INTRODUCTION
Characterizing protein localization in Xenopus laevis embryos is an important aspect of developmental and regenerative studies that use this advantageous model system. Although whole-mount immunohistochemistry is an efficient and powerful way to visualize surface and ectodermal expression, its ability to localize proteins in internal tissues and cells is limited by the incomplete penetration of antibodies through outer layers of the embryo. Microtome sections of paraffin-embedded embryos provide good internal resolution, but precise orientation of embryos can be difficult, and sectioning many samples is time intensive. Further, care must be taken with sections to minimize tissue damage, because heat and organic solvents used during the process can render some proteins invisible to antibody detection. The method described here is a short protocol for generating robust sections for use in immunoreactions with as little as two days from collection to visualization, making it useful as a rapid screening process. Advantages of this method include: (1) the durability of the sections produced (which can be treated as if they were wholemounts and processed by fluid aspiration in vials rather than mounted onto slides); (2) the ability to examine multiple antibody targets in tandem, in tissue that is never heated or extracted with harsh reagents; (3) the lack of autofluorescence as occurs in glutaraldehyde-containing media; and (4) the ease of orientation of embryos in a fully transparent block.
RELATED INFORMATION
Related protocols include A Rapid Protocol for Whole-Mount In Situ Hybridization on Xenopus Embryos (Monsoro-Burq 2007) and Whole-Mount Fluorescence Immunocytochemistry on Xenopus Embryos (Lee et al. 2008). Embedding and sectioning Xenopus embryos in agarose is described in Preparation of Fixed Xenopus Embryos for Confocal Imaging (Wallingford 2010). For an earlier version of this protocol that makes use of glutaraldehyde, see Levin (2004).
MATERIALS
CAUTIONS AND RECIPES: Please see Appendices for appropriate handling of materials marked with <!>, and recipes for reagents marked with <R>.
Reagents
Agarose solution (low melting point [LMP], 4% [w/v])
<R>Alkaline phosphatase buffer with levamisole (AP buffer with levamisole; for AP reactions only)
<R>AP reaction solution (for AP reactions only)
Horseradish peroxidase (HRP) substrate (available commercially)
<R>Hybridization solution for Xenopus (for AP reactions only)
<R>Hydrogen peroxide (3% in methanol) (for HRP reactions only)
<R>MEMFA
-
<!>Methanol (25%, 50%, 75%, and 100%)
<R>Prepare dilutions of methanol in PBT buffer.
<R>PBT buffer
<R>PBT buffer containing 10% heat-inactivated normal goat serum (Blocking buffer)
Tyramide amplification kit (e.g., Invitrogen) (for tyramide detection)
-
Vectamount
<R>50% glycerol in PBT buffer can be used as an alternative (see Step 36).
Xenopus laevis embryos
Table 1.
Sample primary antibodies that can be used in Xenopus
| Target | Source information | Dilution |
|---|---|---|
| Cardiac troponin T: heart | University of Iowa Developmental Studies Hybridoma Bank (DSHB), Antibody CT3 | 1:10 |
| Tor70: notochord | See http://mcb.berkeley.edu/labs/harland/pages/tor70.html for details. Note that special secondary is needed. | 1:5 |
| Caspase-3: apoptosis | Abcam ab13847 | 1:500 |
| 12/101: somite/muscle | http://dshb.biology.uiowa.edu/skeletal-muscle-marker http://xtropicalis.cpsc.ucalgary.ca/xenwiki/index.php/Monoclonal_12/101_-_somites | 1:3 |
| 2G9: neural tube | http://dshb.biology.uiowa.edu/LAMP-gene-symbol-Lsamp | 1:5 |
| H3P: proliferating cells | Millipore 06-570. Note that while the rabbit version works well in Xenopus, the mouse version does not seem to. | 1:1000 |
| Acetylated α-tubulin: cilia and neurons | Sigma T6793 | 1:1000 |
Table 2.
Sample secondary antibodies that can be used in Xenopus
| Detection | Source information | Dilution |
|---|---|---|
| Fluorescence | Invitrogen Alexa Fluor 546 or Alexa Fluor 555, anti-rabbit or anti-mouse | 1:500 |
| Alkaline phosphatase | Jackson ImmunoResearch | 1:1000–1:2500 |
| HRP (horseradish peroxidase) | Jackson ImmunoResearch | 1:750 using TrueBlue peroxidase substrate (KPL) |
Equipment
Coverslips (optional)
<!>Cyanoacrylate adhesive (e.g., Super Glue)
Forceps, fine
Freezer preset to −20°C
Hybridization oven preset to 65°C
Microscope (with appropriate cubes for visualizing fluorescently conjugated secondary antibodies)
Microwave
Mixer (Nutator)
Paintbrush
Paper towel (optional; see Step 13)
Pipettes, disposable transfer
Micropipettor and tips
Molds, plastic disposable biopsy (15 × 15 × 5 mm; e.g., Tissue-Tek Cryomold 4565)
Parafilm
Petri dishes
Razor blade
Refrigerator preset to 4°C
Reservoir (provided with Vibratome; e.g., Leica buffer tray 14046327408)
Sectioning blocks (provided with Vibratome; e.g., Leica specimen discs 14046327406)
Slides, glass
Tissue (e.g., KimWipe)
Vibratome (e.g., Leica VT1000S)
Vials, scintillation (for embryos and sections, e.g., 4-mL volume)
METHOD
Perform all washes and incubations with gentle rocking on a Nutator at room temperature unless otherwise specified. For all washes, use enough buffer to fill the scintillation vial.
Fixing Embryos
-
1
Fix the embryos in scintillation vials using an appropriate protocol for the epitope of interest.
For most antibodies, fixing embryos in 1X MEMFA for 1.5 h at room temperature (or overnight at 4°C) is sufficient. -
2
Wash the embryos three times with PBT buffer for 10 min each.
Preparing Embryos for Sectioning
Dehydration of embryos helps keep the tissue from popping out of the agarose sections and reduces tissue damage in early-stage embryos.
-
3
Dehydrate the embryos through sequential 5-min washes of 25%, 50%, and 75% methanol in PBT buffer, followed by a 5-min wash in 100% methanol.
-
4
Store the embryos for at least 4 h at −20°C.
-
5
Before embedding the embryos in agarose, rehydrate them through sequential 5-min washes of 75%, 50%, and 25% methanol in PBT buffer.
-
6
Wash the embryos three times in PBT buffer for 5 min each.
Orienting Embryos in Agarose
-
7
Heat a 4% LMP agarose solution in a microwave until the solution is clear. Fill a plastic mold with the melted agarose.
-
8
Use a disposable pipette to transfer a single Xenopus embryo to a flat, dry lab tissue.
The lab tissue will remove all excess liquid, increasing binding between the agarose and the sample. Avoid damaging the surface of the embryo. -
9
With fine forceps, lift the embryo off the lab tissue from below and place it in the agarose-filled mold. Orient the embryo into any position desired.
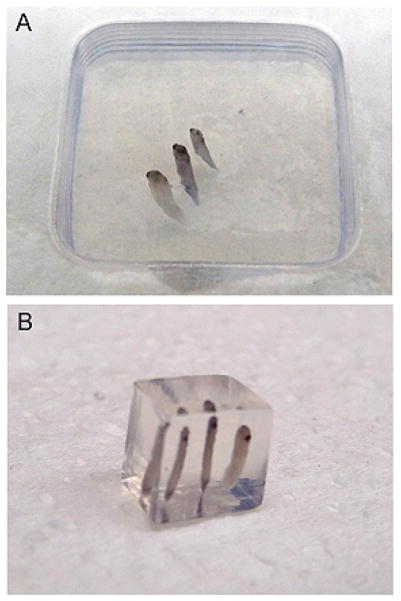
Up to four mid-tailbud stage embryos or 10 early cleavage-stage embryos can be placed in the same block side by side (see Fig. 1A). -
10
Allow the agarose to cool and harden, ~5–10 min.
-
11
Remove the agarose block from the mold and cut away the excess with a razor blade (see Fig. 1B).
-
12
Repeat Steps 7–11 with all samples.
-
13
Store cut blocks in labeled, Parafilm-sealed Petri dishes at 4°C until sectioning. For storage longer than 48 h, add a small section of wet paper towel to the Petri dish before sealing with Parafilm.
FIGURE 1.
Example of embryos oriented in agarose. (A) Using standard molds and LMP agarose, numerous embryos can be positioned in the same block. Orientation with respect to the bottom of the mold typically yields the best results, because the surface is flat and can easily be attached to sectioning plates. (B) Once blocks are cooled, excess agarose can be cut away, leaving less material to section and decreasing the time spent sectioning.
Sectioning Tissue
-
14
Attach each block of agarose to individual sectioning blocks using a droplet of Super Glue. Allow the glue to dry, ~5–10 min.
-
15
Section the blocks in a water-filled reservoir using the appropriate speed and frequency for the Vibratome.
Sections ranging in size from 150 to 300 μm are easy to manipulate with forceps and hold up well through multiple washes. However, sections as thin as 40 μm can be used with this protocol.See Troubleshooting. -
16
Collect the sections using a paintbrush and transfer them to scintillation vials filled with PBT buffer.
Quenching, Blocking, and Incubating with Primary Antibody
It is not necessary to remove the agarose from the sections; it does not affect the ability to image the sections and it makes the samples easier to manipulate. For all steps, use enough solution to keep all sections well covered with liquid during rocking stages so that the sections do not experience the air-water interface.
-
17
Quench endogenous peroxide activity in sections that will be incubated with HRP-conjugated secondary antibodies:
-
Incubate the sections in 3% hydrogen peroxide in methanol for 15 min at room temperature. Do not rock.
Be mindful that during bleaching (HRP inactivation), gas pressure can cause dangerous explosions of glass scintillation vials. Be sure to vent; do not shake and do not increase peroxide content of this solution (increase incubation time if additional bleaching/inactivation is desired). Continue to Step 19.
-
-
18
Quench endogenous AP activity in sections that will be incubated with AP-conjugated secondary antibodies:
Inactivate endogenous AP activity by incubating the sections in hybridization solution for Xenopus with gentle rocking on a Nutator for 3 h at 65°C.
Wash the sections in PBT buffer on a Nutator at room temperature until the formamide is removed completely (at least 4 times for 15 min each).
Continue to Step 19.
-
19
Wash the sections with PBT buffer on a Nutator for 15 min at room temperature.
-
20
Block the sections in ~1 mL of blocking buffer on a Nutator for 1 h at room temperature.
-
21
Prepare the primary antibody at the desired concentration in blocking buffer.
See Table 1 for some common antibodies that work well in Xenopus. -
22
Incubate the embryos in 1 mL of primary antibody solution overnight at 4°C on a Nutator.
Whenever possible, make sure to process a sample without secondary antibody (to control for endogenous autofluorescence) and another sample without primary antibody (to control for nonspecific binding of secondary antibody). This is especially important when using AP detection.
Incubating with Secondary Antibody
Perform all washes and incubations with gentle rocking on a Nutator at room temperature unless otherwise specified. For all washes, use enough buffer to fill the scintillation vial. See Discussion for further information about secondary antibody options.
-
23
Remove the primary antibody solution, and wash the sections three times in PBT buffer for 15 min each.
-
24
Prepare a solution of an appropriate secondary antibody at the desired concentration in PBT buffer.
See Table 2 for some common antibodies that work well in Xenopus. -
25
Incubate the sections with 1 mL of secondary antibody solution for 1 h.
Do not rock the vials during this incubation, because of the small volume of antibody solution. Keep the vials in darkness if using a fluorescent secondary antibody. -
26
Remove the secondary antibody solution, and wash the sections three times in PBT buffer for 15 min each.
Keep the vials in darkness if using a fluorescent secondary antibody.
For fluorescent secondary antibodies
-
27
Proceed to Imaging, Step 32.
For AP-conjugated secondary antibodies
-
28
Wash the sections twice with AP buffer with levamisole on a Nutator in darkness for 5 min each.
-
29
Remove the AP buffer with levamisole and replace with 1–2 mL of AP reaction solution. Incubate the vials in darkness on a Nutator until the signal is visible.
The reaction can take from 10 min up to several hours. -
30
After the signal is visible, wash the sections three times with PBT buffer on a Nutator for 15 min.
For HRP-conjugated secondary antibodies
-
31
Process samples that have been treated with HRP-conjugated antibodies as follows:
For tyramide detection of HRP-conjugated antibodiesPrepare a tyramide working solution according to the manufacturer’s instructions.
-
Add 1 mL of tyramide working solution to each vial. Incubate the vials in the dark for 10 min.
Do not rock the vials. -
Wash the samples three times with PBT buffer on a Nutator in the dark for 15 min.
For other HRP substrates Prepare a working solution of HRP substrate according to the manufacturer’s instructions.
-
Add 1 mL of HRP substrate to each vial. Incubate the vials in the dark until the signal is visible.
Do not rock the vials. This reaction can take 10 min to several hours. Wash the sections three times with PBT buffer on a Nutator for 15 min.
Preparing Sections for Imaging
-
32
Empty the sections into a small Petri dish.
-
33
Transfer the sections to a glass slide as follows:
For thick sections (150 μm or larger)-
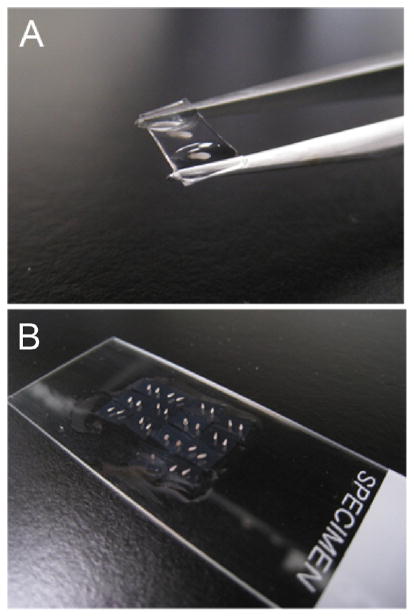
Slide the tips of fine forceps (slightly opened) under each section, raise it out of the PBT buffer, and deposit it on a glass slide (see Fig. 2A).
For thin sections (40–150 μm) Use a disposable pipette to transfer sections to a glass slide.
Suction off excess PBT buffer with the pipette.
If sections stick to the inside of the pipette, pull PBT buffer up and down in the pipette.
-
-
34
Repeat Step 33 with all sections. Arrange the sections in a grid-like fashion for easier imaging (see Fig. 2B).
-
35
Image the sections with a microscope as they are, or place a coverslip over them for higher magnification photographs.
-
36
Seal the slides with Vectamount or 50% glycerol in PBT buffer and store for up to 1 mo at 4°C.
-
37. Alternatively, return each set of sections to their original scintillation vial in fresh PBT buffer:
Angle the slide into the vial.
Pipette fresh PBT buffer over the top of the sections.
-
Store for up to 2 wk at 4°C.
The sections should wash into the vial without any damage and can be used for a second reaction at a later point.
-
38
For long-term storage, dehydrate the sections into methanol as in Step 3. Store indefinitely at 4°C.
Ethanol can be substituted for methanol when dehydrating the sections.
FIGURE 2.
Example of technique used for transferring sections to slides for imaging. (A) Thick sections (150 μm+) can be lifted out of a Petri dish individually, using forceps, without directly touching the embedded tissue. (B) Each sample can then be placed on a slide in a grid fashion for ease of imaging, with or without a coverslip as necessary. When imaging is complete, simply tilt the slide into a scintillation vial and pipette PBT over the surface to wash the samples back into the vial.
TROUBLESHOOTING
Problem: Tissue falls out of agarose slices after sectioning.
[Step 15]
Solution: Consider fixing the embryos and/or dehydrating the embryos in methanol for a longer period, especially for one- to 32-cell embryos. Sections that fall out can still be processed if care is taken when washing.
Problem: Sections through tadpole eyes look distorted.
Solution: Developed Xenopus eyes contain hardened tissue and thick lenses. Increasing the frequency setting of the Vibratome may be necessary.
DISCUSSION
We regularly use this protocol to analyze localization of proteins and quantitatively assay for the presence of specific tissues (e.g., nerve or muscle) or distinct cell states (e.g., apoptosis or mitosis). Not only is it useful in an exploratory fashion, such as when screening numerous antibodies for expression profiles, but it also produces publication-ready images of quality similar to that produced by other methods of sectioning and processing (although Vibratome sectioning in a soft medium is not ideal for obtaining subcellular resolution). In addition, the samples hold up very well over time. Embryos ranging from cleavage stages up to stage 45 can be easily oriented to achieve sections in any plane. We have processed sections through immunohistochemistry, stored them for up to 4 wk at −20°C in 100% methanol, and then run them through a subsequent round of immunohistochemistry with a second antibody, without any damage seen to the tissue.
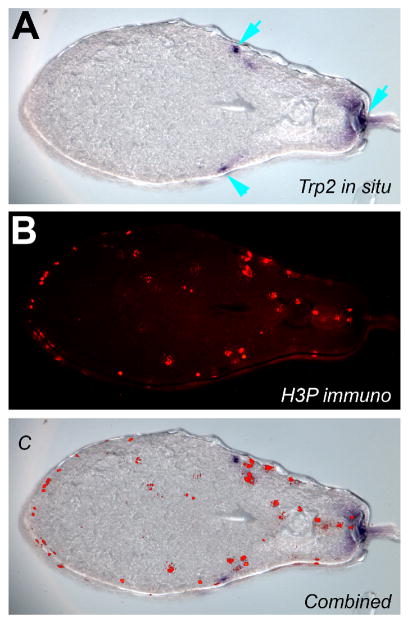
We have found that fluorescent secondary antibodies typically yield the best images, especially those fluorescing under long wavelengths (647 nm being the cleanest). Further, fluorescent secondary antibodies are highly useful for embryos processed after whole-mount in situ hybridization (see Fig. 3 for an example, and A Rapid Protocol for Whole-Mount In Situ Hybridization on Xenopus Embryos [Monsoro-Burq 2007]), allowing expression of RNA and protein to be visualized in the same section without cross-signal interference. Fluorescent antibodies offer speed and the availability of multiple colors that can be imaged simultaneously, whereas AP-conjugated antibodies have the advantage of a fixable (permanent), dark purple color that is directly visible on tissue (not requiring fluorescence) and offers a degree of amplification due to enzyme processivity.
FIGURE 3.
Detection of mRNA and protein in the same section. (A) Embryos that underwent whole-mount in situ hybridization with an antisense probe to the melanocyte marker Trp-2 were sectioned using the method described. Blue arrows indicate signal revealing the position of melanocytes. (B) The sections were processed for immunohistochemistry with an antibody to the phosphorylated form of Histone 3B (a mitosis marker) and an Alexa-555-conjugated secondary antibody (red signal on section). (C) The two images can be easily overlaid, allowing simultaneous detection of both signals, in this case revealing what percentage of mitotic cells is Trp-2-positive. (For color figure, see doi: 10.1101/pdb.prot5532 online at www.cshprotocols.org.)
For HRP-conjugated secondary antibodies, an HRP substrate can be used, or a tyramide amplification kit if needed. The tyramide kit uses enzymatic activity to amplify positive signals and uses covalent bonds that minimize signal diffusion (Gerth et al. 2005). Thus, this kit is most appropriate for detection of low-abundance proteins or when primary antibodies are only available in small quantities. It is also appropriate when sections will be stored post-treatment for imaging at a later time. The tyramide amplification kits available from Invitrogen have a wide range of Alexa Fluor conjugates, allowing different fluorescent wavelengths to be selected as needed. However, the tyramide detection kits are much more expensive than other detection systems. Thus, other HRP substrates can be used successfully for detection of abundant proteins when imaging can take place immediately or when fluorescence microscopes are not available.
Acknowledgments
We thank members of the Levin lab and the Xenopus community for their help and advice on immunohistochemistry. D.B. is supported by National Institues of Health (NIH) T32 grant 5T32DE007327-07. L.N.V. is supported by NIH National Research Service Award (NRSA) grant 1F32GM087107. M.L. gratefully acknowledges support from American Heart Association (Established Investigator Grant 0740088N), NIH (EY018168 and GM078484), and award W81XWH-10-2-0058 from the Telemedicine and Advanced Technology Research Center (TATRC) at the U.S. Army Medical Research and Materiel Command (USAMRMC).
References
- Gerth VE, Zhou X, Vize PD. Nephrin expression and three-dimensional morphogenesis of the Xenopus pronephric glomus. Dev Dyn. 2005;233:1131–1139. doi: 10.1002/dvdy.20415. [DOI] [PubMed] [Google Scholar]
- Lee C, Kieserman E, Gray RS, Park TJ, Wallingford J. Whole-mount fluorescence immunocytochemistry on Xenopus embryos. Cold Spring Harb Protoc. 2008 doi: 10.1101/pdb.prot4957. [DOI] [PubMed] [Google Scholar]
- Levin M. A novel immunohistochemical method for evaluation of antibody specificity and detection of labile targets in biological tissue. J Biochem Biophys Methods. 2004;58:85–96. doi: 10.1016/s0165-022x(03)00149-0. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq AH. A rapid protocol for whole-mount in situ hybridization on Xenopus embryos. Cold Spring Harb Protoc. 2007 doi: 10.1101/pdb.prot4809. [DOI] [PubMed] [Google Scholar]
- Wallingford JB. Preparation of fixed Xenopus embryos for confocal imaging. Cold Spring Harb Protoc. 2010 doi: 10.1101/pdb.prot5426. [DOI] [PubMed] [Google Scholar]