.1128/AEM.02288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manteca A, Fernández M, Sánchez J. A death round affecting a young compartmentalized mycelium precedes aerial mycelium dismantling in confluent solid cultures of Streptomyces antibioticus. Microbiology. 2005;151:3689–3697. doi: 10.1099/mic.0.28045-0. [DOI] [PubMed] [Google Scholar]
- Manteca A, Fernández M, Sánchez J. Cytological and biochemical evidence for an early cell dismantling event in solid cultures of Streptomyces antibioticus. Res Microbiol. 2006a;157:143–152. doi: 10.1016/j.resmic.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Manteca A, Mader U, Connolly BA, Sánchez J. A proteomic analysis of Streptomyces coelicolor programmed cell death. Proteomics. 2006b;6:6008–6022. doi: 10.1002/pmic.200600147. [DOI] [PubMed] [Google Scholar]
- Manteca A, Claessen D, Lopez-Iglesias C, Sánchez J. Aerial hyphae in solid cultures of Streptomyces lividans and Streptomyces coelicolor originate from viable segments surviving an earlyprogrammed cell death event. FEMS Microbiol lett. 2007;274:118–125. doi: 10.1111/j.1574-6968.2007.00825.x. [DOI] [PubMed] [Google Scholar]
- Manteca A, Alvarez R, Salazar N, Yagüe P, Sánchez J. Mycelium differentiation and antibiotic production in liquid cultures of Streptomyces coelicolor. Appl Environ Microbiol. 2008;74:3877–3886. doi: 10.1128/AEM.02715-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manteca A, Sánchez J, Jung HR, Schwämmle V, Jensen ON. Quantitative proteomic analysis of Streptomyces coelicolor development demonstrates the switch from primary to secondary metabolism associated with hyphae differentiation. Mol Cell Proteomics. 2010a;9:1423–1436. doi: 10.1074/mcp.M900449-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manteca A, Jung HR, Schwämmle V, Jensen ON, Sánchez J. Quantitative proteomic analysis of Streptomyces coelicolor in liquid cultures reveals the switch in metabolism associated with hyphae differentiation. J Proteome Res. 2010b;9:4801–4811. doi: 10.1021/pr100513p. [DOI] [PubMed] [Google Scholar]
- Martín JF. Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: an unfinished story. J Bacteriol. 2004;186:5197–5201. doi: 10.1128/JB.186.16.5197-5201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride MJ, Ensign JC. Metabolism of endogenous trehalose by Streptomyces griseus spores and by spores or cells of other actinomycetes. J Bacteriol. 1987;169:5002–5007. doi: 10.1128/jb.169.11.5002-5007.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick JR, Flärdh K. Signals and regulators that govern Streptomyces development. FEMS Microbiol Rev. 2012;36:206–231. doi: 10.1111/j.1574-6976.2011.00317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick JR, Su EP, Driks A, Losick R. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol Microbiol. 1994;14:243–254. doi: 10.1111/j.1365-2958.1994.tb01285.x. [DOI] [PubMed] [Google Scholar]
- Mendez C, Braña AF, Manzanal MB, Hardisson C. Role of substrate mycelium in colony development in Streptomyces. Can J Microbiol. 1985;31:446–450. doi: 10.1139/m85-083. [DOI] [PubMed] [Google Scholar]
- Metsä-Ketelä M, Halo L, Munukka E, Hakala J, Mäntsälä P, Ylihonko K. Molecular evolution of aromatic polyketides and comparative sequence analysis of polyketide ketosynthase and 16S ribosomal DNA genes from various Streptomyces species. Appl Environ Microbiol. 2002;68:4472–4479. doi: 10.1128/AEM.68.9.4472-4479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguelez EM, Rueda B, Hardisson C, Manzanal MB. Colony development in Streptomyces carpinensis: a streptomycete with substrate mycelium spores. FEMS Microbiol Lett. 1997;157:103–107. [Google Scholar]
- Miguelez EM, Hardisson C, Manzanal MB. Hyphal death during colony development in Streptomyces antibioticus: morphological evidence for the existence of a process of cell deletion equivalent to apoptosis in a multicellular prokaryote. J Cell Biol. 1999;145:515–525. doi: 10.1083/jcb.145.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulík K, Janda I, Weiser J, Stastná J, Jiránová A. RNA and ribosomal protein patterns during aerial spore germination in Streptomyces granaticolor. Eur J Biochem. 1984;145:381–388. doi: 10.1111/j.1432-1033.1984.tb08565.x. [DOI] [PubMed] [Google Scholar]
- Neumann T, Piepersberg W, Distler J. Decision phase regulation of streptomycin production in Streptomyces griseus. Microbiology. 1996;142:1953–1963. [Google Scholar]
- Nicieza GR, Huergo J, Connolly BA, Sánchez J. Purification, characterization, and role of nucleases and serine proteases in Streptomyces differentiation. J Biol Chem. 1999;274:20366–20375. doi: 10.1074/jbc.274.29.20366. [DOI] [PubMed] [Google Scholar]
- Ning SB, Guo HL, Wang L, Song YC. Salt stress induces programmed cell death in prokaryotic organism Anabaena. J Appl Microbiol. 2002;93:15–28. doi: 10.1046/j.1365-2672.2002.01651.x. [DOI] [PubMed] [Google Scholar]
- Nishio Y, Nakamura Y, Usuda Y, Sugimoto S, Matsui K, Kawarabayasi Y, Kikuchi H, Gojobori T, Ikeo K. Evolutionary process of amino acid biosynthesis in Corynebacterium at the whole genome level. Mol Biol Evol. 2004;21:1683–1691. doi: 10.1093/molbev/msh175. [DOI] [PubMed] [Google Scholar]
- Noens EE, Mersinias V, Willemse J, Traag BA, Laing E, Chater KF, Smith CP, Koerten HK, van Wezel GP. Loss of the controlled localization of growth stage-specific cell wall synthesis pleiotropically affects developmental gene expression in an ssgA mutant of Streptomyces coelicolor. Mol Microbiol. 2007;64:1244–1259. doi: 10.1111/j.1365-2958.2007.05732.x. [DOI] [PubMed] [Google Scholar]
- Novella IS, Barbés C, Sánchez J. Sporulation of Streptomyces antibioticus ETHZ 7451 in submerged culture. Can J Microbiol. 1992;38:769–773. doi: 10.1139/m92-125. [DOI] [PubMed] [Google Scholar]
- Novotna J, Vohradsky J, Berndt P, Gramajo H, Langen H, Li XM, Minas W, Orsaria L, Roeder D, Thompson CJ. Proteomic studies of diauxic lag in the differentiating prokaryote Streptomyces coelicolor reveal a regulatory network of stress-induced proteins and central metabolic enzymes. Mol Microbiol. 2003;48:1289–1303. doi: 10.1046/j.1365-2958.2003.03529.x. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y, Seo JW, Horinouchi S. Deprogrammed sporulation in Streptomyces. FEMS Microbiol Lett. 2002;216:1–7. doi: 10.1111/j.1574-6968.2002.tb11406.x. [DOI] [PubMed] [Google Scholar]
- Okanishi M, Suzuki K, Umezawa H. Formation and reversion of Streptomycete protoplasts: cultural condition and morphological study. J Gen Microbiol. 1974;80:389–400. doi: 10.1099/00221287-80-2-389. [DOI] [PubMed] [Google Scholar]
- Petersen F, Zähner H, Metzger JW, Freund S, Hummel RP. Germicidin, an autoregulative germination inhibitor of Streptomyces viridochromogenes NRRL B-1551. J Antibiot (Tokyo) 1993;46:1126–1138. doi: 10.7164/antibiotics.46.1126. [DOI] [PubMed] [Google Scholar]
- Petrickova K, Petricek M. Eukaryotic-type protein kinases in Streptomyces coelicolor: variations on a common theme. Microbiology. 2003;149:1609–1621. doi: 10.1099/mic.0.26275-0. [DOI] [PubMed] [Google Scholar]
- Piette A, Derouaux A, Gerkens P, et al. From dormant to germinating spores of Streptomyces coelicolor A3(2): new perspectives from the crp null mutant. J Proteome Res. 2005;4:1699–1708. doi: 10.1021/pr050155b. [DOI] [PubMed] [Google Scholar]
- Rho YT, Lee KJ. Kinetic characterization of sporulation in Streptomyces albidoflavus SMF301 during submerged culture. Microbiology. 1994;140:2061–2065. doi: 10.1099/13500872-140-8-2061. [DOI] [PubMed] [Google Scholar]
- Rigali S, Nothaft H, Noens EE, et al. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol Microbiol. 2006;61:1237–1251. doi: 10.1111/j.1365-2958.2006.05319.x. [DOI] [PubMed] [Google Scholar]
- Rueda B, Miguélez EM, Hardisson C, Manzanal MB. Mycelial differentiation and spore formation by Streptomyces brasiliensis in submerged culture. Can J Microbiol. 2001;47:1042–1047. [PubMed] [Google Scholar]
- Salas JA, Guijarro JA, Hardisson C. High calcium content in Streptomyces spores and its release as an early event during spore germination. J Bacteriol. 1983;155:1316–1323. doi: 10.1128/jb.155.3.1316-1323.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima K, Earnshaw WC. Trashing the genome: the role of nucleases during apoptosis. Nat Rev Mol Cell Biol. 2005;6:677–688. doi: 10.1038/nrm1715. [DOI] [PubMed] [Google Scholar]
- Sánchez L, Braña AF. Cell density influences antibiotic biosynthesis in Streptomyces clavuligerus. Microbiology. 1996;142:1209–1220. doi: 10.1099/13500872-142-5-1209. [DOI] [PubMed] [Google Scholar]
- Sánchez S, Chávez A, Forero A, et al. Carbon source regulation of antibiotic production. J Antibiot (Tokyo) 2010;63:442–459. doi: 10.1038/ja.2010.78. [DOI] [PubMed] [Google Scholar]
- Sevillano L, Díaz M, Yamaguchi Y, Inouye M, Santamaría RI. Identification of the first functional toxin-antitoxin system in Streptomyces. PLoS ONE. 2012;7:e32977. doi: 10.1371/journal.pone.0032977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SP, Montgomery BL. Determining cell shape: adaptive regulation of cyanobacterial cellular differentiation and morphology. Trends Microbiol. 2011;19:278–285. doi: 10.1016/j.tim.2011.03.001. [DOI] [PubMed] [Google Scholar]
- Søgaard-Andersen L, Yang Z. Programmed cell death: role for MazF and MrpC in Myxococcus multicellular development. Curr Biol. 2008;18:337–339. doi: 10.1016/j.cub.2008.02.060. [DOI] [PubMed] [Google Scholar]
- Song L, Barona-Gomez F, Corre C, Xiang L, Udwary DW, Austin MB, Noel JP, Moore BS, Challis GL. Type III polyketide synthase beta-ketoacyl-ACP starter unit and ethylmalonyl-CoA extender unit selectivity discovered by Streptomyces coelicolor genome mining. J Am Chem Soc. 2006;128:14754–14755. doi: 10.1021/ja065247w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strakova E, Bobek J, Zikova A, Rehulka P, Benada O, Rehulkova H, Kofronova O, Vohradsky J. Systems insight into the spore germination of Streptomyces coelicolor. J Proteome Res. 2013;12:525–536. doi: 10.1021/pr300980v. [DOI] [PubMed] [Google Scholar]
- Süsstrunk U, Pidoux J, Taubert S, Ullmann A, Thompson CJ. Pleiotropic effects of cAMP on germination, antibiotic biosynthesis and morphological development in Streptomyces coelicolor. Mol Microbiol. 1998;30:33–46. doi: 10.1046/j.1365-2958.1998.01033.x. [DOI] [PubMed] [Google Scholar]
- Thoma L, Muth G. Conjugative DNA transfer in Streptomyces by TraB: is one protein enough? FEMS Microbiol Lett. 2012;337:81–88. doi: 10.1111/1574-6968.12031. [DOI] [PubMed] [Google Scholar]
- Ueda K, Seki T, Kudo T, Yoshida T, Kataoka M. Two distinct mechanisms cause heterogeneity of 16S rRNA. J Bacteriol. 1999;181:78–82. doi: 10.1128/jb.181.1.78-82.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SL, Fan KQ, Yang X, Lin ZX, Xu XP, Yang KQ. CabC, an EF-hand calcium-binding protein, is involved in Ca2+-mediated regulation of spore germination and aerial hypha formation in Streptomyces coelicolor. J Bacteriol. 2008;190:4061–4068. doi: 10.1128/JB.01954-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellington EM, Cresswell N, Saunders VA. Growth and survival of Streptomycete inoculants and extent of plasmid transfer in sterile and nonsterile soil. Appl Environ Microbiol. 1990;56:1413–1419. doi: 10.1128/aem.56.5.1413-1419.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener P, Egan S, Huddleston AS, Wellington EM. Evidence for transfer of antibiotic-resistance genes in soil populations of streptomycetes. Mol Ecol. 1998;7:1205–1216. doi: 10.1046/j.1365-294x.1998.00450.x. [DOI] [PubMed] [Google Scholar]
- Wildermuth H. Development and organization of the aerial mycelium in Streptomyces coelicolor. J Gen Microbiol. 1970;60:43–50. doi: 10.1099/00221287-60-1-43. [DOI] [PubMed] [Google Scholar]
- Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 2011;25:89–99. doi: 10.1101/gad.600211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ST. Oligotrophy in soil: fact or fiction? In: Fletcher M, Floodgate GD, editors. Bacteria in their Natural Environment. Academic Press; London: 1985. pp. 81–110. [Google Scholar]
- Yagüe P, Rodriguez-Garcia A, Lopez-Garcia MT, Martin JF, Rioseras B, Sanchez J, Manteca A. Transcriptomic analysis of Streptomyces coelicolor differentiation in solid sporulating cultures: first compartmentalized and second multinucleated mycelia have different and distinctive transcriptomes. PLoS ONE. 2013;8:e60665. doi: 10.1371/journal.pone.0060665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CC. Bacterial signalling involving eukaryotic-type protein kinases. Mol Microbiol. 1996;20:9–15. doi: 10.1111/j.1365-2958.1996.tb02483.x. [DOI] [PubMed] [Google Scholar]
- Zhou LH, Li YQ, Li YQ, Wu D. Spatio-temporal expression of the pathway-specific regulatory gene redD in S. coelicolor. J Zhejiang Univ Sci. 2005;6:464–469. doi: 10.1631/jzus.2005.B0464. [DOI] [PMC free article] [PubMed] [Google Scholar]