Abstract
Phenotypic variation in genetically identical malaria parasites is an emerging topic. Although antigenic variation is only part of a more global parasite strategy to create adaptation through epigenetically controlled transcriptional variability, it is the central mechanism enabling immune evasion and promoting pathogenesis. The var gene family is the best-studied example in a wide range of clonally variant gene families in Plasmodium falciparum. It is unique in its strict selection of a single member for activation, a process termed monoallelic expression. The conceptual advances that have emerged from studying var genes show striking common epigenetic features with many other clonally variant gene families or even single-copy genes that show a variegated expression in parasite populations. However, major mechanistic questions, such as the existence of a potential expression site and the identity of transcription factors or genetic elements driving singular gene choice, are still unanswered. In this review we discuss the recent findings in the molecular processes essential for clonal variation, namely silencing, activation, poising and switching. Integrating findings about all clonally variant gene families and other mutually exclusive expression systems will hopefully drive mechanistic understanding of antigenic variation.
Introduction
Immune evasion is critical for pathogens in order to establish long-lasting infection and ensure effective transmission. The sites of host–parasite interaction are constantly exposed to recognition by the host immune system. Protozoan pathogens have developed a wide range of sophisticated immune evasion strategies such as antigenic variation (Deitsch et al., 2009). Parasites actively modify the expression of variant surface proteins in order to remain invisible to the adaptive immune system.
The protozoan pathogen Plasmodium falciparum, which causes human malaria, uses antigenic variation to establish chronic blood stage infections in malaria patients. P. falciparum merozoites invade red blood cells and undergo multiple rounds of replication before releasing newly formed merozoites into the blood stream (Fig. 1A). During intracellular development, proteins encoded by distinct clonally variant gene families are transported to the red blood cell surface. Only a small subset of these genes is expressed at a given time. The most extreme form of clonal variation is mutually exclusive expression, also called monoallelic expression. Here, only a single gene of the entire family is transcribed whereas all the others are silenced, as it is the case of the var gene family (Scherf et al., 2008). The var gene family has 60 members and codes for the immunodominant variant adhesion surface molecule ‘P. falciparum Erythrocyte Membrane Protein 1’ (PfEMP1). Most var genes are in subtelomeric regions, whereas others are arranged in more chromosome central positions. Var genes consist of an Exon1, coding polymorphic sequences forming the extracellular domain, an Exon2, coding the semi-conserved intracellular domain, connected by a single highly conserved intron.
Fig. 1.
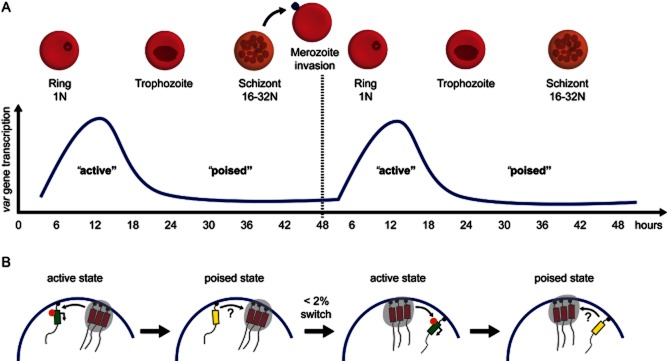
Var gene activation and silencing throughout asexual blood stage development of P. falciparum.
A. The single var gene is transcribed at the beginning of the asexual blood stage cycle (ring) soon after merozoites invasion of a red blood cells. Shortly before parasite DNA replication starts, var gene transcription ceases but it remains in a poised state (trophozoite and schizont) ready to be activated at the next blood stage cycle.
B. All var genes, central as well as subtelomeric ones, are tethered to the nuclear periphery (blue) and silent var genes (red) cluster in repressive centers (grey) (Lopez-Rubio et al., 2009). A single active var gene (green) segregates away from repressive centres and forms a perinuclear expression site containing required transcription factors (orange). The transition from the active to the poised state (yellow) is still poorly defined and it is unknown whether positional memory establishes throughout late stages. However, recently a putative methyltransferase PfSet10 has been specifically associated with the poised var gene (Volz et al., 2012). Variable var gene switching rates have been measured in vitro (Horrocks et al., 2004), but it is unclear at which point in the cell cycle switching occurs.
Understanding antigenic variation is critical in the fight against human pathogens. In this review we discuss potential mechanisms regulating mutual exclusive expression while focusing on very recent findings. The results from var gene expression studies have wide implications for other plasmodial gene families and single-copy genes controlled by similar epigenetic mechanisms (Rovira-Graells et al., 2012). Another context where mutual exclusion occurs is the expression of olfactory receptor genes (> 2000 members) in mice (Serizawa et al., 2003). Mechanisms investigated there have served as models for P. falciparum and vice versa.
By default, var genes are silenced, which requires the coordination of several distinct epigenetic processes. A single var gene is selectively activated for a period of about 10–14 h during the early phase of the 48 h blood stage development (Scherf et al., 1998; Schieck et al., 2007). Transcription of the activate var gene ceases in late blood stage, but despite being silent is epigenetically marked for re-activation during the next erythrocytic cycle, a state called poised (Fig. 1A). Finally, var gene inactivation occurs at a low frequency in some cells, leading to the switch to another member of the gene family. Hence, silencing, activation, poising and switching are the basic ingredients that make antigenic variation a successful instrument for immune evasion. We will discuss key factors such as genetic elements, epigenetic chromatin modifications and spatial regulation involved in these four steps. Further, we will highlight outstanding questions in the field and propose future research directions.
Default silencing
Epigenetic silencing marks
Though the silent state of var genes is maintained over many blood stage generations, it is critical to antigenic variation that each member can be re-activated. In the absence of programmed DNA rearrangements in the activation process (Scherf et al., 1998), reversible histone modifications were demonstrated to be decisive to the process of var gene regulation (Freitas-Junior et al., 2005; Lopez-Rubio et al., 2007).
The first functional studies showing an effect on var gene silencing involved the inactivation of NAD-dependent histone deacetylase Sir2A (Duraisingh et al., 2005). Knocking out Sir2A causes de-repression of a subset of var genes, especially of upstream promoter sequence (ups) A and B subtypes, as well as some members of the rifins, another clonally variant gene family. Inactivation of a second member of the Sir2 family, called Sir2B, showed some complementary de-repression effect on other, mostly upsC, var gene members (Tonkin et al., 2009), indicating that both plasmodial Sir2 genes act on chromatin of clonally variant gene families. Histone deacetylation by Sir2 presumably enables establishment of the silencing heterochromatin mark Histone 3 lysine 9 trimethylation (H3K9me3), which is enriched in promoter regions of repressed var genes (Chookajorn et al., 2007), including exon1 (Lopez-Rubio et al., 2007). Detailed genome-wide Chromatin ImmunoPrecipitation (ChIP) analysis of H3K9me3 distribution in P. falciparum has demonstrated an association of the H3K9me3 chromatin mark with multiple gene families including the silent var genes and other genes families known to be involved in immune evasion, such as stevor and rifins (Lopez-Rubio et al., 2009). Recently, silencing of the olfactory receptor gene family has also been associated with the heterochromatin mark H3K9me3, suggesting that epigenetic regulation could be a common feature in monoallelic expression (Magklara et al., 2011).
H3K9me3 histone modification promotes heterochromatin formation by specific recruitment of Heterochromatin Protein 1 (PfHP1) to silent but not active var genes (Perez-Toledo et al., 2009). Genome-wide ChIP analysis of PfHP1 showed a strict association with H3K9me3 linking it to the formation of heterochromatin (Flueck et al., 2009). To date there is no functional data on how the repressive mark is established but PfKMT1, a member of the SET domain containing family, is a potential lysine methyl transferases candidate (Cui et al., 2008).
Epigenetic silencing in multiple clonally variant gene families
Importantly, the large number of gene families showing containing the H3K9me3 mark indicates that clonally variant gene expression is much more widely used by P. falciparum than was anticipated (Lopez-Rubio et al., 2009; Salcedo-Amaya et al., 2010). Unexpectedly, this included also a member of the putative transcriptional regulator gene family AP2 (Apetala 2). Clonally variant transcription in genes involved in processes other than immune evasion has been shown for a receptor protein functioning in erythrocyte invasion (Jiang et al., 2010). Further, a detailed genome-wide transcript analysis of freshly cloned parasites revealed the potential for tremendous transcriptional plasticity in genetically identical parasite lines (Rovira-Graells et al., 2012). Transcriptional differences could all be linked to genes that show the epigenetic silencing mark H3K9me3 (Fig. 2). Thus, in addition to immune evasion, the epigenetically determined variegated transcription may provide the parasite with another level of adaptation to the variable environments that may occur during infection (bet-hedging strategy). It will be interesting to see whether deciphering var gene regulation can help uncover a more generalized transcriptional variation mechanism in other clonally variant gene families and vice versa.
Fig. 2.
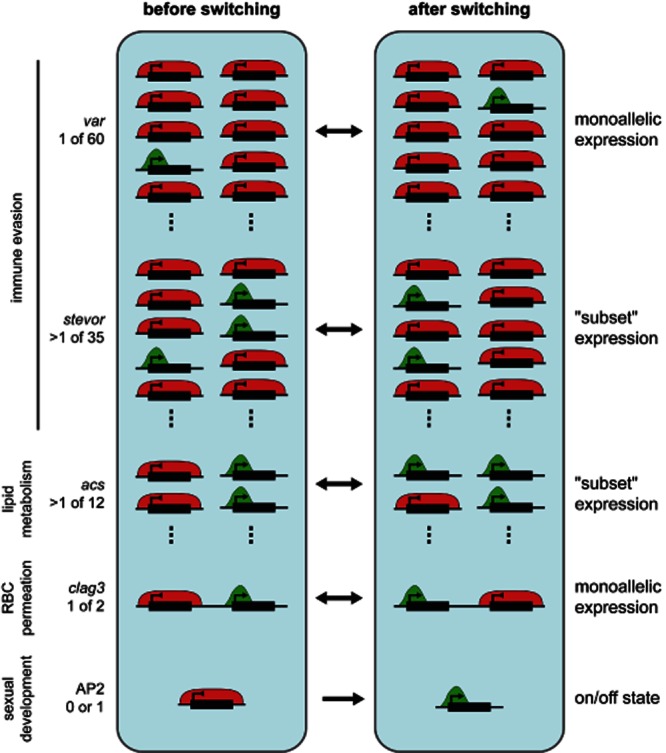
Epigenetic switching in different clonally variant gene families of P. falciparum. Several gene families can be differentially expressed in genetically identical parasites (Rovira-Graells et al., 2012). The number of active genes (green) varies but silent genes are widely associated with epigenetic silencing mark H3K9me3 (red) (Lopez-Rubio et al., 2009). Combinatorial switching of the different gene families can result in a tremendous range of phenotypic variation within genetically identical populations. Gene families involved in immune evasion have different numbers, switching rates and strictness in gene counting. While var genes undergo strict monoallelic expression, multiple stevor genes can be transcribed in the same cell (Kaviratne et al., 2002). Gene families not involved in immune evasion can also undergo clonal variation. Here three examples with different ‘switching modes’ are shown. From the 12 members of the acs gene family, encoding for acyl-CoA synthetases 4 members have been shown to be clonally variant in blood stages (Rovira-Graells et al., 2012). The clag3 family, containing two members important for red blood cell permeation pathways, is the only other gene family besides var for which mutually exclusive expression has been shown (Cortes et al., 2007). A single member of the putative AP2 transcription family (PF3D7_1222600) is associated with H3K9me3. It is thought to be involved in inducing gametocytogenesis (Kafsack et al., MPM meeting 2012 Woods Hole). In this simple form epigenetically regulated activation at low frequency could constitute a developmental switch in the life cycle.
Spatial organization is critical to var gene silencing
Nuclear architecture plays a significant role in the biology of virulence genes in P. falciparum (Scherf et al., 2008). Telomeres, and consequently subtelomeric var genes, cluster at the nuclear periphery by an unknown mechanism. Interestingly, chromosome central var genes are also tethered to the nuclear periphery (Ralph et al., 2005; Lopez-Rubio et al., 2009). A repeat region within the intron of central var genes was recently identified as being able to recruit episomes to the nuclear periphery (Zhang et al., 2011a). This var intron region interacts with a complex of proteins including actin and a member of the AP2 DNA-binding protein family. Pharmacologically induced F-actin formation impairs perinuclear localization of intron-carrying episomes and var genes and causes partial de-repression. Hence, tethering to the nuclear periphery seems to be an intrinsic requirement for the default-silencing pathway of var genes. Antibodies that label heterochromatin associated molecular markers such as H3K9me3 and PfHP1 are enriched in perinuclear clusters and silent olfactory receptor genes in mice regroup into heterochromatic foci (Clowney et al., 2012), supporting the concept of Perinuclear Repressive Centers (PERCs) (Lopez-Rubio et al., 2009) (Fig. 1B).
Var intron as silencing element
Functional gene expression assays using reporter genes highlighted the central role of two var genetic elements in expression. The 5′ upstream promoter region and the intron control silencing and activation independently of antigen production (Dzikowski et al., 2006; Voss et al., 2006). The intron silences expression of transgenes under the control of an episomal var promoter, but not other promoters (Deitsch et al., 2001; Frank et al., 2006). Loss of one-to-one pairing of var promoter and intron by episomal recombination in the parasite causes activation of the respective promoter (Deitsch et al., 2001; Frank et al., 2006). Silencing by strict intron-promoter pairing works downstream as well as upstream of the var promoter. Integrating an unpaired promoter into a silent var gene cluster leads to its activation, suggesting that the var intron plays a direct role, upstream of chromatin spreading, in regulation of var gene silencing (Swamy et al., 2011). The intron has bidirectional promoter activity transcribing non-coding RNA (Calderwood et al., 2003), which may be controlled by the putative Apetala 2 (AP2) domain transcription factor that binds to the central intron region (Zhang et al., 2011a). The intron promoter activity is intriguing but the role of this non-coding RNA in var gene expression remains unknown.
Activation
Epigenetic activation marks
In order to activate a var gene, repressive histone marks need to be removed. In the promoter region of the active var gene, H3K9me3 is replaced by Histone 3 lysine 4 bi- and tri-methylation (H3K4me2/3) and Histone 3 lysine 9 acetylation (H3K9ac), which renders it permissive for transcription (Lopez-Rubio et al., 2007). Consistently, the 5′ ups is no longer associated with PfHP1 when active (Perez-Toledo et al., 2009). Histone 4 lysine acetylation is another modification involved in transcriptional activation and potentially regulated by the histone acetyltransferase PfMYST, which is enriched at the active var promoter (Miao et al., 2010). However, it can be assumed that PfMYST association is not var gene specific since overexpression also induces cell cycle defects.
Incorporation of the histone variant H2A.Z around the transcription start site has been associated with increased transcriptional activity (Bartfai et al., 2010). In contrast to other developmentally regulated genes, H2A.Z is incorporated in active var genes only during ring stages, while it is depleted in later stages (Petter et al., 2011). Periodic H2A.Z removal could allow the activation of another var gene in the next cycle.
Var gene promoter region
An episomal var promoter expressing a selection marker under drug pressure causes silencing of endogenous var genes, demonstrating its infiltration into the mutual exclusive counting mechanism (Voss et al., 2006). A similar study, where the construct was integrated into the genome, concluded that the var promoter can drive monoallelic expression only when paired with the intron (Dzikowski et al., 2006). Discrepancies between those findings have been discussed elsewhere (Dzikowski et al., 2007). Nevertheless, those studies suggest that regulatory elements within each var gene contain all the necessary information for the singular gene choice upon activation. There seems to be a limiting activation factor for which the active var gene is competing (Dzikowski et al., 2006). Recently, an eight-base pair sequence motif has been identified in the 5′ UTR of most var genes, which apparently is critical for mutually exclusive expression (Brancucci et al., 2012). Deleting this mutually exclusive element (MEE) from an episomal active var promoter revokes silencing of endogenous var genes. This suggests that this element could be the binding site for a limited activating factor.
Var expression site
Mutual exclusive expression of variant surface glycoproteins (VSGs) in Typanosoma brucei occurs at the expression site body, a singular spot within the nucleus defined by the presence of the VSG-specific RNA Polymerase I (Navarro and Gull, 2001). The model claims that this site can only accommodate a single active VSG. This observation has brought forward the hypothesis that a unique and defined expression site for var gene transcription could exist in P. falciparum. Var gene transcription is driven by RNA Polymerase II (Kyes et al., 2007), and spatial segregation of the active var gene locus and its transcripts from perinuclear repressive centers upon activation has been shown by DNA and RNA fluorescence in situ hybridization (FISH) (Freitas-Junior et al., 2005; Ralph et al., 2005; Voss et al., 2006; Lopez-Rubio et al., 2009) (Fig. 1B).
Further insights come from exceptional cases where more than one var gene is active within a single cell. The HB3 P. falciparum strain contains two nearly identical copies of the var2csa gene. Despite the fact that one copy lies on chromosome 12 and the other on chromosome 1, RNA FISH analysis showed that simultaneous expression of both copies occurs at the same site in the nucleus (Brolin et al., 2009). The Scherf laboratory has recently generated a parasite mutant strain expressing multiple endogenous var genes simultaneously at the same spatial expression site (Q. Zhang and A. Scherf, unpubl. data). Interestingly, an episomal promoter of the rifin gene families also colocalizes with the active var gene expression site when activated through drug selection (Howitt et al., 2009). Altogether these observations suggest that there might indeed be a committed nuclear site for expression of clonally variant genes. They prove, however, that this site can, under specific conditions, accommodate more than one active promoter. In yeast, nuclear pores take part in enhanced gene expression (Akhtar and Gasser, 2007). It was tested whether nuclear pores define the var gene expression site, or the perinuclear clusters highly expressing 18S ribosomal RNA (Mancio-Silva et al., 2010), by colocalization studies combining RNA FISH with immunofluorescence. However, the perinuclear expression site of the active var gene, or 18S ribosomal RNA, showed no association with nuclear pores (J. Guizetti et al., manuscript submitted). An alternative view is that an activation complex is recruited to the var gene in situ. This initiates the transcriptional activation cascade including its relocation away from the repressive zone. This scenario predicts that any perinuclear site outside the repressive centres may form an expression site. Depending on which model turns out to be accurate, var gene activation would occur before segregation from the repressive cluster or segregation would be a precondition for activation. In either case, the recent discovery of nuclear actin associated to var gene intron regions opens a new exciting avenue. Nuclear actin could provide a mechanical framework for spatial organization of active and silent genes (Zhang et al., 2011a).
Enhancer element as monoallelic activator
A different hypothesis for a mutual exclusive expression mechanism postulates the existence of a unique trans-acting enhancer element. The discovery of the H-element involved in activation of olfactory receptor genes provides a blueprint for how a single-copy genetic element activates only a single gene through genome-wide interaction in trans (Serizawa et al., 2003; Lomvardas et al., 2006). It is, however, debated whether the H-element does, indeed, mediate activation interchromosomally. Development and adaptation of different genome-wide chromosome conformation capture (3C) analysis for P. falciparum would allow testing for such a hypothesis (Simonis et al., 2006). A recent study from the Newbold laboratory using genome-wide 3C technology could not detect an enhancer element in monoallelic expression of var genes (Lemieux et al., MPM meeting 2012 Woods Hole).
Poising
Var genes are only transcribed during the early stages of the blood stage cycle and intrinsic switching rates determined from in vitro cultured parasites can reach up to 2% per cycle (Roberts et al., 1992). Hence, most of the time the same var genes will be re-activated after the next round of invasion (Fig. 1A). ChIP analysis has shown that the activating mark H3K4me2, remains associated with the poised var promoter in the late blood cell stages, while H3K9me3 is prevented from spreading into the promoter region (Lopez-Rubio et al., 2007). For those marks to facilitate reactivation in the next cycle, they must be maintained throughout S-Phase. Indeed, a H3K4 methyltransferase called PfSet10 colocalizes with the previously active var gene locus in post-ring stages, whereas repressing marks are excluded from this site, presumably contributing to the poised state (Volz et al., 2012) (Fig. 1B). It is intriguing to speculate that poising might involve nuclear positional memory. In yeast there is evidence for transcriptional memory involving repositioning of genes to the nuclear envelope (Brickner, 2009). The relevance of this topic in P. falciparum awaits further investigation.
Switching
Var gene switching patterns
Switching between the clonally variant genes is central to antigenic variation and must be adapted to the host so the variant gene repertoire is not depleted too fast whilst effective immune evasion is still possible. It is important to keep in mind that the parasite needs to control its own proliferation so the host is not severely harmed by high parasitemia and effective transmission is guaranteed.
To this day, no genes modifying switch rates have been identified and no functional data is available on the molecular mechanism of switching. Nevertheless, descriptive studies demonstrated that transition rates can vary between var members (Horrocks et al., 2004). Epigenetic poising of var genes during the non-transcribed phase can be experimentally erased by promoter titration (Dzikowski and Deitsch, 2008; Fastman et al., 2012). Once memory is erased, the re-activation pattern of var genes is independent of previous stages. Another recent study on switch pathways used experimental evidence combined with mathematical modelling to conclude that var switching is non-random and necessitates a balanced process of parasite-intrinsic switching and immune-mediated selection (Recker et al., 2011). A more detailed transcriptional analysis of var gene switching patterns using silencing with drug-selectable markers in multiple isolates suggests that a var gene switching pattern is conserved throughout different genetic backgrounds (Enderes et al., 2011). Further modelling approaches confirmed that highly structured switching patterns could optimize infection length and robustness (Recker et al., 2011). Rigorous statistical analysis and short-term transcriptional data can be used to further improve our models of antigenic switching networks (Noble and Recker, 2012). Parasites having a high dynamic range of on- and off- probabilities within their variant gene families can improve their survival in naïve as well as re-infected hosts.
Switching and external factors
Another critical question is whether the observed intrinsic switch rate is influenced by external factors, i.e. does the parasite react to its environment, or is the switching pattern hard-wired into the parasite's genome? Although no longitudinal data from human patients are available to support this concept directly, the epigenetic machinery controlling var gene silencing may be modulated by external factors. For example, Sir2 depends on NAD levels for its activity. Low nutrition states of patients may influence Sir2 activity and hence alter the switch rate of distinct var subtypes. Experimental data from in vitro cultured parasites using peroxide or starvation stress can lead to a slight up-regulation of central var genes (Rosenberg et al., 2009). Elevated PfSir2 levels, which correlate with high temperature and lactate levels, are linked to subtle changes in the var gene expression pattern in patients (Merrick et al., 2012).
The relative paucity of patient data related to this topic makes it difficult to investigate switching rates in the human host. Two recently published var expression studies from malaria patients, however, support the idea that host factors contribute to var gene transcriptional control. Unlike in vitro cultured parasites, ups A-type var genes were frequently expressed in P. falciparum-infected patients (Bachmann et al., 2011; Zhang et al., 2011b). Subsequent cultivation resulted in random expression of many var genes mostly of the ups B and C-type. It will be crucial to investigate whether host factors modify the intrinsic switch rate of ups A-type var genes.
Conclusions
Mutually exclusive expression relies on distinct layers of genetic and epigenetic control factors. Genetic elements, such as the promoter and the intron of var genes cooperate with chromatin modifying enzymes in a complex interplay. On top of this, spatial regulation creates specific nuclear sub-compartments most likely critical for default silencing and monoallelic expression. Other factors such as non-coding RNA produced in subtelomeric regions adjacent or within var genes may contribute as well to antigenic variation (Epp et al., 2009; Sierra-Miranda et al., 2012; N. Siegel and A. Scherf, unpubl. data).
Properties and composition of the hypothetical var gene expression site remain to be explored. The biggest puzzle, however, in understanding mutual exclusive expression, is the absence of a putative activation factor. The concept of a limiting factor driving monoallelic var transcription has been postulated on many occasions but remains to be demonstrated. The notion that a unique enhancer element is the activating factor or expression site body would seem plausible, since there is exactly one copy in the nucleus of this haploid parasite. A transcription factor, whose expression is restricted to extremely low levels, may be considered but it seems difficult to really make sure that exactly one transcription factor is present. Considering that under special conditions more than one promoter can be activated we favour a model where potential positive feedback loops within the activating complex could ensure aggregation around the expressed var gene. In the absence of candidate genes for a monoallelic activating factor, identification of mutant parasites that have lost var gene expression remains the biggest challenge in the field. Traditional biochemical and mass spectrometric analysis of factors interacting with key genetic elements (Zhang et al., 2011a; Brancucci et al., 2012; Volz et al., 2012) have advanced our knowledge, but technical breakthroughs are needed that will allow identification of molecular complexes of the expression site within the cellular context. Forward genetic screens based on transposition mutagenesis (Balu and Adams, 2006) may reveal unsuspected mechanisms in monoallelic expression. Further, small molecule inhibitors against methyltransferases, as recently developed (Malmquist et al., 2012), could provide useful tools to study epigenetically controlled expression of clonally variant genes.
It remains enigmatic how one gene is active while others are silenced, when and how a specific gene is poised rather switched, but many exciting research avenues are left to explore in the field of mutually exclusive expression.
Acknowledgments
We thank Christophe Zimmer, Jose Juan Lopez-Rubio and Nick Malmquist for critical reading and comments. This work was supported by the ERC Advanced Grant (PlasmoEscape 250320), the French Parasitology Network grant ParaFrap and the Human Frontier Science Program fellowship to J.G. We apologize for not citing all relevant publications due to limitations in the allowed number of references.
Conflict of interest
The authors declare that they have no conflicts of interest.
References
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Bachmann A, Predehl S, May J, Harder S, Burchard GD, Gilberger TW, et al. Highly co-ordinated var gene expression and switching in clinical Plasmodium falciparum isolates from non-immune malaria patients. Cell Microbiol. 2011;9:1397–1409. doi: 10.1111/j.1462-5822.2011.01629.x. [DOI] [PubMed] [Google Scholar]
- Balu B, Adams JH. Functional genomics of Plasmodium falciparum through transposon-mediated mutagenesis. Cell Microbiol. 2006;8:1529–1536. doi: 10.1111/j.1462-5822.2006.00776.x. [DOI] [PubMed] [Google Scholar]
- Bartfai R, Hoeijmakers WA, Salcedo-Amaya AM, Smits AH, Janssen-Megens E, Kaan A, et al. H2A.Z demarcates intergenic regions of the plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLoS Pathog. 2010;6:e1001223. doi: 10.1371/journal.ppat.1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancucci NM, Witmer K, Schmid CD, Flueck C, Voss TS. Identification of a cis-acting DNA-protein interaction implicated in singular var gene choice in Plasmodium falciparum. Cell Microbiol. 2012;12:1836–1848. doi: 10.1111/cmi.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JH. Transcriptional memory at the nuclear periphery. Curr Opin Cell Biol. 2009;21:127–133. doi: 10.1016/j.ceb.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brolin KJ, Ribacke U, Nilsson S, Ankarklev J, Moll K, Wahlgren M, Chen Q. Simultaneous transcription of duplicated var2csa gene copies in individual Plasmodium falciparum parasites. Genome Biol. 2009;10:R117. doi: 10.1186/gb-2009-10-10-r117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood MS, Gannoun-Zaki L, Wellems TE, Deitsch KW. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J Biol Chem. 2003;278:34125–34132. doi: 10.1074/jbc.M213065200. [DOI] [PubMed] [Google Scholar]
- Chookajorn T, Dzikowski R, Frank M, Li F, Jiwani AZ, Hartl DL, Deitsch KW. Epigenetic memory at malaria virulence genes. Proc Natl Acad Sci USA. 2007;104:899–902. doi: 10.1073/pnas.0609084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney EJ, Legros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, et al. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes A, Carret C, Kaneko O, Yim Lim BY, Ivens A, Holder AA. Epigenetic silencing of Plasmodium falciparum genes linked to erythrocyte invasion. PLoS Pathog. 2007;3:e107. doi: 10.1371/journal.ppat.0030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Fan Q, Cui L, Miao J. Histone lysine methyltransferases and demethylases in Plasmodium falciparum. Int J Parasitol. 2008;38:1083–1097. doi: 10.1016/j.ijpara.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch KW, Calderwood MS, Wellems TE. Malaria. Cooperative silencing elements in var genes. Nature. 2001;412:875–876. doi: 10.1038/35091146. [DOI] [PubMed] [Google Scholar]
- Deitsch KW, Lukehart SA, Stringer JR. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol. 2009;7:493–503. doi: 10.1038/nrmicro2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, et al. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- Dzikowski R, Deitsch KW. Active transcription is required for maintenance of epigenetic memory in the malaria parasite Plasmodium falciparum. J Mol Biol. 2008;382:288–297. doi: 10.1016/j.jmb.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikowski R, Frank M, Deitsch K. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog. 2006;2:e22. doi: 10.1371/journal.ppat.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikowski R, Li F, Amulic B, Eisberg A, Frank M, Patel S, et al. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 2007;8:959–965. doi: 10.1038/sj.embor.7401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enderes C, Kombila D, Dal-Bianco M, Dzikowski R, Kremsner P, Frank M. Var Gene promoter activation in clonal Plasmodium falciparum isolates follows a hierarchy and suggests a conserved switching program that is independent of genetic background. J Infect Dis. 2011;204:1620–1631. doi: 10.1093/infdis/jir594. [DOI] [PubMed] [Google Scholar]
- Epp C, Li F, Howitt CA, Chookajorn T, Deitsch KW. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA. 2009;15:116–127. doi: 10.1261/rna.1080109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fastman Y, Noble R, Recker M, Dzikowski R. Erasing the epigenetic memory and beginning to switch – the onset of antigenic switching of var genes in Plasmodium falciparum. PLoS ONE. 2012;7:e34168. doi: 10.1371/journal.pone.0034168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flueck C, Bartfai R, Volz J, Niederwieser I, Salcedo-Amaya AM, Alako BT, et al. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 2009;5:e1000569. doi: 10.1371/journal.ppat.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Dzikowski R, Costantini D, Amulic B, Berdougo E, Deitsch K. Strict pairing of var promoters and introns is required for var gene silencing in the malaria parasite Plasmodium falciparum. J Biol Chem. 2006;281:9942–9952. doi: 10.1074/jbc.M513067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, et al. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- Horrocks P, Pinches R, Christodoulou Z, Kyes SA, Newbold CI. Variable var transition rates underlie antigenic variation in malaria. Proc Natl Acad Sci USA. 2004;101:11129–11134. doi: 10.1073/pnas.0402347101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt CA, Wilinski D, Llinas M, Templeton TJ, Dzikowski R, Deitsch KW. Clonally variant gene families in Plasmodium falciparum share a common activation factor. Mol Microbiol. 2009;73:1171–1185. doi: 10.1111/j.1365-2958.2009.06846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Lopez-Barragan MJ, Jiang H, Mu J, Gaur D, Zhao K, et al. Epigenetic control of the variable expression of a Plasmodium falciparum receptor protein for erythrocyte invasion. Proc Natl Acad Sci USA. 2010;107:2224–2229. doi: 10.1073/pnas.0913396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaviratne M, Khan SM, Jarra W, Preiser PR. Small variant STEVOR antigen is uniquely located within Maurer's clefts in Plasmodium falciparum-infected red blood cells. Eukaryot Cell. 2002;1:926–935. doi: 10.1128/EC.1.6.926-935.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyes S, Christodoulou Z, Pinches R, Kriek N, Horrocks P, Newbold C. Plasmodium falciparum var gene expression is developmentally controlled at the level of RNA polymerase II-mediated transcription initiation. Mol Microbiol. 2007;63:1237–1247. doi: 10.1111/j.1365-2958.2007.05587.x. [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez Rivas R, Scherf A. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol. 2007;66:1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rubio JJ, Mancio-Silva L, Scherf A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe. 2009;5:179–190. doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmquist NA, Moss TA, Mecheri S, Scherf A, Fuchter MJ. Small-molecule histone methyltransferase inhibitors display rapid antimalarial activity against all blood stage forms in Plasmodium falciparum. Proc Natl Acad Sci USA. 2012;109:16708–16713. doi: 10.1073/pnas.1205414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancio-Silva L, Zhang Q, Scheidig-Benatar C, Scherf A. Clustering of dispersed ribosomal DNA and its role in gene regulation and chromosome-end associations in malaria parasites. Proc Natl Acad Sci USA. 2010;107:15117–15122. doi: 10.1073/pnas.1001045107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick CJ, Huttenhower C, Buckee C, Amambua-Ngwa A, Gomez-Escobar N, Walther M, et al. Epigenetic Dysregulation of virulence gene expression in severe Plasmodium falciparum malaria. J Infect Dis. 2012;205:1593–1600. doi: 10.1093/infdis/jis239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Fan Q, Cui L, Li X, Wang H, Ning G, et al. The MYST family histone acetyltransferase regulates gene expression and cell cycle in malaria parasite Plasmodium falciparum. Mol Microbiol. 2010;78:883–902. doi: 10.1111/j.1365-2958.2010.07371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro M, Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414:759–763. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- Noble R, Recker M. A statistically rigorous method for determining antigenic switching networks. PLoS ONE. 2012;7:e39335. doi: 10.1371/journal.pone.0039335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Toledo K, Rojas-Meza AP, Mancio-Silva L, Hernandez-Cuevas NA, Delgadillo DM, Vargas M, et al. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res. 2009;37:2596–2606. doi: 10.1093/nar/gkp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petter M, Lee CC, Byrne TJ, Boysen KE, Volz J, Ralph SA, et al. Expression of P. falciparum var genes involves exchange of the histone variant H2A.Z at the promoter. PLoS Pathog. 2011;7:e1001292. doi: 10.1371/journal.ppat.1001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph SA, Scheidig-Benatar C, Scherf A. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc Natl Acad Sci USA. 2005;102:5414–5419. doi: 10.1073/pnas.0408883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recker M, Buckee CO, Serazin A, Kyes S, Pinches R, Christodoulou Z, et al. Antigenic variation in Plasmodium falciparum malaria involves a highly structured switching pattern. PLoS Pathog. 2011;7:e1001306. doi: 10.1371/journal.ppat.1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DJ, Craig AG, Berendt AR, Pinches R, Nash G, Marsh K, Newbold CI. Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature. 1992;357:689–692. doi: 10.1038/357689a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E, Ben-Shmuel A, Shalev O, Sinay R, Cowman A, Pollack Y. Differential, positional-dependent transcriptional response of antigenic variation (var) genes to biological stress in Plasmodium falciparum. PLoS ONE. 2009;4:e6991. doi: 10.1371/journal.pone.0006991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira-Graells N, Gupta AP, Planet E, Crowley VM, Mok S, Ribas de Pouplana L, et al. Transcriptional variation in the malaria parasite Plasmodium falciparum. Genome Res. 2012;22:925–938. doi: 10.1101/gr.129692.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo-Amaya AM, Hoeijmakers WA, Bartfai R, Stunnenberg HG. Malaria: could its unusual epigenome be the weak spot? Int J Biochem Cell Biol. 2010;42:781–784. doi: 10.1016/j.biocel.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Scherf A, Hernandez-Rivas R, Buffet P, Bottius E, Benatar C, Pouvelle B, et al. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 1998;17:5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A, Lopez-Rubio JJ, Riviere L. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol. 2008;62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- Schieck E, Pfahler JM, Sanchez CP, Lanzer M. Nuclear run-on analysis of var gene expression in Plasmodium falciparum. Mol Biochem Parasitol. 2007;153:207–212. doi: 10.1016/j.molbiopara.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Sierra-Miranda M, Delgadillo DM, Mancio-Silva L, Vargas M, Villegas-Sepulveda N, Martinez-Calvillo S, et al. Two long non-coding RNAs generated from subtelomeric regions accumulate in a novel perinuclear compartment in Plasmodium falciparum. Mol Biochem Parasitol. 2012;185:36–47. doi: 10.1016/j.molbiopara.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Swamy L, Amulic B, Deitsch KW. Plasmodium falciparum var gene silencing is determined by cis DNA elements that form stable and heritable interactions. Eukaryot Cell. 2011;10:530–539. doi: 10.1128/EC.00329-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin CJ, Carret CK, Duraisingh MT, Voss TS, Ralph SA, Hommel M, et al. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 2009;7:e84. doi: 10.1371/journal.pbio.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz JC, Bartfai R, Petter M, Langer C, Josling GA, Tsuboi T, et al. PfSET10, a Plasmodium falciparum methyltransferase, maintains the active var gene in a poised state during parasite division. Cell Host Microbe. 2012;11:7–18. doi: 10.1016/j.chom.2011.11.011. [DOI] [PubMed] [Google Scholar]
- Voss TS, Healer J, Marty AJ, Duffy MF, Thompson JK, Beeson JG, et al. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature. 2006;439:1004–1008. doi: 10.1038/nature04407. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Huang Y, Zhang Y, Fang X, Claes A, Duchateau M, et al. A critical role of perinuclear filamentous actin in spatial repositioning and mutually exclusive expression of virulence genes in malaria parasites. Cell Host Microbe. 2011a;10:451–463. doi: 10.1016/j.chom.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhang Y, Huang Y, Xue X, Yan H, Sun X, et al. From in vivo to in vitro: dynamic analysis of Plasmodium falciparum var gene expression patterns of patient isolates during adaptation to culture. PLoS ONE. 2011b;6:e20591. doi: 10.1371/journal.pone.0020591. [DOI] [PMC free article] [PubMed] [Google Scholar]


