Abstract
Recent advances in next-generation DNA sequencing and proteomics provide an unprecedented ability to survey mRNA and protein abundances. Such proteome-wide surveys are illuminating the extent to which different aspects of gene expression help to regulate cellular protein abundances. Current data demonstrate a substantial role for regulatory processes occurring after mRNA is made — that is, post-transcriptional, translational and protein degradation regulation — in controlling steady-state protein abundances. Intriguing observations are also emerging in relation to cells following perturbation, single-cell studies and the apparent evolutionary conservation of protein and mRNA abundances. Here, we summarize current understanding of the major factors regulating protein expression.
Production and maintenance of cellular protein requires a remarkable series of linked processes, spanning the transcription, processing and degradation of mRNAs to the translation, localization, modification and programmed destruction of the proteins themselves. Protein abundances reflect a dynamic balance among these processes. It has long been an open question of how this balance is achieved and to what extent each of these processes contributes to the regulation of cellular protein abundances. Until recently, such questions have been difficult to address, as it was largely impossible to estimate protein concentrations at a large scale. It has been common practice to use mRNA concentrations as proxies for the concentrations and activities of the corresponding proteins, thereby assuming that transcript abundances are the main determinant of protein abundances. However, recent technological advances, in particular in mass spectrometry and high-throughput cell imaging, have allowed for large-scale surveys of the proteome. The advent of next-generation sequencing has complemented these new findings with a detailed description of the transcriptome. These studies are changing our understanding of protein-expression regulation. In particular, proteomics has now advanced sufficiently to allow for the systematic quantification of the absolute abundances of thousands of proteins (BOX 1).
“Current data demonstrate a substantial role for regulatory processes occurring after mRNA is made”
Box 1. Key concepts for analyses of protein abundances.
Absolute versus relative concentrations
Absolute concentrations are defined by an amount of protein (or RNA) per unit — for example, molecules per cell — and they can be used independently of a reference data set.
Relative concentrations are not proper concentrations but are ratios of two absolute concentrations (that is, fold changes), such as intensity ratios from dual-channel DNA microarray measurements or from stable isotopic labelling with amino acids in cell culture (SILAC). Each measurement represents a concentration relative to a reference sample and can report either steady-state or non-steady-state conditions.
Rates versus concentrations
We can easily distinguish between the rate (or speed) at which a process happens, and the concentrations of the participating molecules at a given time but, in practice, these concepts are often confused. For example, abundant proteins are often assumed to exhibit high rates of translation or transcription. However, such proteins may be slowly translated but very stable, producing high final concentrations. The opposite scenario applies as well.
Steady-state versus non-steady-state (perturbed) systems
The ‘steady state’ is defined by a zero net change of a parameter in a system. For example, the abundance of a protein might not vary during the time of observation because rates of translation and degradation are balanced. The cell is said to be at the steady state with respect to the concentration of this protein. Cells may encounter different steady states: for example, under normal conditions or when a gene is mutated.
A population of cells growing in log phase is often said to be at the steady state. Protein concentrations in individual cells may change with cell division, but the average concentration of a protein across the population is roughly constant and thus fulfils the steady-state condition. If a population of cells has been subjected to a stimulus — for example, stress — concentrations of proteins across the population are changing over a specific period of time, and thus are not at the steady state, until the population reaches the steady state again. This steady state might differ to that before the perturbation.
Single cell versus populations
Single-cell observations are an important method for distinguishing single-cell observations from population averages, especially in the context of gene expression noise, as well as steady-state conditions such as those described above. The same test (for example, changes in protein concentration over time) may produce entirely different results when focusing on a single cell or the population.
Emerging evidence is changing our view of the role for the many regulatory mechanisms occurring after mRNAs are manufactured (FIG. 1). In almost every organism that has been examined to date, steady-state transcript abundances only partially predict protein abundances1, suggesting that after experimental errors have been eliminated, other modes of regulation must be invoked to explain how the levels of proteins are set within cells. In this Progress article, we will summarize recent technological advances, describe examples in which these technologies have enabled novel studies of protein and mRNA regulation in the steady state and in perturbed systems and describe how such studies are informing models of protein expression regulation.
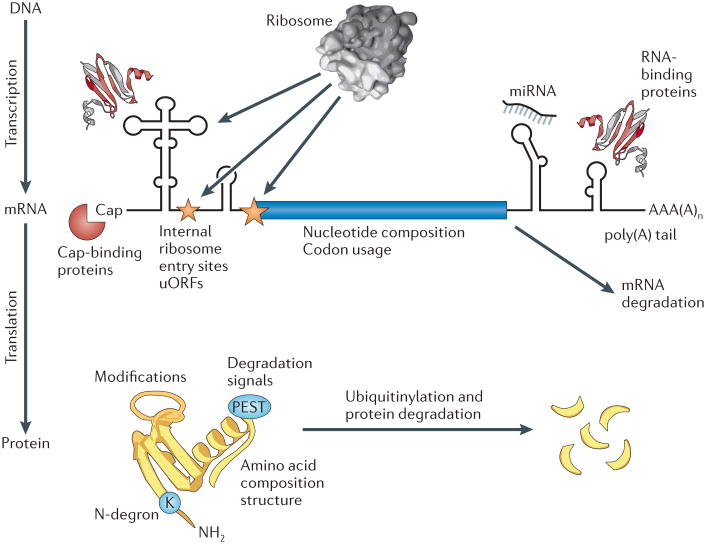
Figure 1. Modes of translation and protein-degradation regulation.
Protein abundances are determined by a balance of regulation of both RNA and protein production and turnover, and some of the major determinants of protein abundance are illustrated here. The figure focuses on major mechanisms of the regulation of translation and transcript stability (upper panel) and protein degradation (lower panel). Mechanisms of transcription regulation are not discussed in this article. miRNA, microRNA; uORF, upstream open reading frame. Figure adapted, with permission, from REF. 1© (2009) Royal Society of Chemistry.
Recent technological advances
A major technological driver in proteomics has been the development of the Orbitrap mass detector2, which made rapid, high-sensitivity protein mass spectrometry more affordable and more widely available. Experiments typically involve ‘shotgun proteomics’, in which cellular proteins are enzymatically digested, and the resulting peptides are analysed by nanoflow chromatography and high-resolution tandem mass spectrometry (see REF. 3 for a recent review). Shotgun proteomics experiments can be made quantitative: for example, by measuring ion intensities in the mass spectrometer or by counting spectra that are derived from each peptide. Both techniques enable the absolute quantification of proteins in the sample, providing that the resulting intensities or counts are suitably calibrated to molecular concentrations4–6. In methods such as AQUA, spiked-in labelled peptides in known concentrations provide such absolute concentration reference standards7. Alternatively, it is possible to compare protein samples that incorporate different isotopes of carbon and nitrogen. Different isotopes can be incorporated by feeding cells or organisms isotopically labelled amino acids through the medium — as in the stable isotopic labelling with amino acids in cell culture (SILAC) method8 — or by isotopically labelling peptides during sample preparation in a method such as isobaric tag for relative and absolute quantification (iTRAQ)9. The differentially labelled peptides from these samples can then be directly compared in the mass spectrometer, resulting in relative quantification of protein concentrations between two or more samples. If isotopic labelling is feasible for the biological system, then SILAC and related methods are highly useful owing to their high accuracy and sensitivity. By such means, thousands of proteins from a sample can now be routinely quantified10.
In parallel, next-generation-sequencing advances now allow for routine large-scale quantification of RNA abundances in any organism11. The number of sequenced ‘reads’ per sequence (after accounting for sequence length) is used as a proxy of mRNA abundance. In a clever twist on this approach that has been termed ribosome footprinting, the mRNA sequence stretches bound by ribosomes were analysed12. Such data provide readouts of the efficiency of translation initiation and elongation, giving insights into the regulation of protein abundance12.
At the same time, there has been a growth in the automation of microscopy and, for example, of the generation of libraries of modified yeast or human cells that express proteins fused to fluorescent proteins or to other detectable epitopes. Such strains provide a direct readout of protein abundance and have led to large-scale surveys of protein expression within single cells13,14 and cell populations14,15. These approaches have also enabled the measurement of dynamic changes in protein levels, such as rates of protein degradation16–18. Perhaps most importantly, the comparison of data collected from these three distinct technologies — mass spectrometry, sequencing and microscopy — has provided a valuable opportunity of cross-validation. In the case of protein abundances, such comparisons have broadly confirmed results from each approach, whereas analyses of protein degradation rates, for example, have highlighted discrepancies that need to be resolved5,16–18.
mRNA and protein measurements
Concentrations, production and turnover rates of mRNA and proteins at steady state
From the above technologies, we now have measurements of the absolute concentrations of mRNAs and proteins from various organisms, including mammalian cells, worms, flies, yeast and a few species of bacteria1,5,19. In general, in both bacteria and eukaryotes, the cellular concentrations of proteins correlate with the abundances of their corresponding mRNAs, but not strongly. They often show a squared Pearson correlation coefficient of ∼0.40, which implies that ∼40% of the variation in protein concentration can be explained by knowing mRNA abundances1,19 (FIG. 2a). Higher correlations have also been observed1,19. To explain the remaining ∼60% of the variation, some combination of post-transcriptional regulation and measurement noise needs to be invoked.
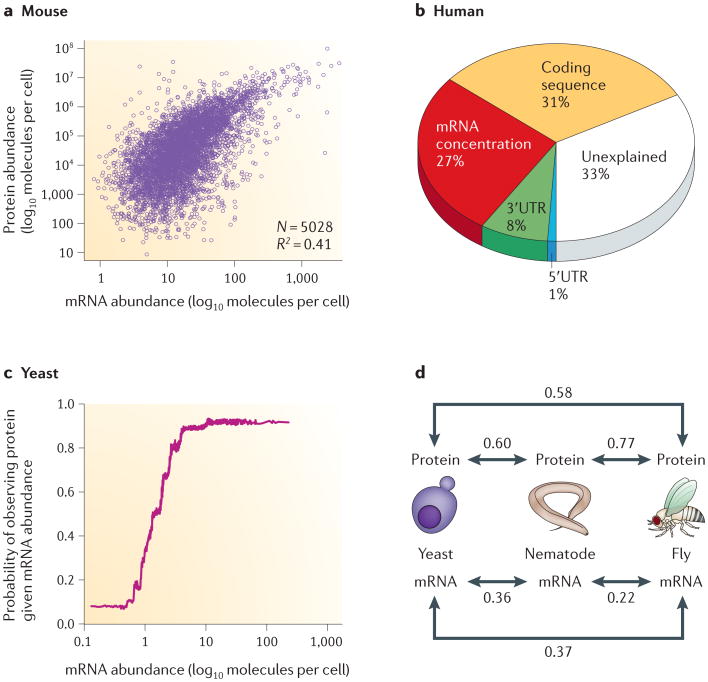
Figure 2. Relationships between mRNA and protein abundances, as observed in large-scale proteome- and transcriptome-profiling experiments.
a | mRNA transcript abundances only partially correlate with protein abundances, typically explaining approximately one- to two-thirds of the variance in steady-state protein levels, depending on the organism. This trend is evident in data from NIH3T3 mouse fibroblast cells. b | In mammalian cells, as shown here for a human DAOY medulloblastoma cell line, ∼30–40% of the variance in protein abundance is explained by mRNA abundance. A similarly large fraction of variance can be explained by other factors, which is indicative of post-transcriptional and translational regulation and protein degradation5,24. c | Nonetheless, mRNA levels are an excellent proxy (in general) for the presence of a protein — or, more precisely, for its detectability using current proteomics technologies. The resulting ‘lazy step function’ has been observed in bacteria, yeast and human cell culture: beyond a certain mRNA concentration, the probability of detecting a protein in the sample does not increase any further. d | Preliminary evidence also suggests that, when considering orthologues across highly divergent species, abundances of proteins are more conserved than abundances of the corresponding mRNAs39,40, suggesting that protein abundances may be evolutionarily favoured. (Numbers indicate Spearman rank correlation coefficients between molecular abundances.) Data such as these support an important role for regulatory mechanisms occurring downstream from the setting of mRNA levels. Panel a of this figure is adapted, with permission, from REF. 5 © (2011) Macmillan Publishers Ltd. All rights reserved. Panel b of this figure is adapted from REF. 24. Panel c of this figure is adapted, with permission, from REF. 42 © (2009) Oxford University Press. Panel d of this figure is adapted, with permission, from REF. 40 © (2010) Wiley.
The concentration of proteins in steady-state cell populations under different growth conditions may vary. This variation is often expressed as a log ratio of the measured abundances and is denoted here as relative abundance (BOX 1). Relative abundances of proteins may or may not occur in proportion to their relative mRNA levels. For example, in haploid versus diploid yeast cells, a moderate correlation (R = 0.46 to 0.68) between the relative abundance in proteins and mRNAs (at least the well-measured ones) was observed20. Similarly, relative abundances were only partially predicted by relative mRNA abundances across three human cell lines (Spearman correlation = 0.63)21.
The above-mentioned protein and mRNA abundances are determined by the relationships between the rates of the processes producing and degrading the participating molecules. Initial large-scale estimates have now also become available for these rates of the different processes of protein expression (FIG. 1). In mammalian cells, mRNAs are produced at a much lower rate than proteins are; on average, a mammalian cell produces two copies of a given mRNA per hour, whereas it produces dozens of copies of the corresponding protein per mRNA per hour. Similarly, mRNAs are less stable than proteins (with an average half-life of 2.6–7 hours versus 46 hours, respectively)5,22. The long half-lives of proteins have been confirmed by independent studies in other systems17,18 and suggest a potentially large role of protein ‘dilution’ — that is, the decrease in protein concentration owing to cell division17. The long half-lives of mammalian mRNAs are in strong contrast to measured bacterial mRNA half-lives, which averaged at roughly 7 minutes23.
All such comparisons summarize average properties and global trends among genes. Any particular protein may have rates or abundances that are very different from average; investigation of its particular rates and abundances may thus help to illuminate interesting biology that is relevant to the protein, perhaps pointing to extremely strong transcriptional or post-transcriptional regulation. For example, RNAs and proteins from mammalian metabolic genes tend to be very stable5 and have high proteinper-mRNA ratios24; by contrast, proteins that are involved in chromatin organization and transcriptional regulation tend to be rapidly degraded5. Protein abundance regulation thus mirrors specific biological roles: regulatory proteins may have to be produced and degraded very rapidly to react to a stimulus, whereas structural or housekeeping proteins would be much longer-lived.
mRNA and protein levels in response to perturbation
In addition to steady-state measurements, efforts have now also turned to perturbed systems (BOX 1). To characterize dynamic changes in proteomes, time-dependent measurements are necessary. Such studies had been limited until recently by factors such as measurement noise, a fairly small number of available isotopic labels and the sequential nature — as opposed to easily parallelizable nature — of mass-spectrometry experiments, which reduces the numbers of samples that can be analysed. However, recent work has provided us with detailed data sets that deliver paradoxical results. For example, yeast was subjected to osmolarity stress and time-dependent expression changes were measured25. The authors found that for upregulated genes, the maximum mRNA and maximum protein concentrations are well-correlated, but this trend was not true for downregulated genes. In a different time-series study, yeast that had been subjected to oxidative stress did not display this behaviour but indicated substantial post-transcriptional regulation of a large fraction of the genes independently of their up- or downregulation26,27. The same appears to be true for bacteria in which time-course analyses of perturbed systems reveal large differences between protein and mRNA abundance changes28,29. Clearly, our understanding of perturbed systems is still incomplete and requires further analyses.
mRNA and protein levels in single cells
Single-cell methods have advanced enormously over the past few years and are now capable of detailed high-throughput analysis of many genes, and the findings from these assays contrast those of population-wide analyses (BOX 1). (For an excellent review of single-cell analyses, see REF. 30.) A recent large-scale survey of single Escherichia coli cells suggests that abundances of bacterial proteins and mRNAs are entirely uncorrelated13, although these measurements have not yet been comprehensively collected nor have they been independently verified. Nonetheless, the population average of the signals from many single-cell measurements produces correlations that are comparable to bulk measurements on cell populations, and mRNA concentrations explain ∼54–77% of the variance in average protein levels13. The lack of correlation between RNA and protein concentration in single cells can be explained by the different lifetimes of the molecules: bacterial mRNAs are short-lived and few copies per cell are produced, causing their concentrations in single cells to fluctuate much more than those of the longer-lived corresponding proteins. When averaging across populations, these fluctuations disappear, and mRNA and protein concentrations correlate.
In addition, translation regulation has a role in the lack of correlation: when comparing different classes of genes across yeast cells, concentrations of proteins in the same complex are less noisy compared with proteins that are not within one complex31, although this is not true at the level of mRNAs32. The contributions of both intra- and intercellular biological noise are an active area of research and are reviewed elsewhere (for example, REF. 33).
Biological interpretations
Understanding steady-state mRNA and protein levels
The evidence above suggests a strong regulatory role for processes downstream of transcription, and therefore the next question to consider is how much each aspect of regulation contributes to setting protein abundances. This question has recently been addressed in mammalian cell lines, both computationally and experimentally5,24: these efforts target the remaining ∼60% of the variation in protein concentration that cannot be explained by measuring mRNAs alone.
To decode the contributions of different regulatory processes, two strategies have proved to be useful thus far. First, from direct measurements of mRNA and protein abundances and mRNA and protein degradation, it is possible to estimate transcription and translation rates (using rate equations) and then to integrate the relative contributions of these different rates in mathematical models of gene expression regula-tion5. Second, using statistical techniques, including regression, it is possible to relate deviations in protein abundance to protein and mRNA sequence features that are characteristic of different modes of regulation (for example, PEST sequences that suggest regulation by protein degradation)24. Such strategies provide estimates of the relative parts played by different regulatory steps.
After the contributions of mRNA abundances (namely, transcription and mRNA degradation) have been factored out, the strongest remaining contributions to the setting of protein abundances appear to come from either translation or protein degradation (FIG. 1) and not from biological or experimental noise. Experimental noise (or measurement errors) can be estimated from replicate experiments and are surprisingly low: for both transcriptomic and proteomic data, replicate measurements of concentrations can correlate with Pearson correlation coefficients >0.95 (REF. 24), although this does not rule out factors such as platform- or macromolecule-specific measurement biases. Experiments in mammalian cells found that variation in protein-expression levels are primarily determined by regulation of translation5, although our own computational analyses also suggest substantial contributions of protein degradation24. Importantly, the analysis accounted for nonlinear relationships and different dynamic ranges of the contributing measures. Both analyses agree that regulation of post-transcription, translation and protein degradation contribute as much to variation in protein concentrations as transcription and transcript degradation do (as for the example in FIG. 2b). Similar results have been obtained from bacteria28. Thus, it has been stated that “transcription regulation is only half the story”34.
These analyses highlight features that correlate with post-transcriptional regulation, such as protein and 3′ untranslated region (UTR) lengths. Parts of these observations can be explained. For example, in yeast cells, there is a strong inverse relationship between protein abundance and coding-sequence length35 that probably derives partially from length-dependent differences in ribosome densities. Using ribosome footprinting, a roughly threefold higher ribosome density was observed for the first ∼30–40 codons following the translation start of mRNAs, followed by reduced ribo-some densities for the remaining codons12. However, the same may not be true for mammalian cells36. Further, there is a trend in cancer cells for highly expressed genes to exhibit shorter 3′UTRs with fewer micro-RNA (miRNA)-binding sites, decreasing miRNA-mediated translation repression37,38. The observation of shorter 3′UTRs for more highly expressed mammalian proteins corresponds well with this.
Conservation of protein abundances
Another recently observed trend is noteworthy. Preliminary evidence suggests that steady-state protein abundances of orthologues are well-conserved across large evolutionary distances: for example, across worms, flies, bacteria, yeast and a human cancer cell line39–41. The observation appears to hold true even when accounting for biases from different technological platforms. Caveats abound with these data, most importantly that the abundance measurements are often compared across platforms and laboratories and that, for the case of data collected from tissues and organisms, averaging of measurements across cell types may bias the observations towards the most abundant proteins. Nonetheless, it is an intriguing observation with an obvious explanation: the steady-state abundances of proteins are determined by their functions and are based on, for example, matching stoichiometries between interacting proteins in the same physical complexes. Conservation of function between orthologues therefore implies conservation of protein abundances.
Perhaps more surprisingly, protein abundances appear to be more evolutionarily conserved than the levels of their corresponding mRNAs are across bacteria, yeast, worms, flies, human cells and plants39,40 (FIG. 2d). The observation holds true even if accounting for the different dynamic ranges of protein and mRNA data — that is, by using a rank-based correlation coefficient. Again, observations are scarce and have associated caveats: for example, proteomics and transcriptomics methods have very different levels of sensitivity and measurement error. Nonetheless, it appears that mRNA levels of conserved genes diverge across time, but post-transcriptional, translational and protein-degradational regulation help to compensate for this drift and bring protein abundances back to evolutionarily preferred levels. These observations are consistent with the large role for post-transcriptional regulation discussed above and, in combination with these data, they suggest the following model.
A model for understanding regulation of protein abundance
Although the expression level of an mRNA only explains a fraction of variation in protein abundance, the abundance of an mRNA is often an excellent proxy for the presence of a protein: that is, for whether or not that protein is detectable within the cells42 (FIG. 2c). We thus propose the following model to explain the observations mentioned above. RNA expression may act in a switch-like fashion: proteins are undetectable (at least by typical mass spectrometry experiments) when mRNA levels are low, but the ability to detect proteins rises sharply at higher mRNA levels. Graphically, this corresponds to a ‘lazy step function’, as is plotted for yeast in FIG. 2c. The same function has also been found in E. coli and humans42. A stochastic switch between ‘on’ and ‘off’ states has been suggested for transcription, resulting in bursts of gene expression occurring from bacteria to humans43,44. Regulation of transcript abundance could be thought of as controlling the on or off state of each gene and setting the order of magnitude of protein abundances. A combination of post-transcriptional, translational and degradative regulation, acting through miRNAs45 or other mechanisms, then fine-tunes protein abundances to their preferred levels, acting both at immediate and evolutionary timescales. Indeed, miRNAs have been found to fine-regulate protein expression levels, rather than to cause large expression changes46,47. In a simple sense, regulation at the level of mRNA thus serves as a switch, whereas regulation downstream functions as a rheostat for further tuning of protein abundances.
Consistent with this model, proteins exhibit a larger dynamic range of concentrations than transcripts do5,21,24,48; such differential signal amplification must occur by post-transcriptional mechanisms. Similarly, across transcriptome data sets from different Metazoa (but not yeast), there is a class of low-expression mRNAs that do not appear to be functional but are rather halted in an off state49. Transcription of these mRNAs can be ‘switched on’ through regulatory factors to express the mRNAs at higher levels, which then also have detectable protein concentrations.
Such a model is, of course, simplistic given that transcription, translation and degradation are often extensively coupled and may frequently regulate each other through feedback loops, as described in an excellent review by Dahan et al.50. Some links are better understood than others: for example, the interaction between the proteasome and chromatin is still largely unclear, as this would link protein degradation regulation to processes that affect chromatin structure and thus affect the efficiency of transcription initiation51,52. For example, in bacteria, co-transcriptional translation offers several mechanisms for coupling transcription and translation, but this is more difficult in eukaryotes. Recent views, however, hypothesize that coupling in eukaryotes can be enabled through proteins that are associated with nascent mRNAs that later regulate translation. Such interdependencies complicate both the model of gene expression and the assessment of the relative contributions of different modes of regulation of protein abundance.
Conclusions
In conclusion, recent studies suggest a perhaps undervalued role for post-transcriptional, translational and degradation regulation in the determination of protein concentrations, contributing at least as much as transcription itself. Future work must almost certainly focus on a deeper understanding of the rates of protein production and turnover, on how these rates change under different cellular conditions and on the principles that govern their regulation. Advances in mass spectrometry provide a clear path towards addressing these issues, especially the ability to survey proteome turnover by pulse–chase experiments on continuously growing cells (for example, as in REF. 5). Recent years have also seen a number of advances in methods that analyse translation efficiency through, for example, the above-mentioned ribosome profiling12. However, we still often do not understand the kinetics of the participating processes, in particular, those of translation. We cannot yet measure the kinetics of translation at the single-cell level, and such experiments would be essential for understanding, for example, translation bursts or the relative contributions of noise in transcription and translation. Considerable work still lies in store to understand the apparent coupling between the different processes that are required to synthesize proteins and to maintain expression levels. Finally, there are still questions that are entirely open regarding the specificity of translation regulation, feedback and coupling between regulatory processes (such as, transcription, translation and degradation), the roles of miRNAs and other translation regulators, such as RNA-binding proteins, and undoubtedly new mechanisms of protein abundance regulation that verify or disprove current observations.
Acknowledgments
The authors acknowledge support from the US National Institutes of Health and the Welch Foundation (F1515, to E.M.M.). We also thank T. Lionnet for helpful discussions.
Glossary
- High-resolution tandem mass spectrometry
The use of two consecutive mass spectrometry steps to measure mass-to-charge ratios for peptides and their fragment ions, respectively. Modern technology enables a mass accuracy of <0.01 Da
- Nanoflow chromatography
In the context of peptides, this method separates a peptide mixture by differences in biophysical properties. It operates at flow rates of nanolitres per minute to increase separation efficiency and decrease sample volumes
- PEST sequences
Protein sequence motif enriched for proline (P), glutamate (E), serine (S) and threonine (T) that serves as a protein degradation signal
- Ribosome footprinting
Identification of ribosomal binding sites on mRNA through ribosome stalling and next-generation sequencing of the bound RNA fragments
- Stable isotopic labelling with amino acids in cell culture
(SILAC). A widely used technique for estimating relative protein concentrations by mass spectrometry
Footnotes
Competing interests statement: The authors declare no competing financial interests.
Further Information: Christine Vogel's homepage: http://www.nyu.edu/projects/vogel, Edward M. Marcotte's homepage: http://www.marcottelab.org/index.php/main_page
Contributor Information
Christine Vogel, Email: cvogel@nyu.edu, Center for Genomics and Systems Biology, New York University, New York 10003, USA.
Edward M. Marcotte, Email: marcotte@icmb.utexas.edu, Center for Systems and Synthetic Biology, Institute for Cellular and Molecular Biology, Department of Chemistry and Biochemistry, University of Texas at Austin, Austin, Texas 78712, USA.
References
- 1.de Sousa Abreu R, Penalva LO, Marcotte E, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu Q, et al. The Orbitrap: a new mass spectrometer. J Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 3.Mallick P, Kuster B. Proteomics: a pragmatic perspective. Nature Biotech. 2010;28:695–709. doi: 10.1038/nbt.1658. [DOI] [PubMed] [Google Scholar]
- 4.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nature Biotech. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 5.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 6.Malmstrom J, et al. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature. 2009;460:762–765. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 9.Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 11.Martin JA, Wang Z. Next-generation transcriptome assembly. Nature Rev Genet. 2011;12:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- 12.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 15.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 16.Belle A, Tanay A, Bitincka L, Shamir R, O'Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci USA. 2006;103:13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eden E, et al. Proteome half-life dynamics in living human cells. Science. 2011;331:764–768. doi: 10.1126/science.1199784. [DOI] [PubMed] [Google Scholar]
- 18.Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 19.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 20.de Godoy LM, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 21.Lundberg E, et al. Defining the transcriptome and proteome in three functionally different human cell lines. Mol Syst Biol. 2010;6:450. doi: 10.1038/msb.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharova LV, et al. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selinger DW, Saxena RM, Cheung KJ, Church GM, Rosenow C. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 2003;13:216–223. doi: 10.1101/gr.912603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogel C, et al. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol. 2010;6:400. doi: 10.1038/msb.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee MV, et al. A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol Syst Biol. 2011;7:514. doi: 10.1038/msb.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fournier ML, et al. Delayed correlation of mRNA and protein expression in rapamycin-treated cells and a role for Ggc1 in cellular sensitivity to rapamycin. Mol Cell Proteomics. 2009;9:271–284. doi: 10.1074/mcp.M900415-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogel C, Silva GM, Marcotte EM. Protein expression regulation under oxidative stress. Mol Cell Proteomics. 2011 Sep 20; doi: 10.1074/mcp.M111.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maier T, et al. Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol Syst Biol. 2011;7:511. doi: 10.1038/msb.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jayapal KP, et al. Uncovering genes with divergent mRNA-protein dynamics in Streptomyces coelicolor. PLoS ONE. 2008;3:e2097. doi: 10.1371/journal.pone.0002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li GW, Xie XS. Central dogma at the single-molecule level in living cells. Nature. 2011;475:308–315. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. Noise minimization in eukaryotic gene expression. PLoS Biol. 2004;2:e137. doi: 10.1371/journal.pbio.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gandhi SJ, Zenklusen D, Lionnet T, Singer RH. Transcription of functionally related constitutive genes is not coordinated. Nature Struct Mol Biol. 2011;18:27–34. doi: 10.1038/nsmb.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pilpel Y. Noise in biological systems: pros, cons, and mechanisms of control. Methods Mol Biol. 2011;759:407–425. doi: 10.1007/978-1-61779-173-4_23. [DOI] [PubMed] [Google Scholar]
- 34.Plotkin JB. Transcriptional regulation is only half the story. Mol Syst Biol. 2010;6:406. doi: 10.1038/msb.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coghlan A, Wolfe KH. Relationship of codon bias to mRNA concentration and protein length in Saccharomyces cerevisiae. Yeast. 2000;16:1131–1145. doi: 10.1002/1097-0061(20000915)16:12<1131::AID-YEA609>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 36.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of Mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schrimpf SP, et al. Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol. 2009;7:e48. doi: 10.1371/journal.pbio.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laurent J, et al. Protein abundances are more conserved than mRNA abundances across diverse taxa. Proteomics. 2010;10:4209–4212. doi: 10.1002/pmic.201000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiss M, Schrimpf S, Hengartner MO, Lercher MJ, von Mering C. Shotgun proteomics data from multiple organisms reveals remarkable quantitative conservation of the eukaryotic core proteome. Proteomics. 2010;10:1297–1306. doi: 10.1002/pmic.200900414. [DOI] [PubMed] [Google Scholar]
- 42.Ramakrishnan SR, et al. Integrating shotgun proteomics and mRNA expression data to improve protein identification. Bioinformatics. 2009;25:1397–1403. doi: 10.1093/bioinformatics/btp168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 44.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mukherji S, et al. MicroRNAs can generate thresholds in target gene expression. Nature Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 48.Jacobs JM, et al. Utilizing human blood plasma for proteomic biomarker discovery. J Proteome Res. 2005;4:1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- 49.Hebenstreit D, et al. RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol Syst Biol. 2011;7:497. doi: 10.1038/msb.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dahan O, Gingold H, Pilpel Y. Regulatory mechanisms and networks couple the different phases of gene expression. Trends Genet. 2011;27:316–322. doi: 10.1016/j.tig.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 51.Auld KL, Silver PA. Transcriptional regulation by the proteasome as a mechanism for cellular protein homeostasis. Cell Cycle. 2006;5:1503–1505. doi: 10.4161/cc.5.14.2979. [DOI] [PubMed] [Google Scholar]
- 52.Kodadek T. No splicing, no dicing: non-proteolytic roles of the ubiquitin-proteasome system in transcription. J Biol Chem. 2010;285:2221–2226. doi: 10.1074/jbc.R109.077883. [DOI] [PMC free article] [PubMed] [Google Scholar]