Abstract
Nuclear transfer (NT) remains the most effective method to reprogram somatic cells to totipotency. Somatic cell nuclear transfer (SCNT) efficiency however remains low, but recurrent problems occurring in partially reprogrammed cloned embryos have recently been identified and some remedied. In particular, the trophectoderm has been identified as a lineage whose reprogramming success has a large influence on SCNT embryo development. Several interspecific hybrid and cybrid reprogramming systems have been developed as they offer various technical advantages and potential applications, and together with SCNT, they have led to the identification of a series of reprogramming events and responsible reprogramming factors. Interspecific incompatibilities hinder full exploitation of cross-species reprogramming systems, yet recent findings suggest that these may not constitute insurmountable obstacles.
Keywords: Nuclear reprogramming, Somatic cell nuclear transfer (SCNT), Interspecies SCNT (iSCNT), Hybrid, Nucleocytoplasmic hybrid (Cybrid), Nucleocytoplasmic incompatibility, Microcell-mediated chromosome transfer (MMCT), Interspecies intra-cytoplasmic sperm injection (iICSI), Cell fusion
The recent finding that non-enucleated human oocytes can efficiently reprogram transferred human somatic nuclei to pluripotency (21% triploid blastocyst formation and ~3% ES-like cell line derivation) [1] renews the interest in NT. SCNT using enucleated oocytes in other mammalian species seems to remain the most effective method to reprogram somatic cells to pluripotency [2,3]. In addition, since NT is the only available method to reprogram cells to totipotency, and chimera formation in primates is exclusively possible via aggregation of totipotent cells [4], the eventual generation of chimeric primates containing cells genetically-modified in culture would require NT. The mechanisms of reprogramming by NT are being investigated both directly in intraspecies SCNT embryos and in a variety of cross-species hybrid and cybrid systems. Interspecific hybrid cells or organisms comprise genomes, or parts of genomes, that originate from more than one species, while interspecific cybrids, also known as nucleocytoplasmic hybrids, originate from the combination of the nucleus or genome of one species with cytoplasm of another species.
The viability of cross-species (hybrid and cybrid) embryos tends to be inversely correlated with the genetic or evolutionary distance between the species that are combined [5,6]. A better understanding of the boundaries between species is not only fundamentally interesting, but it may also enable researchers, through intervention, to extend the actual range of viable hybrids and cybrids. This could be useful for various zoological, agricultural and biomedical purposes, including the identification of conserved reprogramming mechanisms. In addition to highlighting major advances in SCNT research, we review here the current hybrid and cybrid systems, the mechanistic insights they have recently provided into nuclear reprogramming, as well as the nature of interspecific incompatibilities.
Recent progress in somatic cell nuclear transfer
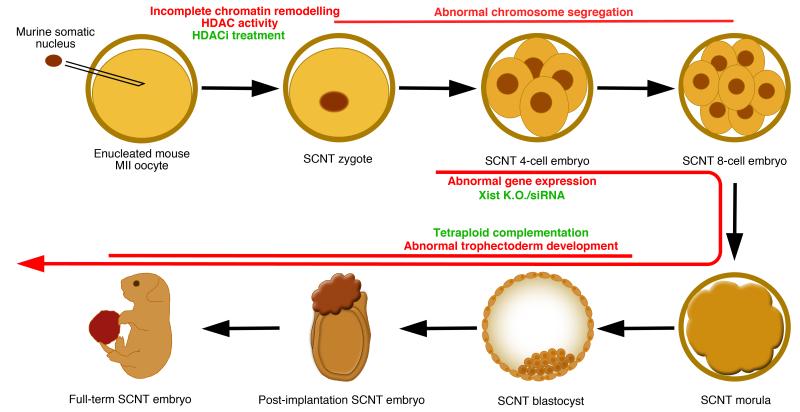
In mice, using cumulus cells as nuclear donors, up to ~65% of SCNT embryos develop to the morula/blastocyst stage, but only <3% of the transferred embryos develop to term [7]. With such a low implantation and post-implantation development efficiency, several investigators have asked whether recurrent defects could explain the lethality of SCNT embryos. Inhibition of histone deacetylases, such as with Trichostatin A (TSA) treatment, increases full-term development of cloned embryos [8] likely through improving several early events of nuclear reprogramming, including chromatin remodelling, histone modifications, DNA replication and transcriptional activity [9,10]. In pigs, DJ-1 is an ooplasm component that is, specifically in SCNT embryos, required for survival to the blastocyst stage through negatively regulating p53 signalling [11]. Inhibition of p53 also improves trophectoderm function of SCNT blastocysts in mice [12]. Since p53 normally acts to suppress cell division in response to DNA damage, these results suggest that DNA damage may often occur during the early cleavages in cloned embryos, activating p53, which then induces cell cycle arrest and/or apoptosis, thereby explaining some of the developmental delay and death of SCNT embryos. Perhaps in connection with this, errors in chromosome segregation often occur (>90% of the cases) during early cleavages in murine SCNT embryos, which appear not to affect survival until the blastocyst stage, but to impair only post-implantation development [13]. Interestingly, if the extra-embryonic lineage of a cloned blastocyst is replaced by one generated by in vitro fertilization (IVF) in a process called tetraploid complementation, the frequency of full-term development of cloned mice increases by about 6-fold [14]. In the converse experiment, where an IVF-derived inner cell mass is combined with cloned trophectoderm lineage, post-implantation development is reduced to a level near that of regular SCNT, suggesting that defects in the trophectoderm lineage may explain a large proportion of the post-transfer lethality observed in SCNT embryos [14]. Genome wide transcriptional analysis of cloned embryos revealed specific and recurrent decreased X-linked expression due to persistent Xist (a non-coding RNA that inactivates one of the two X chromosomes in females) expression from the active X chromosome (Xa) of transferred nuclei [15]. Inhibition of Xist function before the morula stage restored X-linked expression and increased implantation and post-implantation development of cloned embryos by more than 8-fold [15,16]. An overlapping set of genes (including many X-linked downregulated genes) is also often inappropriately reprogrammed in SCNT blastocysts across different nuclear donor cell types [15,17]. These results together suggest that multiple defects occur during early cleavage in SCNT embryos, including abnormal chromatin remodelling and chromosome segregation. When combined with persisting Xist expression from Xa and the resulting globally reduced X-linked expression, as well as recurrent failure to reprogram a relatively specific set of non X-linked genes, this may often impair trophectoderm function, and hence, implantation and post-implantation development of cloned blastocysts (Figure 1). This does not imply that cloned embryonic tissues are without flaws; SCNT-derived post-implantation stage epiblast stem cells and post-natal individuals often have aberrant transcriptional and/or epigenetic statuses [18,19], although whether these observed defects are cell/tissue-autonomous, or secondary to trophectoderm defects remains uncertain.
Figure 1. Recurrent reprogramming defects in SCNT embryos.
In this example, a murine somatic cell nucleus is transferred into a mitotically-enucleated oocyte, containing cytoplasmic and nuclear reprogramming factors. After NT, the chromatin of the somatic nucleus is often not completely remodelled due to persistent histone deacetylase (HDAC) activity. This, and other reprogramming aspects, can be improved by HDAC inhibitor (HDACi) treatment. Abnormal chromosome segregation often occurs during the early cleavages and appears to be a major cause of developmental failure when it happens before the 8-cell stage. Following zygotic genome activation, abnormal gene expression, including Xist RNA from Xa and the subsequent under-expression of X-linked genes, further inhibits SCNT embryo development. This can be improved by removing Xist from Xa in donor nuclei, or injection of Xist siRNA in SCNT zygotes. Finally, incomplete reprogramming of the trophectoderm lineage, and the resulting defects in trophectoderm development are a major cause of the lethality of post-implantation stage SCNT embryos. This can be rescued by replacing the trophectoderm lineage with one generated from in vitro fertilized embryos through tetraploid complementation.
In mice, recent experiments [20] solidified the concept that essential reprogramming factors are located in the zygote nucleoplasm [21,22]. In order to leave these factors in the egg/zygote, enucleation must be performed during mitosis, when the nuclear envelope is broken down. In primates, SCNT is more problematic, and reproducible derivation of monkey SCNT ES cells required the development of a less invasive mitotic egg enucleation protocol [23,24]. In humans however, even with extreme care, enucleated mitotic eggs or zygotes were unable to support the development of somatic cell nuclei beyond the 6-10 cell stage [1,25]. Interestingly, the investigators asked what happens if the oocyte haploid genome was left intact, and development of the resulting NT embryos proceeded to the blastocyst stage, enabling the derivation of triploid human ES-like cells [1]. This suggests that in humans, as opposed to all the other species in which SCNT has been tried so far, essential reprogramming factors are tightly associated with, or even comprised within, the oocyte genome, such that even mitotic enucleation removes these factors. SCNT in humans, at least with current technology, is therefore unlikely to generate clinically relevant diploid ES cells.
Hybrid experimental systems and reprogramming insights
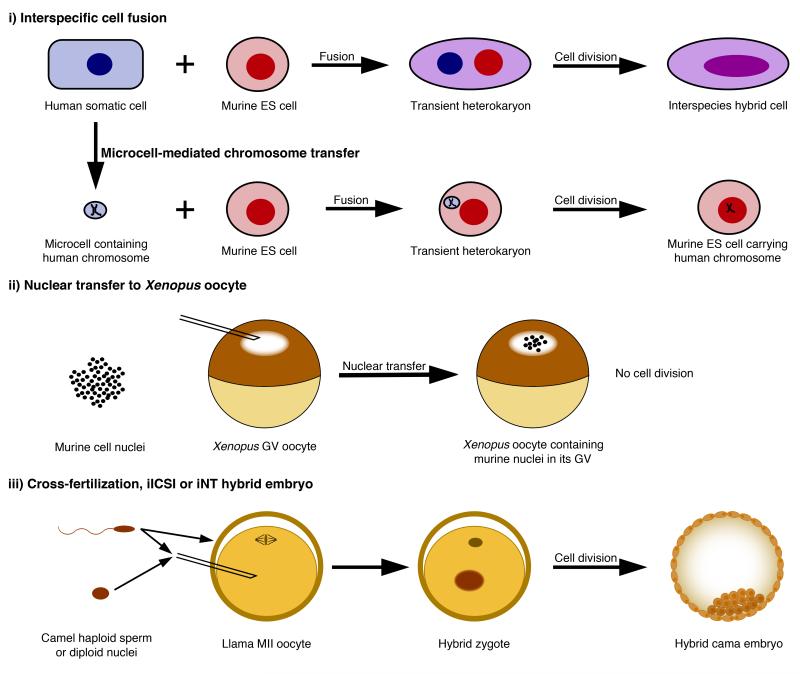
Three interspecies hybrid experimental systems currently exist: (i) hybrid cells generated by the fusion of cells or microcells from two species (we ignore here hybrids containing less than one chromosome from another species), (ii) interspecies NT (iNT) to Xenopus oocyte (at the first meiotic prophase), and (iii) hybrid embryos (Figure 2). When undifferentiated cells are fused with differentiated cells, differentiated cells are usually reprogrammed towards an undifferentiated state, even if the two cell types originate from different species [26,27]. Transcriptional analyses are greatly facilitated in interspecific hybrids due to genetic differences between species that make it possible to distinguish expression from each species’ genome by RT-PCR [28]. Using interspecific cell fusion, it was shown that the Oct4 transcription factor, Polycomb repressive complex (PRC), and AID-dependent DNA demethylation activity are required for ES cells to successfully reactivate pluripotency gene transcription in somatic cells [28-30] (See review by A. Fisher in this issue). A cell fusion variant called microcell-mediated chromosome transfer interestingly enables the fusion of only one to a few chromosomes from one cell type/species to another cell [31]. Chromosome-wide analysis of murine cells that carried human chromosome 21 revealed that murine transcription factors were bound to and transcribed genes from human chromosome 21 in a human manner [32]. Thus, genetic sequence primarily directs transcriptional programs, even in an interspecific cellular environment. Xenopus oocytes are intensely active in transcription. It is possible to transplant up to hundreds of mammalian cell nuclei directly into the Xenopus oocyte nucleus, referred to as the germinal vesicle (GV), and this procedure induces the reactivation of transcriptionally silent mammalian loci [33]. Transcriptional reprogramming of mammalian somatic nuclei upon nuclear transfer to a Xenopus oocyte GV is inhibited by the presence of a histone variant called Macro H2A in somatic nuclei, and requires oocyte linker histone B4 and nuclear actin polymerization [34-36]. Interspecies intra-cytoplasmic sperm injection (iICSI) has been used mainly to study the early reprogramming effect of non-enucleated oocytes on interspecific sperm nuclei, including DNA demethylation, chromatin structure (HP1 levels), and several histone modifications (changes in H3 methylation, H3 and H4 acetylation) [37-39]. Interspecific cell fusion, iICSI and iNT to Xenopus oocyte experiments have thus significantly contributed to our current knowledge of reprogramming factors and mechanisms (reviewed in [40]), while they also suggest that these mechanisms are highly conserved.
Figure 2. Interspecific hybrid reprogramming systems.
(i) In interspecific cell fusion, cultured cells of one species are fused to cells of another species, resulting in the formation of transient heterokaryons in which one cell type is transcriptionally reprogrammed towards the other cell type. A heterokaryon may undergo mitosis, in which case the two nuclei will fuse to generate a proliferating hybrid cell. Interspecies microcell-mediated chromosome transfer is similar to cell fusion, except that the nuclei of the cells from one species are fragmented into microcells containing only one to a few chromosomes prior to fusing with interspecific cells. (ii) In NT to Xenopus oocyte GV, a few hundreds of mammalian nuclei are injected into the oocyte GV, which induces the transcriptional reactivation of previously silenced genes within a day or two in the absence of cell division. (iii) Hybrid embryos can be generated either by cross-fertilization, iICSI or iNT using non-enucleated interspecies oocytes.
Cybrid experimental systems and reprogramming insights
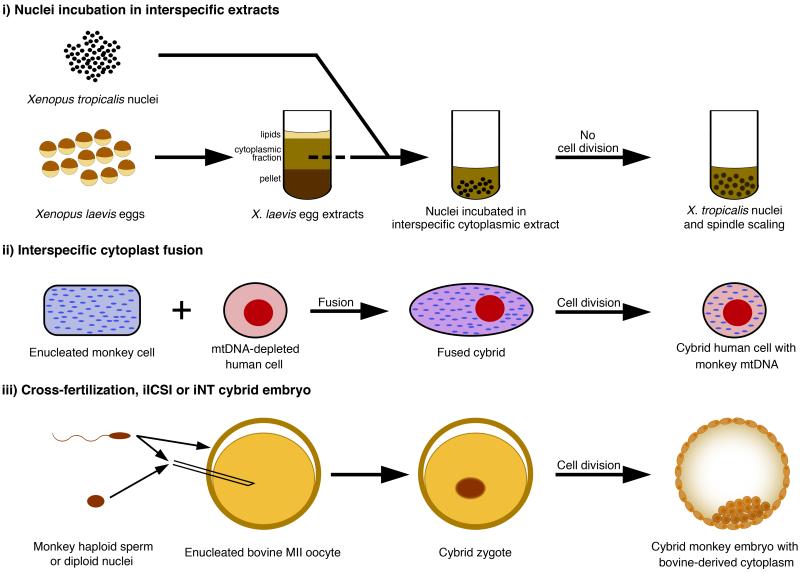
Current cybrid experimental systems can be subdivided into three categories: those that involve (i) nuclei incubated in interspecific cellular extract, (ii) cybrid cell lines generated through cytoplast fusion, and (iii) cybrid embryos (Figure 3). Xenopus laevis egg and oocyte extracts can be easily prepared in large quantities and their ability to reprogram nuclei from other species has been investigated. Using this system, the chromatin remodelling ATPases ISWI and BRG1, as well as nucleoplasmin, were found as egg components required for reprogramming of somatic nuclei [41-43]. Extracts from a distantly related frog species, Xenopus tropicalis, whose embryonic cells and nuclei are smaller in size than those of X. laevis, have also been prepared, and the incubation of sperm nuclei from one species in egg extracts of another species and vice-versa, demonstrated that the magnitude of nuclear swelling, as well as spindle length, were dependent on egg cytoplasmic factors/species rather than on nuclear content/species. This was due to differences in the relative concentrations of two key nuclear transport factors in cells of the two species in the case of nuclear swelling, and to the gain of an inhibitory phosphorylation site in a microtubule-severing factor homolog in X. laevis in the case of spindle scaling [44-46]. Thus, there seem to exist quantitative and functional differences in relatively few key factors between species that may account for their divergent nuclear remodelling phenotypes.
Figure 3. Interspecific cybrid reprogramming systems.
(i) Nuclei from one species are incubated into M-phase egg extracts prepared from another species, resulting in nuclear and spindle scaling. (ii) In interspecific cytoplast fusion, cultured cells from one species are enucleated and fused with cells from another species. This can be combined with mtDNA-depletion of the other species cells to generate xenomitochondrial cybrid cells. (iii) Cybrid embryos can be generated either by cross-fertilization, iICSI or iNT using enucleated interspecies oocyte.
Experiments in which cultured mtDNA-less human cells were fused with various cytoplasts isolated from other primate species showed that human nuclei are compatible with mtDNA of some of their closest non-human relatives, such as chimpanzees and gorillas, but not with mtDNA of species separated by 18 million years (MY) of evolution (orangutans) or more [47]. Interestingly, intraspecific fusion of human fibroblasts or hepatocytes with ES cell cytoplasts did not fully reprogram the somatic nuclei, suggesting that reprogramming to pluripotency by ES cell fusion requires the ES cell nucleus [48,49]. Many investigators have tested the ability of enucleated eggs to reprogram interspecific somatic nuclei and generate viable cybrid embryos/progeny, largely using interspecies SCNT (iSCNT) [5,6]. In most cases, a fraction of iSCNT embryos developed until the blastocyst stage, although reprogramming defects were observed in some instances, including a failure to properly activate pluripotency genes [50-52]. Several pluripotency genes were nonetheless properly activated in many cybrids [50,53,54], and ES-like cells have been isolated from iNT blastocysts in a few occasions, although the efficiency was low and the reproducibility of the results remains uncertain [55-57]. These results are nonetheless important as they suggest that homologous egg factors needed to reprogram cells to pluripotency may function across many mammalian species.
The nature of hybrid and cybrid incompatibilities
A long-standing question is whether the lethality and developmental failure of distantly related hybrid and cybrid embryos are due to cellular incompatibilities, reprogramming defects, and/or developmental failure. Hybrids generally survive better than cybrids of the same interspecific combinations, indicating that the ooplasm most often supports better the presence of an interspecific genome than the lack of an intraspecific genome [58,59]. Indeed, most cybrids made from divergent species could only develop until the blastocyst stage, and they were characterized by a range of defects, including developmental delay or failure, reduced cell numbers, nuclear/genome damage, structural disorganization, aberrant gene expression and energy levels, as well as defects in nucleologenesis [50-52,54,59-66]. Despite all these observed defects in cybrid embryos, very little is known about the nature of the underlying nucleocytoplasmic incompatibility. In iSCNT, treatment with TSA improves aspects of reprogramming in some cybrid embryos, as in same-species SCNT, but it does not improve survival [67-69]. This suggests that cybrid incompatibility may not be primarily due to reprogramming defects, at least of the types that are improved by TSA treatment [9,10]. Alternatively, cellular and developmental defects may underlie cybrid lethality. An interesting report in which ES cell extracts were transferred along with murine somatic nuclei into enucleated, mtDNA-depleted pig oocytes, showed that these changes (mtDNA replacement and addition of ES cell extract components) improved cybrid development, although it is not known exactly how these changes were beneficial [70]. As discussed earlier, it was found that differences in the concentration or regulation of relatively few proteins between two Xenopus species could explain their different nuclear and spindle sizes but it remains unclear whether such differences affect the viability of interspecific cybrids [45,46]. Yet, a difference in the concentration of an embryonic transcription factor between the same two frog species may be responsible for some of the incapacity of cybrid embryos to undergo efficient gastrulation movements [59]. Specifically, the Xenopus cybrid had a relatively insufficient, cytoplasmic species-like, transcription factor concentration, which likely contributed to the inefficient induction response and subsequent gastrulation defects in embryos with nuclei that normally have a higher concentration of that same transcription factor. Viable cells have been isolated from cybrid embryos, but in most cases they could not be expanded normally in vitro, suggesting that their viability or ability to proliferate is reduced [56,57,71]. In an in vivo study, cybrid cell viability was improved if these cells were transplanted into embryos of the cytoplasmic species, suggesting that inter-cellular signalling with cytoplasmic species cells may rescue some of the cellular incompatibility of cybrids [72]. The cellular incompatibility of cybrids could result from nucleo-mitochondrial incompatibilities causing respiratory defects, as occurs in cybrids cells generated by cytoplast fusion in culture [47,73,74], or from other kinds of cellular nucleocytoplasmic incompatibilities, such as chromosomal loss/damage. More studies will be necessary to determine the full spectrum of possible nucleocytoplasmic incompatibilities between species, and whether it may be possible to correct them.
Conclusion
Analyses of nuclear reprogramming in SCNT and interspecific systems have led to the identification of several reprogramming factors and recurrent defects occurring in SCNT embryos, as well as some differentiation marks that restrict reprogramming in somatic nuclei. On the other hand, they have also led to the discovery of a nucleo-mitochondrial incompatibility between highly divergent species in culture in vitro [47,73,74], although the in vivo relevance of this phenomenon remains unclear [59,75]. Also, quantitative and functional differences in relatively few key factors between species may account for their divergent phenotypes, while these differences may underlie hybrid and cybrid lethality [45,46,59]. Comparative genome-wide analyses of chromatin and transcription in divergent species [76,77], as well as in hybrids and cybrids [54,78,79], are being carried out and may help to identify some of these disparities. A better understanding of the differences and incompatibilities between species may indeed help to develop a variety of widely relevant new tools and systems, including iNT-generated functional ES cells and interspecific chimeras [80,81].
An interesting point is that, at the moment, it appears that enucleated animal oocytes are better than their human counterpart at reprogramming intraspecies somatic cells [1,7,24,25]. For reasons of egg availability, and enucleated reprogramming capacity, we therefore wonder whether it would be most advantageous to investigate further the possibility of reprogramming human somatic nuclei using interspecific systems, including closely related non-human primate species with compatible mitochondrial genomes [47]. Obviously, the ethical implications of these experiments would need careful examination. Identifying and rescuing cross-species incompatibilities also becomes a priority.
Acknowledgements
We are grateful to J. Jullien, S. Wang and M. Teperek-Tkacz for their critical reading of the manuscript, and to S. Wang for his access, through NIH, to the PubMed library. PN is a Junior Research Fellow of Wolfson College supported by a postdoctoral fellowship from the Human Frontier Science Program. KM has been supported by the International Fellowship Program of the Japan Society for the Promotion of Science and is now a Herchel Smith postdoctoral Fellow. Research in the Gurdon laboratory is funded by the Wellcome Trust and the Medical Research Council.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1••.Noggle S, Fung H-L, Gore A, Martinez H, Satriani KC, Prosser R, Oum K, Paull D, Druckenmiller S, Freeby M, et al. Human oocytes reprogram somatic cells to a pluripotent state. Nature. 2011;478:70–75. doi: 10.1038/nature10397. [DOI] [PubMed] [Google Scholar]; This paper unequivocally shows that non-enucleated human eggs can reprogram human somatic nuclei towards pluripotency, leading to the derivation of NT triploid blastocysts and ES-like cells lines. Using genome-wide allelotyping and gene expression profiling, the authors did not find any evidence of epigenetic memory in the reprogrammed somatic alleles.
- 2••.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LIR, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]; Here, the authors directly compare the differentiation potential of NT-derived ES cells to that of iPS cells. They detected a more pronounced bias in iPS cells towards differentiating along lineages related to the donor cell type, leading them to conclude that nuclear transfer is more effective than transcription factor-based reprogramming at establishing pluripotency.
- 3.Byrne J. Global transcriptional analysis of oocyte-based and factor-based nuclear reprogramming in the nonhuman primate. Cell Reprogram. 2011;13:473–481. doi: 10.1089/cell.2011.0033. [DOI] [PubMed] [Google Scholar]
- 4••.Tachibana M, Sparman M, Ramsey C, Ma H, Lee H-S, Penedo MCT, Mitalipov S. Generation of Chimeric Rhesus Monkeys. Cell. 2012;148:285–295. doi: 10.1016/j.cell.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors here show that aggregation of totipotent cells from 4-cell monkey embryos can lead to chimeric primate formation. Their data also suggest that the inner cell mass in monkey blastocysts already possesses differentiation characteristics that prevent chimera formation at this later stage.
- 5.Beyhan Z, Iager AE, Cibelli JB. Interspecies nuclear transfer: implications for embryonic stem cell biology. Cell Stem Cell. 2007;1:502–512. doi: 10.1016/j.stem.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Loi P, Modlinski JA, Ptak G. Interspecies somatic cell nuclear transfer: a salvage tool seeking first aid. Theriogenology. 2011;76:217–228. doi: 10.1016/j.theriogenology.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Wakayama T, Perry ACF, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- 8.Kishigami S, Mizutani E, Ohta H, Hikichi T, Thuan NV, Wakayama S, Bui H-T, Wakayama T. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem Biophys Res Commun. 2006;340:183–189. doi: 10.1016/j.bbrc.2005.11.164. [DOI] [PubMed] [Google Scholar]
- 9•.Bui H-T, Wakayama S, Kishigami S, Park K-K, Kim J-H, Thuan NV, Wakayama T. Effect of Trichostatin A on Chromatin Remodeling, Histone Modifications, DNA Replication, and Transcriptional Activity in Cloned Mouse Embryos. Biol Reprod. 2010;83:454–463. doi: 10.1095/biolreprod.109.083337. [DOI] [PubMed] [Google Scholar]; This paper describes how TSA treatment improves some of the early aspects of nuclear reprogramming.
- 10.Bui H-T, Seo H-J, Park M-R, Park J-Y, Van Thuan N, Wakayama T, Kim J-H. Histone Deacetylase Inhibition Improves Activation of Ribosomal RNA Genes and Embryonic Nucleolar Reprogramming in Cloned Mouse Embryos. Biol Reprod. 2011;85:1048–1056. doi: 10.1095/biolreprod.110.089474. [DOI] [PubMed] [Google Scholar]
- 11•.Miyamoto K, Nagai K, Kitamura N, Nishikawa T, Ikegami H, Binh NT, Tsukamoto S, Matsumoto M, Tsukiyama T, Minami N, et al. Identification and characterization of an oocyte factor required for development of porcine nuclear transfer embryos. Proc Natl Acad Sci U S A. 2011;108:7040–7045. doi: 10.1073/pnas.1013634108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Here, an oocyte factor (DJ-1) that is specifically required for the survival of SCNT embryos, but not for in vitro fertilized embryos, is identified. The factor is shown to act through suppressing the p53 pathway.
- 12.Esteves TC, Balbach ST, Pfeiffer MJ, Araúzo-Bravo MJ, Klein DC, Sinn M, Boiani M. Somatic cell nuclear reprogramming of mouse oocytes endures beyond reproductive decline. Aging Cell. 2011;10:80–95. doi: 10.1111/j.1474-9726.2010.00644.x. [DOI] [PubMed] [Google Scholar]
- 13•.Mizutani E, Yamagata K, Ono T, Akagi S, Geshi M, Wakayama T. Abnormal chromosome segregation at early cleavage is a major cause of the full-term developmental failure of mouse clones. Dev Biol. 2012;364:56–65. doi: 10.1016/j.ydbio.2012.01.001. [DOI] [PubMed] [Google Scholar]; In this analysis, chromosome segregation during early cleavage stages of SCNT embryo development was monitored before implantation. The authors then linked SCNT embryo developmental potential to the faithfulness of chromosome segregation during early cleavages to find that no SCNT embryo that showed abnormal chromosome segregation before the 8-cell stage could develop to term.
- 14••.Lin J, Shi L, Zhang M, Yang H, Qin Y, Zhang J, Gong D, Zhang X, Li D, Li J. Defects in Trophoblast Cell Lineage Account for the Impaired In Vivo Development of Cloned Embryos Generated by Somatic Nuclear Transfer. Cell Stem Cell. 2011;8:371–375. doi: 10.1016/j.stem.2011.02.007. [DOI] [PubMed] [Google Scholar]; Authors here show that fusing inner cell masses isolated from SCNT blastocysts with in vitro fertilization-derived tetraploid blastocysts improves SCNT embryo post-implantation development, indicating that defects in the trophectoderm lineage are responsible for the lethality of a large number of clones.
- 15••.Inoue K, Kohda T, Sugimoto M, Sado T, Ogonuki N, Matoba S, Shiura H, Ikeda R, Mochida K, Fujii T, et al. Impeding Xist Expression from the Active X Chromosome Improves Mouse Somatic Cell Nuclear Transfer. Science. 2010;330:496–499. doi: 10.1126/science.1194174. [DOI] [PubMed] [Google Scholar]; Here, genome-wide expression profiling showed that expression from the X chromosome is globally decreased in cloned blastocysts. The authors went on to show that this happens because of persistent Xist RNA from Xa after NT, and that knocking out Xist from Xa in donor cells improves post-embryonic development.
- 16.Matoba S, Inoue K, Kohda T, Sugimoto M, Mizutani E, Ogonuki N, Nakamura T, Abe K, Nakano T, Ishino F, et al. RNAi-Mediated Knockdown of Xist Can Rescue the Impaired Postimplantation Development of Cloned Mouse Embryos. Proc Natl Acad Sci U S A. 2011;108:20621–20626. doi: 10.1073/pnas.1112664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Fukuda A, Cao F, Morita S, Yamada K, Jincho Y, Tane S, Sotomaru Y, Kono T. Identification of Inappropriately Reprogrammed Genes by Large-Scale Transcriptome Analysis of Individual Cloned Mouse Blastocysts. PLoS ONE. 2010;5:e11274. doi: 10.1371/journal.pone.0011274. [DOI] [PMC free article] [PubMed] [Google Scholar]; Genome-wide expression profiling of individual SCNT blastocysts enabled the authors to identify here an overlapping set of commonly-misexpressed genes in SCNT embryos across different donor cell types. Consistent with [15], many of the commonly downregulated genes were located on the X chromosome.
- 18•.Maruotti J, Dai XP, Brochard V, Jouneau L, Liu J, Bonnet-Garnier A, Jammes H, Vallier L, Brons IGM, Pedersen R, et al. Nuclear Transfer-Derived Epiblast Stem Cells Are Transcriptionally and Epigenetically Distinguishable from Their Fertilized-Derived Counterparts. Stem Cells. 2010;28:743–752. doi: 10.1002/stem.400. [DOI] [PubMed] [Google Scholar]; In this paper, SCNT-derived epiblast stem cells are shown to have a transcriptome that is distinguishable from those derived from fertilized embryos.
- 19.Shen C-J, Cheng WTK, Wu S-C, Chen H-L, Tsai T-C, Yang S-H, Chen C-M. Differential Differences in Methylation Status of Putative Imprinted Genes among Cloned Swine Genomes. PLoS ONE. 2012;7:e32812. doi: 10.1371/journal.pone.0032812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Egli D, Eggan K. Recipient Cell Nuclear Factors Are Required for Reprogramming by Nuclear Transfer. Development. 2010;137:1953–1963. doi: 10.1242/dev.046151. [DOI] [PMC free article] [PubMed] [Google Scholar]; Here, the authors asked why interphase zygote enucleation is incompatible with SCNT. Their results overall suggest that the problem with interphase enucleation is that it removes essential nuclear reprogramming factors that otherwise become cytoplasmic during mitosis.
- 21.Grȩda P, Karasiewicz J, Modliński JA. Mouse zygotes as recipients in embryo cloning. Reproduction. 2006;132:741–748. doi: 10.1530/rep.1.01204. [DOI] [PubMed] [Google Scholar]
- 22.Egli D, Rosains J, Birkhoff G, Eggan K. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature. 2007;447:679–685. doi: 10.1038/nature05879. [DOI] [PubMed] [Google Scholar]
- 23.Sparman ML, Tachibana M, Mitalipov SM. Cloning of non-human primates: the road “less traveled by”. Int J Dev Biol. 2010;54:1671–1678. doi: 10.1387/ijdb.103196ms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byrne JA, Pedersen DA, Clepper LL, Nelson M, Sanger WG, Gokhale S, Wolf DP, Mitalipov SM. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450:497–502. doi: 10.1038/nature06357. [DOI] [PubMed] [Google Scholar]
- 25•.Egli D, Chen AE, Saphier G, Ichida J, Fitzgerald C, Go KJ, Acevedo N, Patel J, Baetscher M, Kearns WG, et al. Reprogramming within hours following nuclear transfer into mouse but not human zygotes. Nat Commun. 2011;2:488. doi: 10.1038/ncomms1503. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors here show that, contrary to what happens in mouse, mitotically-enucleated human zygotes are unable to reprogram somatic nuclei.
- 26.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tada M, Tada T, Lefebvre L, Barton SC, Surani MA. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 1997;16:6510–6520. doi: 10.1093/emboj/16.21.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pereira CF, Terranova R, Ryan NK, Santos J, Morris KJ, Cui W, Merkenschlager M, Fisher AG. Heterokaryon-Based Reprogramming of Human B Lymphocytes for Pluripotency Requires Oct4 but Not Sox2. PLoS Genet. 2008;4:e1000170. doi: 10.1371/journal.pgen.1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that the activation-induced cytidine deaminase AID is required for OCT4 and NANOG promoter demethylation and transcriptional re-activation following interspecific cell fusion of human fibroblasts to murine embryonic stem cells.
- 30••.Pereira CF, Piccolo FM, Tsubouchi T, Sauer S, Ryan NK, Bruno L, Landeira D, Santos J, Banito A, Gil J, et al. ESCs Require PRC2 to Direct the Successful Reprogramming of Differentiated Cells toward Pluripotency. Cell Stem Cell. 2010;6:547–556. doi: 10.1016/j.stem.2010.04.013. [DOI] [PubMed] [Google Scholar]; Polycomb-group proteins prevent ES cell from expressing differentiation genes, and the authors find here that these proteins are also required for mouse ES cells to transcriptionally reprogram human lymphocyte upon interspecific cell fusion. Their data suggest that these proteins act by suppressing expression of genes that otherwise interfere with the reprogramming process, rather than by inducing the reprogramming process themselves.
- 31.Fournier RE, Ruddle FH. Microcell-mediated transfer of murine chromosomes into mouse, Chinese hamster, and human somatic cells. Proc Natl Acad Sci U S A. 1977;74:319–323. doi: 10.1073/pnas.74.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VLJ, Fisher EMC, Tavaré S, Odom DT. Species-Specific Transcription in Mice Carrying Human Chromosome 21. Science. 2008;322:434–438. doi: 10.1126/science.1160930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jullien J, Pasque V, Halley-Stott RP, Miyamoto K, Gurdon JB. Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process? Nat Rev Mol Cell Biol. 2011;12:453–459. doi: 10.1038/nrm3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•.Jullien J, Åstrand C, Halley-Stott RP, Garrett N, Gurdon JB. Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation. Proc Natl Acad Sci U S A. 2010;107:5483–5488. doi: 10.1073/pnas.1000599107. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that an important early step in nuclear reprogramming by nuclear transfer to Xenopus oocytes, consists in the replacement of the transferred nuclei linker histones with the recipient oocyte’s kind of linker histone.
- 35•.Pasque V, Gillich A, Garrett N, Gurdon JB. Histone variant macroH2A confers resistance to nuclear reprogramming. EMBO J. 2011;30:2373–2387. doi: 10.1038/emboj.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]; Here, a histone variant (macroH2A) that associates with silenced chromatin during differentiation, is shown to contribute to the resistance to transcriptional reprogramming by nuclear transfer to Xenopus oocytes.
- 36•.Miyamoto K, Pasque V, Jullien J, Gurdon JB. Nuclear actin polymerization is required for transcriptional reprogramming of Oct4 by oocytes. Genes & Dev. 2011;25:946–958. doi: 10.1101/gad.615211. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show here that actin polymerizes in somatic nuclei transplanted into Xenopus oocytes and that this is required for the transcriptional reactivation of Oct4.
- 37.Beaujean N, Taylor JE, McGarry M, Gardner JO, Wilmut I, Loi P, Ptak G, Galli C, Lazzari G, Bird A, et al. The effect of interspecific oocytes on demethylation of sperm DNA. Proc Natl Acad Sci U S A. 2004;101:7636–7640. doi: 10.1073/pnas.0400730101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barnetova I, Fulka H, Fulka J., Jr Epigenetic characteristics of paternal chromatin in interspecies zygotes. J Reprod Dev. 2010;56:601–606. doi: 10.1262/jrd.09-172a. [DOI] [PubMed] [Google Scholar]
- 39.Fulka H, Barnetova I, Mosko T, Fulka J. Epigenetic analysis of human spermatozoa after their injection into ovulated mouse oocytes. Hum Reprod. 2008;23:627–634. doi: 10.1093/humrep/dem406. [DOI] [PubMed] [Google Scholar]
- 40.Pasque V, Jullien J, Miyamoto K, Halley-Stott RP, Gurdon JB. Epigenetic factors influencing resistance to nuclear reprogramming. Trends Genet. 2011;27:516–525. doi: 10.1016/j.tig.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamada H, Thuan NV, Reed P, Nelson D, Katoku-Kikyo N, Wudel J, Wakayama T, Kikyo N. Chromatin Decondensation and Nuclear Reprogramming by Nucleoplasmin. Mol Cell Biol. 2006;26:1259–1271. doi: 10.1128/MCB.26.4.1259-1271.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansis C, Barreto G, Maltry N, Niehrs C. Nuclear Reprogramming of Human Somatic Cells by Xenopus Egg Extract Requires BRG1. Curr Biol. 2004;14:1475–1480. doi: 10.1016/j.cub.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 43.Kikyo N, Wade PA, Guschin D, Ge H, Wolffe AP. Active remodeling of somatic nuclei in egg cytoplasm by the nucleosomal ATPase ISWI. Science. 2000;289:2360–2362. doi: 10.1126/science.289.5488.2360. [DOI] [PubMed] [Google Scholar]
- 44.Brown KS, Blower MD, Maresca TJ, Grammer TC, Harland RM, Heald R. Xenopus tropicalis egg extracts provide insight into scaling of the mitotic spindle. J Cell Biol. 2007;176:765–770. doi: 10.1083/jcb.200610043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Levy DL, Heald R. Nuclear Size Is Regulated by Importin α and Ntf2 in Xenopus. Cell. 2010;143:288–298. doi: 10.1016/j.cell.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; X. tropicalis are smaller frogs than X. laevis and they also have smaller nuclei. Here, by incubating sperm nuclei from one species into egg extracts generated from the other species and vice-versa, the authors find that nuclear size depends on cytoplasmic factors rather than nuclear content or species. They show that two nuclear transport proteins exist at different concentrations in the cells of the two species, and that this is responsible for their different nuclear sizes.
- 46••.Loughlin R, Wilbur JD, McNally FJ, Nédélec FJ, Heald R. Katanin Contributes to Interspecies Spindle Length Scaling in Xenopus. Cell. 2011;147:1397–1407. doi: 10.1016/j.cell.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; In addition to having bigger nuclei, the mitotic spindle of X. laevis is longer than that of X. tropicalis. Using interspecific extract nuclei incubation, the authors show here that the gain of an inhibitory phosphorylation site in a microtubule-severing factor is responsible for the increased spindle length in X. laevis.
- 47.Kenyon L, Moraes CT. Expanding the functional human mitochondrial DNA database by the establishment of primate xenomitochondrial cybrids. Proc Natl Acad Sci U S A. 1997;94:9131–9135. doi: 10.1073/pnas.94.17.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo J, Tecirlioglu RT, Nguyen L, Koh K, Jenkin G, Trounson AO. Reprogramming factors involved in hybrids and cybrids of human embryonic stem cells fused with hepatocytes. Cell Reprogram. 2010;12:529–541. doi: 10.1089/cell.2009.0054. [DOI] [PubMed] [Google Scholar]
- 49.Hasegawa K, Zhang P, Wei Z, Pomeroy JE, Lu W, Pera MF. Comparison of reprogramming efficiency between transduction of reprogramming factors, cell-cell fusion, and cytoplast fusion. Stem Cells. 2010;28:1338–1348. doi: 10.1002/stem.466. [DOI] [PubMed] [Google Scholar]
- 50.Wang K, Beyhan Z, Rodriguez RM, Ross PJ, Iager AE, Kaiser GG, Chen Y, Cibelli JB. Bovine ooplasm partially remodels primate somatic nuclei following somatic cell nuclear transfer. Cloning Stem Cells. 2009;11:187–202. doi: 10.1089/clo.2008.0061. [DOI] [PubMed] [Google Scholar]
- 51.Chung Y, Bishop CE, Treff NR, Walker SJ, Sandler VM, Becker S, Klimanskaya I, Wun W-S, Dunn R, Hall RM, et al. Reprogramming of human somatic cells using human and animal oocytes. Cloning Stem Cells. 2009;11:213–223. doi: 10.1089/clo.2009.0004. [DOI] [PubMed] [Google Scholar]
- 52.Lagutina I, Fulka H, Brevini TAL, Antonini S, Brunetti D, Colleoni S, Gandolfi F, Lazzari G, Fulka J, Jr, Galli C. Development, embryonic genome activity and mitochondrial characteristics of bovine-pig inter-family nuclear transfer embryos. Reproduction. 2010;140:273–285. doi: 10.1530/REP-09-0578. [DOI] [PubMed] [Google Scholar]
- 53.Lorthongpanich C, Laowtammathron C, Chan AWS, Ketudat-Cairns M, Parnpai R. Development of interspecies cloned monkey embryos reconstructed with bovine enucleated oocytes. J Reprod Dev. 2008;54:306–313. doi: 10.1262/jrd.20049. [DOI] [PubMed] [Google Scholar]
- 54•.Wang K, Otu HH, Chen Y, Lee Y, Latham K, Cibelli JB. Reprogrammed transcriptome in rhesus-bovine interspecies somatic cell nuclear transfer embryos. PLoS ONE. 2011;6:e22197. doi: 10.1371/journal.pone.0022197. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this paper, the authors use microarray analysis to investigate how well the embryonic genome is activated in monkey-to-bovine iSCNT embryos. They find that several hundred monkey genes were transcribed by the bovine cytoplasm, to levels comparable to in vitro fertilized monkey embryos, indicating that embryonic genome activation does take place in these cybrid embryos.
- 55.Chen Y, He ZX, Liu A, Wang K, Mao WW, Chu JX, Lu Y, Fang ZF, Shi YT, Yang QZ, et al. Embryonic stem cells generated by nuclear transfer of human somatic nuclei into rabbit oocytes. Cell Res. 2003;13:251–263. doi: 10.1038/sj.cr.7290170. [DOI] [PubMed] [Google Scholar]
- 56.Sha H, Chen J, Chen J, Zhang P, Wang P, Chen L, Cheng G, Zhu J. Fates of donor and recipient mitochondrial DNA during generation of interspecies SCNT-derived human ES-like cells. Cloning Stem Cells. 2009;11:497–507. doi: 10.1089/clo.2009.0021. [DOI] [PubMed] [Google Scholar]
- 57.Tecirlioglu RT, Guo J, Trounson AO. Interspecies somatic cell nuclear transfer and preliminary data for horse-cow/mouse iSCNT. Stem Cell Rev. 2006;2:277–287. doi: 10.1007/BF02698054. [DOI] [PubMed] [Google Scholar]
- 58.Mastromonaco GF, Favetta LA, Smith LC, Filion F, King WA. The Influence of Nuclear Content on Developmental Competence of Gaur × Cattle Hybrid In Vitro Fertilized and Somatic Cell Nuclear Transfer Embryos. Biol Reprod. 2007;76:514–523. doi: 10.1095/biolreprod.106.058040. [DOI] [PubMed] [Google Scholar]
- 59•.Narbonne P, Simpson DE, Gurdon JB. Deficient Induction Response in a Xenopus Nucleocytoplasmic Hybrid. PLoS Biol. 2011;9:e1001197. doi: 10.1371/journal.pbio.1001197. [DOI] [PMC free article] [PubMed] [Google Scholar]; Here, a nucleocytoplasmic incompatibility between two Xenopus species that results in gastrulation defects in cybrids, is shown to partly occur because of inefficient induction signalling. Interestingly, a component of this signalling cascade exists at different concentrations in the two species, while the concentration in the cybrid was as in the cytoplasmic species, thereby inappropriate for the nuclear species.
- 60.Moore JA. Transplantation of nuclei between Rana pipiens and Rana sylvatica. Exp Cell Res. 1958;14:532–540. doi: 10.1016/0014-4827(58)90159-9. [DOI] [PubMed] [Google Scholar]
- 61.Gurdon JB. The transplantation of nuclei between two species of Xenopus. Dev Biol. 1962;5:68–83. doi: 10.1016/0012-1606(62)90004-0. [DOI] [PubMed] [Google Scholar]
- 62.Kwon DK, Kang JT, Park SJ, Gomez MNL, Kim SJ, Atikuzzaman M, Koo OJ, Jang G, Lee BC. Blastocysts derived from adult fibroblasts of a rhesus monkey ( Macaca mulatta) using interspecies somatic cell nuclear transfer. Zygote. 2011;19:199–204. doi: 10.1017/S0967199411000232. [DOI] [PubMed] [Google Scholar]
- 63.Lagutina I, Zakhartchenko V, Fulka H, Colleoni S, Wolf E, Fulka J, Lazzari G, Galli C. Formation of nucleoli in interspecies nuclear transfer embryos derived from bovine, porcine, and rabbit oocytes and nuclear donor cells of various species. Reproduction. 2011;141:453–465. doi: 10.1530/REP-10-0266. [DOI] [PubMed] [Google Scholar]
- 64.Østrup O, Strejcek F, Petrovicova I, Lucas-Hahn A, Morovic M, Lemme E, Petersen B, Laurincikova N, Niemann H, Laurincik J, et al. Role of ooplasm in nuclear and nucleolar remodeling of intergeneric somatic cell nuclear transfer embryos during the first cell cycle. Cell Reprogram. 2011;13:145–155. doi: 10.1089/cell.2010.0061. [DOI] [PubMed] [Google Scholar]
- 65.Amarnath D, Choi I, Moawad AR, Wakayama T, Campbell KHS. Nuclear-cytoplasmic incompatibility and inefficient development of pig-mouse cytoplasmic hybrid embryos. Reproduction. 2011;142:295–307. doi: 10.1530/REP-11-0044. [DOI] [PubMed] [Google Scholar]
- 66.Song B-S, Lee S-H, Kim S-U, Kim J-S, Park JS, Kim C-H, Chang K-T, Han Y-M, Lee K-K, Lee D-S, et al. Nucleologenesis and embryonic genome activation are defective in interspecies cloned embryos between bovine ooplasm and rhesus monkey somatic cells. BMC Dev Biol. 2009;9:44. doi: 10.1186/1471-213X-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi L-H, Miao Y-L, Ouyang Y-C, Huang J-C, Lei Z-L, Yang J-W, Han Z-M, Song X-F, Sun Q-Y, Chen D-Y. Trichostatin A (TSA) improves the development of rabbit-rabbit intraspecies cloned embryos, but not rabbit-human interspecies cloned embryos. Dev Dyn. 2008;237:640–648. doi: 10.1002/dvdy.21450. [DOI] [PubMed] [Google Scholar]
- 68•.Gómez MC, Pope CE, Biancardi MN, Dumas C, Galiguis J, Morris AC, Wang G, Dresser BL. Trichostatin A modified histone covalent pattern and enhanced expression of pluripotent genes in interspecies black-footed cat cloned embryos but did not improve in vitro and in vivo viability. Cell Reprogram. 2011;13:315–329. doi: 10.1089/cell.2010.0111. [DOI] [PubMed] [Google Scholar]; The authors show here that HDACi treatment improves reprogramming in the context to iSCNT, much like in intraspecies SCNT, but that it does not improve iSCNT embryo development. This result is important in that it suggests that the lethality of cybrid embryos does not result from reprogramming defects of the kinds that are improved by HDACi treatment.
- 69.Lee H-S, Yu X-F, Bang J-I, Cho S-J, Deb GK, Kim B-W, Kong I-K. Enhanced histone acetylation in somatic cells induced by a histone deacetylase inhibitor improved inter-generic cloned leopard cat blastocysts. Theriogenology. 2010;74:1439–1449. doi: 10.1016/j.theriogenology.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 70•.Jiang Y, Kelly R, Peters A, Fulka H, Dickinson A, Mitchell DA, John JC. Interspecies Somatic Cell Nuclear Transfer Is Dependent on Compatible Mitochondrial DNA and Reprogramming Factors. PLoS ONE. 2011;6:e14805. doi: 10.1371/journal.pone.0014805. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors here find that factors present in mouse ES cell extracts can increase the formation of cybrid blastocysts made from mouse fetal fibroblast nuclei transferred into enucleated, mtDNA-depleted, porcine oocytes. They suggest that these factors may be murine-specific reprogramming proteins and mitochondria.
- 71.Narbonne P, Halley-Stott RP, Gurdon JB. On the cellular and developmental lethality of a Xenopus nucleocytoplasmic hybrid. Commun Integr Biol. 2012 doi: 10.4161/cib.20334. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fujimoto T, Saito T, Sakao S, Arai K, Yamaha E. Developmental potential of embryonic cells in a nucleocytoplasmic hybrid formed using a goldfish haploid nucleus and loach egg cytoplasm. Int J Dev Biol. 2010;54:827–835. doi: 10.1387/ijdb.092896tf. [DOI] [PubMed] [Google Scholar]
- 73.McKenzie M, Trounce I. Expression of Rattus norvegicus mtDNA in Mus musculus Cells Results in Multiple Respiratory Chain Defects. J Biol Chem. 2000;275:31514–31519. doi: 10.1074/jbc.M004070200. [DOI] [PubMed] [Google Scholar]
- 74.Yamaoka M, Isobe K, Shitara H, Yonekawa H, Miyabayashi S, Hayashi J-I. Complete Repopulation of Mouse Mitochondrial DNA-less Cells With Rat Mitochondrial DNA Restores Mitochondrial Translation but Not Mitochondrial Respiratory Function. Genetics. 2000;155:301–307. doi: 10.1093/genetics/155.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cannon MV, Dunn DA, Irwin MH, Brooks AI, Bartol FF, Trounce IA, Pinkert CA. Xenomitochondrial mice: investigation into mitochondrial compensatory mechanisms. Mitochondrion. 2011;11:33–39. doi: 10.1016/j.mito.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76•.Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell. 2011;20:483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors examined the embryonic transcriptome of two distantly-related Xenopus species to find that gene expression level and timing are strongly conserved among orthologs. A change in expression levels, early during development, is identified as being the most common expression change between orthologs in embryos of divergent species.
- 77•.Shen-Orr SS, Pilpel Y, Hunter CP. Composition and regulation of maternal and zygotic transcriptomes reflects species-specific reproductive mode. Genome Biol. 2010;11:R58. doi: 10.1186/gb-2010-11-6-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors here performed a genomic comparison of maternal and/or zygotically-expressed genes in animals ranging from worms to humans. They find that maternal genes in mammals are relatively enriched for complex regulatory regions as compared to egg-laying animals.
- 78.Fantini L, Mudry MD, Nieves M. Genome size of two cebus species (primates: platyrrhini) with a fertile hybrid and their quantitative genomic differences. Cytogenet Genome Res. 2011;135:33–41. doi: 10.1159/000330127. [DOI] [PubMed] [Google Scholar]
- 79.Koroma AP, Jones R, Michalak P. Snapshot of DNA methylation changes associated with hybridization in Xenopus. Physiol Genomics. 2011;43:1276–1280. doi: 10.1152/physiolgenomics.00110.2011. [DOI] [PubMed] [Google Scholar]
- 80••.Kobayashi T, Yamaguchi T, Hamanaka S, Kato-Itoh M, Yamazaki Y, Ibata M, Sato H, Lee Y-S, Usui J, Knisely AS, et al. Generation of Rat Pancreas in Mouse by Interspecific Blastocyst Injection of Pluripotent Stem Cells. Cell. 2010;142:787–799. doi: 10.1016/j.cell.2010.07.039. [DOI] [PubMed] [Google Scholar]; By injecting wild-type rat iPS cells into a mouse blastocyst that was deficient for pancreas differentiation, the authors generated a rat pancreas within a mouse’s body. This work paves the way to the generation of human replacement organs within developing animals.
- 81.Shinohara T, Kato M, Takehashi M, Lee J, Chuma S, Nakatsuji N, Kanatsu-Shinohara M, Hirabayashi M. Rats produced by interspecies spermatogonial transplantation in mice and in vitro microinsemination. Proc Natl Acad Sci U S A. 2006;103:13624–13628. doi: 10.1073/pnas.0604205103. [DOI] [PMC free article] [PubMed] [Google Scholar]