Abstract
Animal and plant cytokineses appear morphologically distinct. Recent studies, however, have revealed that these cellular processes have many things in common, including the requirement of co-ordinated membrane trafficking and cytoskeletal dynamics. At the intersection of these two processes are the members of the dynamin family of ubiquitous eukaryotic GTPases. In this review, we highlight the conserved contribution of classical dynamin and dynamin-related proteins during cytokinesis in both animal and plant systems.
Keywords: cell division, cell plate, cytokinesis, dynamin, mitosis, membrane trafficking
Cytokinesis is the final stage of the cell cycle in which a single cell is physically separated into individual daughter cells. In eukaryotes, this process leads to the division and partitioning of chromatin, organelles, cytoplasmic components and the construction of new membrane between daughter cells. Animal cells divide by constricting plasma membrane and targeting membrane along the newly formed cleavage furrow. The cleavage furrow compacts and bundles the microtubules (MTs) found in the spindle midzone into a structure called the midbody (1). On the other hand, cells of higher plants divide by constructing a unique cytoskeletal apparatus, the phragmoplast, across the division plane and targeting new plasma membrane and cell wall components to the center of the cell creating an intermediate membranous network called the cell plate (2). It has long been thought that the mechanisms of cytokinesis in animals and plants were quite different. Recent work, however, has uncovered highly conserved roles for several membrane trafficking and cytoskeletal-associated proteins in these seemingly different modes of cytokinesis (1). Dynamin and dynamin-related proteins have been shown to play essential roles in cell division in plants (3), animals (4,5) and the protist, Dictyostelium discoideum (6). The phylogenetic relationship of Dictyostelium to plant and animal species (7) likely suggests that there is an ancestral role for dynamin in cytokinesis. In this review, we highlight several studies that suggest that dynamin and dynamin-related proteins (DRPs) have conserved functions in a variety of cell division events.
Animal Cytokinesis
Animal cells require the orchestration of both the cytoskeletal and membrane trafficking machinery to complete cytokinesis (8,9). Animal cells rely on the mitotic spindle to specify the position of the cleavage plane (10). The mitotic spindle contains two populations of MTs, asters and the spindle midzone, both of which have been implicated in specifying the position of the cleavage furrow (11,12). As animal cells enter into mitosis, various processes lead to the disassembly of the Golgi (16) and the release of Golgi associated proteins, including myosin II and Cdc42. Once the cleavage furrow is specified, the actin-based contractile ring assembles on the inner surface of the plasma membrane. Ring assembly is mediated by the formins and profilins that act to initiate actin filament formation (13), whereas Cdc42, a highly conserved Rho-type small GTPase, is involved in actin ring organization (14). Non-muscle myosin II drives the constriction of the contractile ring and subsequent ingression of the plasma membrane results in the formation of a cleavage furrow (15).
Membrane trafficking pathways regulate the addition of new membrane along the ingressing cleavage furrow (8), which culminates with the compression of the spindle midzone into a protein-rich structure called the midbody. Using FM1-43, a fluorescent, styryl dye, local membrane accumulation at the late furrow apices has been observed in both Caenorhabditis elegans and Xenopus embryos (17,18). Membrane trafficking to the cleavage furrow is sensitive to Brefeldin A (18,19) and is likely mediated by MTs found in the midzone or along the furrow cortex (17). These data suggest that the local accumulation of membrane is required to separate daughter cells. Endocytosis and membrane recycling are also crucial to cytokinesis, as drugs that inhibit endocytosis, such as chlorpromazine or methyl-beta-cyclodextrin (20), block daughter cell separation. Proteins such as clathrin, syntaxin, endobrevin and dynamin II/DYN-1 are also thought to play an integral role in furrow-specific endocytosis and fusion events during daughter cell separation (8,21). Golgi and Endoplasmic Reticulum (ER) membranes also concentrate along the spindle midzone MTs and within the midbody during late telophase (4,22,23), suggesting that the interaction of these organelles and associated proteins may play a significant role during cytokinesis.
Plant Cytokinesis
Plants also co-ordinate cytoskeletal and membrane traf-ficking machinery to initiate and complete cytokinesis. Cytokinesis in pollen, somatic cells and endosperm syncytia has been visualized in detail and appears morphologically similar in many respects (24–27). During late anaphase in somatic cells, the phragmoplast, a plant-specific cytoskeletal array, composed of interdigitating MTs and actin microfilaments, is formed from the remnants of the spindle. MTs within the barrel-like phragmoplast are organized with their plus ends oriented toward the division plane. The cell plate is presumed to form from Golgi-derived vesicles carrying membrane, protein and cell wall precursors that are transported along MTs to the division plane where they fuse (26). Multiple rounds of vesicle fusion occur until a tubular-vesicular network (TVN) is formed. The polysaccharide 1,3-β-glucan (callose) is synthesized and deposited in the lumen of the tubules at the end of this stage by callose synthases, which are delivered by Golgi-derived vesicles (28). Spreading of the callose deposits is thought to increase the volume of the TVN (24,26), causing it to flatten into a more planar struc-ture (26). The TVN continues to grow outward as the central region of the cell plate matures into a tubular network (TN) and then a fenestrated sheet (FS) as more membrane and callose are deposited at the cell plate. Callose synthesis decreases and the synthesis of the major cell wall polysaccharide 1,4-β-glucan (cellulose) increases (26), as the TVN–TN–FS maturation continues outward toward the parental plasma membrane. The completed membrane system then fuses with the parental plasma membrane separating the two daughter cells. A few components of the molecular fusion machinery required for cytokinesis in plants have been identified. Genetic and biochemical studies have demonstrated that KNOLLE, a syntaxin (29), KUELE, a Sec1 protein (30), SNAP33, a t-SNARE (31), and NSPNII, a v-SNARE (32) interact to promote cell plate vesicle and tubule fusion. KNOLLE, SNAP33 and NSPNII have been localized to the division plane (31–33), and mutant plants deficient in KNOLLE and SNAP33 have characteristic cytokinetic defects with cell wall stubs and large multinucleate cells (29–31,34).
Actin dynamics also plays a significant role in plant cytokinesis (1). Microinjection of profilin causes a delay or an abortion of cytokinesis (35), and an Arabidopsis mutant lacking the formin FH5 has cellularization defects in the endosperm (36). It is thought that an actin–myosin-dependent process directs the growing cell plate to the cortical division site on the parental plasma membrane (37).
Similar to the process of vesicle trafficking to the plasma membrane, bi-directional membrane trafficking processes must be functional for cell plate formation to progress. It has been estimated that 70% of the membrane that is transported to the growing cell plate is removed during the process of cytokinesis (24). Morphological evidence suggests that the excess membrane is recycled by clathrin-dependent endocytosis (27). In tobacco-cultured cells and Arabidopsis meristematic cells, clathrin-coated structures and multivesicular bodies were seen during the TVN and FS stages, respectively (26,27). However, the process of membrane recycling during earlier stages of cell plate formation remains to be defined.
The Dynamin Superfamily
The dynamin superfamily, which is composed of conventional dynamin and DRPs, is conserved throughout eukaryotes. Dynamin I was originally identified as MT-binding protein (38), and since then, a number of dynamin and DRPs have been implicated in various processes such as endocytosis, actin nucleation and dynamics, mitochondrial and chloroplast biogenesis and cytokinesis (39). Despite the participation of dynamin in such broad activities, in vitro studies have characterized the ability of dynamin to bind directly to lipids, oligomerize into spiral structures around lipid bilayers, modulate lipid bilayers into narrow tubules and fragment the tubules in a GTPase-dependent manner (40). The domain structure of all dynamins consists of an N-terminal GTPase domain, a conserved middle domain and a C-terminal GTPase effector domain (GED) (Figure 1). The interaction of the three conserved domains allows for the oligomerization of dynamin (41). Subsequent GTP hydrolysis causes a conformational change of the dynamin rings and spirals (42). Most dynamins contain additional domains, which may account for the great diversity of cellular activities (43). For instance, mammalian dynamins I, II and III contain a plexstrin homology (PH) domain, which allows for phospholipid binding and a proline-rich domain (PRD) that interacts with the SRC-Homology-3 (SH3) domains of various proteins (43).
Figure 1. Domain structure of cytokinesis-associated dynamins.
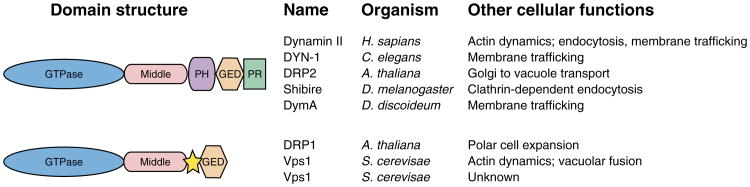
Listed are dynamins that have putative roles during cytokinesis along with their domain structure and other cellular functions. All dynamins contain an N-terminal GTPase domain followed by a conserved middle region and a C-terminal GTPase effector domain (GED). The region between the middle and GED domains has low homology within the superfamily, ranging from a large defined plexstrin homology (PH) domain in bona fide dynamins to a 25–50 undefined amino acid stretch in DRP1 and Vps1 (yellow star). Domains are represented by different shapes and colors. PR = proline rich. See References in text.
Plants have homologs to most dynamin and DRPs (Figure 1) found in animals [see (44) for dynamin nomenclature in plants]. Arabidopsis DRP2A and DRP2B are most similar in domain structure to dynamins I and II from mammals and are involved in vesicular trafficking from the Golgi to the vacuole (45). Plants also have a specific dynamin subfamily (DRP1). Arabidopsis has five genes that encode DRP1 isoforms A–E, three of which appear to have roles in cytokinesis and polar cell growth (3,46). Budding yeast has three DRPs, of which one, Vps1p, functions in actin dynamics and vacuolar fusion (47,48). It is notknown whether Vps1p participates in cytokinesis; however, with its dual role in actin and membrane dynamics, a function in cytokinesis is likely.
Cellular Roles of Animal Dynamin
Membrane trafficking and endocytosis
Dynamin's best-defined role is its involvement in clathrin-mediated endocytosis in mammalian cells (49–51). Dynamin was first identified as having a role in endocytosis when it was shown to be the mammalian homolog of the Shibire protein in Drosophila (52). Initial analysis indicated that a temperature shift in shibire-ts animals gave rise to a rapid and reversible paralysis and accumulation of clathrin-coated pits at the plasma membrane (52–54). The necks of the coated pits were decorated with electrondense collars composed primarily of dynamin. Subsequently, purified dynamin was shown to self-assemble into rings and spirals in low ionic strength buffers (55). The self-assembly of dynamin around the necks of coated pits and other lipid bilayers also stimulates its GTPase activity promoting its function as a mechanoenzyme during the late stages of scission (56–59). Besides its membrane fission activity, dynamin also promotes membrane remodeling through a direct interaction with acidic phospholipids such as PI(4,5)P2 and the lipid bilayer via the PH domain. Interaction with acidic phospholipids also directly stimulates GTPase activity (59,60).
Over the last decade and a half, dynamin has been implicated in numerous membrane trafficking events including phagocytosis (61), trafficking to/from late endosomes and trans-Golgi membranes (62–65) and endocytic mediated pathogen entry into human cells (66). Despite this large amount of work, very little is known about the in vivo function of dynamin during mitosis. In addition, it remains to be determined whether dynamin-dependent endocytosis during cytokinesis uses well-characterized mechanisms such as cytoskeleton clathrin-mediated endocytosis or whether undiscovered processes facilitate the rapid membrane recycling that is needed for cleavage furrow ingression. It is attractive to speculate that dynamin might participate in multiple membrane trafficking events during mitosis, either along the cleavage furrow or during late cytokinesis events. It is also likely that dynamin's function in mitosis is dependent on its interaction with multiple cytoskeletal structures.
Actin nucleation
Dynamin plays a significant role in regulating actin assembly and organization via an interaction between F-actin-containing structures and the PRD domain (67). Dynamin, for example, can regulate actin reorganization via its interaction with cortactin and the Arp2/3 complex (68). Dynamin also interacts directly with two scaffolding proteins Intersectin-1 and Tuba that have been proposed to link dynamin with the actin cytoskeleton (69,70). Both Intersectin-1 and Tuba contain specific domains that catalyze Cdc42 guanine nucleotide exchange. Cdc42 has been implicated in a number of biological processes such as cytokinesis, cell polarity, actin polymerization and endocytosis (71). Most notably, in Xenopus embryos, inhibition or constitutive activation of the GTPase activity of Cdc42 inhibited furrow ingression (72). Lastly, profilins are small actin-binding proteins that regulate actin assembly and dynamics during a variety of cellular processes specifically cell motility, membrane trafficking and cytokinesis (73,74). In mouse brain extracts, profilin I and profilin II bound to a number of proteins required for actin assembly and endocytosis, including dynamin I, clathrin and synapsin (75). Interestingly, profilins were shown to co-localize with dynamin I and synapsin in axonal and dendritic processes (75). The Drosophila profilin homolog chickadee and the C. elegans homolog pfn-1 are required for the stable maintenance of myosin II at the cleavage furrow, and protein depletion results in multinucleated cells (76,77).
Actin is one of the major constituents of the contractile ring in dividing animal cells, yet little is known about how this structure is established. Given recent data, one possibility is that dynamin regulates furrow-specific actin polymerization via interaction with profilin, whereas another possibility is that dynamin interacts with Intersectin-1 and Tuba to regulate Cdc42-specific GTPase activity and actin polymerization associated with the cell cycle. The dynamin –cortactin–Arp2/3 complex may also play a crucial role in remodeling cortical microfilaments outside the cleavage furrow. Future work will shed light on these interesting possibilities.
MT dynamics
Dynamin was originally identified as an MT-binding protein (38); however, there is very little evidence that dynamin and MTs interact in vivo(78). Recent proteomic analyses of mammalian midbodies show significant enrichment for dynamin II, whereas electron microscopy experiments localize dynamin II along MTs in the spindle midzone and midbody (4,5), suggesting that the dynamin and tubulin interaction might be significant. In rat hepatocytes, distinct labeling of dynamin is observed during metaphase along the mitotic spindle (Figure 2, blue) (5). Localization is enhanced at the spindle midzone during the transition from early to late anaphase. At the end of telophase, dynamin II localizes into two distinct areas within the intercellular bridge on each side of the midbody matrix (5). Experiments using glutathione S-transferase-tagged dynamin II identified a protein complex containing actin, alpha-tubulin and gamma-tubulin from rat liver homo-genate. Further analysis revealed that gamma-tubulin and dynamin II interact directly (79). Dynamin II localized to the pericentriolar material and centrioles (Figure 2, blue), and the reduction of dynamin II by siRNAi displayed defects in centrosome splitting, suggesting a novel role for dynamin II in centrosome cohesion (79). Dynamin is likely to be involved in the regulation of MT dynamics during cell division, but the mechanisms have yet to be elucidated. One possibility is that dynamin is required to stabilize and prevent the collapse of the spindle midzone by bundling MTs. Another possibility is that dynamin mediates and stabilizes interactions between MTs, actin and the plasma membrane at the end of telophase. Dynamin is also likely to participate in membrane fission and fusion events that facilitate the separation of the two daughter cells. The ability to interact with the cytoskeleton and the plasma membrane during the late stages of cytokinesis bestows dynamin with the potential to mediate signals to and from the cell surface to and from the spindle.
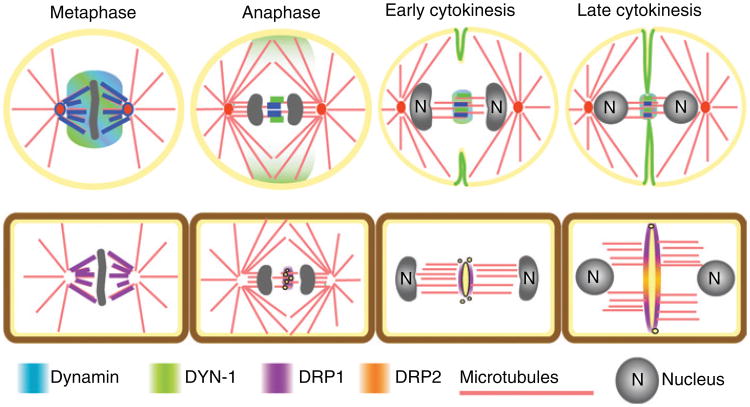
Figure 2. Localization of dynamin and dynamin-related proteins (DRPs) during mitosis and cytokinesis.
Dynamin and DRPs have functional roles throughout the cell cycle but relocalize during mitosis and cytokinesis. In each stage above, the localization of dynamin is presented. In the case of animal cells, the localization was demonstrated by immunofluorescence microscopy (5). The localization of dynamin and DYN-1 differs. Both are found on microtubules, around the chromatin at metaphase and in the midbody in late cytokinesis. However, only DYN-1 is on the ingressing furrow and at the equitorial cortex. In plant cells, DRP1A–DRP1C were found at the cell plate by immunofluorescence and live-cell fluorescence microscopy (3,46,82,84,88) and DRP1A-positive rings were imaged with high-resolution electron tomography (24). DRP2 was localized using live-cell fluorescence microscopy (84).
Dynamin Is Required for Cytokinesis in Animal Cells
Dynamin localizes to cleavage furrow membranes and is required for cytokinesis in a number of organisms (5,6,20,80). The Drosophila dynamin homolog, Shibire, localizes to the sites of membrane invagination while mutations in the GTPase domain disrupt cellularization, an alternate form of cytokinesis in which the cleavage furrow does not complete (80). Like its Drosophila counterpart, the C. elegans dynamin homolog, dyn-1-ts, was isolated in a mutagenesis screen designed to identify genes required for synapse function (81). When shifted to the restrictive temperature, animals displayed a rapid and reversible locomotion defect as well as increased embryonic lethality and sterility (81). Analysis of the C. elegans, dyn-1-ts embryos revealed late cytokinesis failures in which the furrow ingressed but failed to complete cell division. The dyn-1-ts mutants also displayed cellularization defects in the hermaphrodite germline similar to defects observed in Drosophila embryos (5,80). DYN-1 localization is similar to mammalian dynamin ll both during metaphase and at the midbody prior to cell separation (Figure 2, green and blue). Interestingly, DYN-1 localization shifts in anaphase from the metaphase plate to the equatorial cortex and then specifically to the areas of new membrane formation along the cleavage furrow (Figure 2, green) (5). The work presented in both Drosophila and C. elegans embryos suggests that dynamin is involved in membrane trafficking during both phases of cytokinesis. How dynamin regulates these events is unclear, but future work will likely uncover a significant role for dynamin during furrow ingression and completion.
DRPs Are Required for Plant Cytokinesis
The soybean DRP1 homolog, phragmoplastin, was the first dynamin shown to be involved in cytokinesis in any organism (82,83). Subsequent work in the model plant Arabidopsis revealed two subfamilies of dynamins that localize to the division plane during cytokinesis. DRP2A, which is most closely related to mammalian dynamin I (Figure 1), localizes to the cell plate when expressed in dividing tobacco suspension-cultured cells (Figure 2, orange) (84). Of the two isoforms of DRP2, DRP2A has been studied more extensively. DRP2A is thought to be involved in clathrin-dependent (85) trafficking of vesicles between the trans-Golgi network and the vacuole (45). Although DRP2A does not seem to be required for trafficking vesicles to the plasma membrane (45), this does not rule out a possible role in cell plate membrane trafficking. One possible role of DRP2 at the cell plate is recycling of membrane via clathrin-dependent endocytosis during the formation of the TVN, TN and FS.
The function of the plant-specific DRP1 subfamily in cytokinesis has been studied in more detail than DRP2. Members of the DRP1 subfamily are very early markers of the cell plate, appearing at the division plane prior to TVN establishment (Figure 2, violet) (3,84). Live-cell imaging of DRP1A-GFP in roots and leaves indicates that DRP1A localizes to the cell plate before nuclear reassembly (3). In tobacco suspension-cultured cells, GFP-tagged DRP1A and DRP1C also localized to the spindle and phragmoplast (Figure 2, violet) (84); however, localization to these structures has not been observed in other cell types by immunofluorescence microscopy and in cells expressing DRP1-GFP under native conditions (3,46). As the cell plate grows toward the parental plasma membrane, soybean DRP1 and GFP-tagged DRP1A, DRP1C and DRP1E are most strongly associated with the leading edge of the structure, but the proteins also remain associated, albeit at lower levels, with the mature regions of the cell plate (Figure 2, violet) (3,83,84). These localization studies suggest that DRP1s function at several stages of cytokinesis, including the early phases of cell plate formation.
The presence of cell plate-associated dynamin-like proteins has also been observed using high-resolution electron tomography and immunoelectron microscopy (24,27). In syncytial-type cell plates, DRP1A-positive rings are present at sites of tubule constriction where fused vesicles and tubules form dumbbell shapes (24). The tubules in the TVN are initially 50 nm in diameter and, once constricted, are only 20 nm in diameter (24). If DRP1A is involved in generating the constrictions, then DRP1A may not be operating via the canonical dynamin action cycle in which dynamin I severs the tubules (58,86). The rings and spirals are likely transient structures, as there are numerous approximately 20 nm constrictions without the presence of the DRP1A rings (24). Dynamin-like rings and spirals also appear in somatic cell plates at sites of constriction of the fused vesicles before the TVN is formed and around tubules before the FS develops (27). Dynamin-like rings were also seen in association with clathrin-coated structures in the TN and FS. It remains to be determined whether the DRP1 and/or DRP2 proteins are components of these ring-like structures potentially assisting in early cell plate membrane fusion.
In addition to the microscopic and biochemical approaches, genetic studies have provided the most definitive role for DRP1s in plant cytokinesis. drp1a/drp1e double-mutant embryos display defects in cell expansion and cytokinesis and fail to complete embryogenesis (3). The drp1a/drp1e embryos can undergo several rounds of cell division before cytokinetic defects are manifested, which may be due to the presence of another functionally redundant DRP1 or maternal/paternal pools of DRP1 pro-teins. In the cytokinetic defective cells, cell walls are only partially formed, resulting in cell wall stubs (3). This cytokinetic defective phenotype is very similar to that of knolle and kuele mutants which affect cell plate membrane fusion (29,30). In contrast to drp1A/drp1E double mutants, drp1A plants do not show defects in cytokinesis but have polar expansion defects only in select cell types (3). As both plant cytokinesis and anisotropic cell expansion require highly polarized membrane trafficking (87), DRP1A may have a more general role in polar membrane trafficking as well as a specialized role in the cell plate formation and maturation.
Less is known about the roles of the DRP1 isoforms DRP1C and DRP1E during cell plate formation and whether they have similar functions as DRP1A. drp1C mutants display defects in pollen maturation; however, there are no defects in pollen cytokinesis (88). Likewise, drp1E single-mutant plants do not have cytokinetic defects (3). Each DRP1 has a highly variable region between the middle and GED domains, the site of the lipid-interacting PH domain in dynamin I (Figure 1). DRP1A, DRP1C and DRP1E have been shown to interact with soybean DRP1 in a yeast two-hybrid assay. However, as only DRP1A has been verified as a component of the rings and spirals around syncytial-type cell plate tubules (24), it is not known whether the DRP1s can form homoor hetero-rings and spirals in vivo.
Roles of DRPs in plant cytokinesis?
Members of the DRP1 and DRP2 protein subfamilies appear to function throughout the process of cell plate biogenesis; however, their exact biochemical roles remain to be determined. Based on their similarity to other dynamins, DRP1 and DRP2 could function in the formation of Golgi-derived vesicles and/or recycling of the membrane from the nascent cell plate. DRP1s may also be involved in the construction of the intermediate, callose-containing cell wall. Soybean DRP1 was shown to directly interact with components of the cell plate-specific callose– synthase machinery by yeast two-hybrid and in vitro affinity chromatography assays (89). It is hypothesized that callose–synthase complexes are recruited by DRP1 to the cell plate and that the deposition of callose within the initial cell plate tubules leads to the flattening and consolidation of the TVN. In contrast to somatic and syncytial endosperm cytokinesis, callose is not present in the postmeiotic cell plate of developing pollen grains prior to fusion with the parental plasma membrane, and it is interesting that dynamin-like rings or spirals appear to be absent during the formation of this type of cell plate (25).
Another potential function for DRP1 and DRP2 during cytokinesis is that they play a role in the formation and stabilization of dumbbell-shaped intermediates that have been observed on the initial fusion of cell plate vesicles. Dynamin rings have been postulated to constrict the fusing vesicles into tubules (24,27), thereby reducing the enclosed volume that would promote dehydration and gelling of lumenal cell wall polysaccharides (27). The mechanochemical activity of cell plate-associated DRPs could also facilitate the concentration of fusion factors at the ends of the tubules and the formation of the TVN (27).
Interestingly, recent data have revealed that the yeast DRP, Vps1p, has a role in vacuolar fusion in yeast. Vps1p physically interacts with the t-SNARE, Vam3p, on vacuolar membranes (47). This interaction has been suggested to concentrate SNAREs and other membrane fusion components, creating membrane fusion ‘hot spots'. Dynamins could play a similar role during plant cytokinesis in regulating membrane fusions at distinct regions necessary for the formation of the mature cell plate. DRP oligomers may constrict membrane tubules concentrating KNOLLE and other cell plate membrane fusion components at the end of the tubules, whereas another DRP could bind directly to them, limiting their mobility in the membrane. The interaction between DRPs and any of the cell plate membrane fusion machinery remains to be tested.
The interaction of plant dynamins with MTs and actin microfilaments also remains to be fully characterized. DRP2 shares a similar domain structure to mammalian dynamin II (Figure 1), which interacts with actin-binding proteins, suggesting that DRP2 could also modulate the actin cytoskeleton during cytokinesis. In contrast to DRP2, DRP1 isoforms and Vps1p lack the C-terminal proline-rich protein–protein interaction domain of the classical animal dynamins, which is required for cytoskeletal-associated protein interactions (90, see Figure 1). Vps1p has also been shown to help organize the actin cytoskeleton through interaction with Sla1p, an actin-regulatory protein (48). The Sla1-interacting region of Vps1p was mapped to a 50-amino acid region upstream of the GED domain (48), which shares little amino acid homology with the corresponding region of the DRP1 isoforms (Figure 1, star). Interestingly, the localization of DRP1A is altered on treatment of roots with the membrane trafficking inhibitor Brefeldin A and actin cytoskeletal inhibitors cytochalasin D and latrunculin B (CAK and SYB, unpublished results). Thus, DRP1A may interact with actin during polarized expansion and/or cytokinesis.
Conclusions
The essential function for dynamin in a variety of cellular processes that involve both membrane trafficking and actin dynamics is becoming more apparent. These events may require dynamin to oligomerize around lipid bilayers altering membrane morphology or to activate downstream effectors through GTP binding and hydrolysis. These dual functions place dynamin at key positions in cytokinesis where actin dynamics and membrane trafficking are necessary at multiple steps in both plant and animal systems. Membrane addition and actin dynamics are required to initiate and constrict the ingressing cleavage furrow and to align and drive cell plate expansion. In addition, the possible MT-bundling properties of dynamin may play a role at the midbody in animal cells and on phragmoplast MTs in plant cells.
The number of known dynamin-interacting proteins has risen in recent years, as the roles of dynamin and DRPs have been elucidated. This number will most likely grow, as more is known about the roles dynamins play in cytokinesis. In turn, the dynamin-interacting proteins will give us clues as to how dynamin is functioning in cytokinesis: a scaffold for cell wall synthases, a tubulator of cell plate intermediates, a nucleator of actin, a bundler for MTs and/or a recycler of excess membrane at the division plane. The regulatory network of dynamin function in each of these molecular events remains to be determined. There is much to learn about the protein interactions and mechanisms that promote cytokinesis and separate newly formed daughter cells. By elucidating the function of key players in cytokinesis, dynamin and DRPs, we will determine the interplay and relationships between the fundamental events that occur during cytokinesis in higher eukaryotes.
Acknowledgments
We apologize to many authors for the omission of their references owing to restrictions in the length of the review and reference numbers. Research in the authors' laboratories is supported by the Department of Genetics and the University of Wisconsin-Madison Medical School for ARS and grants from the Department of Energy, Division of Energy Biosciences (DE-FG0299ER20332) and United States Department of Agriculture national research initiative competitive grants program (2004-03411) to SYB. CAK is a Howard Hughes Medical Institute Predoctoral Fellow.
References
- 1.Otegui MS, Verbrugghe KJ, Skop AR. Midbodies and phragmoplasts: analogous structures involved in cytokinesis. Trends Cell Biol. 2005;15:404–413. doi: 10.1016/j.tcb.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Staehelin LA, Hepler PK. Cytokinesis in higher plants. Cell. 1996;84:821–824. doi: 10.1016/s0092-8674(00)81060-0. [DOI] [PubMed] [Google Scholar]
- 3.Kang BH, Busse JS, Bednarek SY. Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell. 2003;15:899–913. doi: 10.1105/tpc.009670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skop AR, Liu H, Yates J, III, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 2004;305:61–66. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson HM, Skop AR, Euteneuer U, Meyer BJ, McNiven MA. The large GTPase dynamin associates with the spindle midzone and is required for cytokinesis. Curr Biol. 2002;12:2111–2117. doi: 10.1016/s0960-9822(02)01390-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wienke DC, Knetsch ML, Neuhaus EM, Reedy MC, Manstein DJ. Disruption of a dynamin homologue affects endocytosis, organelle morphology, and cytokinesis in Dictyostelium discoideum. Mol Biol Cell. 1999;10:225–243. doi: 10.1091/mbc.10.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, Tunggal B, Kummerfeld S, Madera M, Konfortov BA, Rivero F, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albertson R, Riggs B, Sullivan W. Membrane traffic: a driving force in cytokinesis. Trends Cell Biol. 2005;15:92–101. doi: 10.1016/j.tcb.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 10.Rappaport R. Cleavage furrow establishment by the moving mitotic apparatus. Dev Growth Differ. 1997;39:221–226. doi: 10.1046/j.1440-169x.1997.t01-1-00010.x. [DOI] [PubMed] [Google Scholar]
- 11.Bringmann H, Hyman AA. A cytokinesis furrow is positioned by two consecutive signals. Nature. 2005;436:731–734. doi: 10.1038/nature03823. [DOI] [PubMed] [Google Scholar]
- 12.Glotzer M. Cleavage furrow positioning. J Cell Biol. 2004;164:347–351. doi: 10.1083/jcb.200310112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol. 2003;161:875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joberty G, Perlungher RR, Sheffield PJ, Kinoshita M, Noda M, Haystead T, Macara IG. Borg proteins control septin organization and are negatively regulated by Cdc42. Nat Cell Biol. 2001;3:861–866. doi: 10.1038/ncb1001-861. [DOI] [PubMed] [Google Scholar]
- 15.Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 16.Diao A, Lowe M. Cell biology. The Golgi goes fission. Science. 2004;305:48–49. doi: 10.1126/science.1100153. [DOI] [PubMed] [Google Scholar]
- 17.Danilchik MV, Bedrick SD, Brown EE, Ray K. Furrow microtubules and localized exocytosis in cleaving Xenopus laevis embryos. J Cell Sci. 2003;116:273–283. doi: 10.1242/jcs.00217. [DOI] [PubMed] [Google Scholar]
- 18.Skop AR, Bergmann D, Mohler WA, White JG. Completion of cytokinesis in C. elegans requires a brefeldin A-sensitive membrane accumulation at the cleavage furrow apex. Curr Biol. 2001;11:735–746. doi: 10.1016/s0960-9822(01)00231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sisson JC, Field C, Ventura R, Royou A, Sullivan W. Lava lamp, a novel peripheral golgi protein, is required for Drosophila melanogaster cellularization. J Cell Biol. 2000;151:905–918. doi: 10.1083/jcb.151.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng B, Schwarz H, Jesuthasan S. Furrow-specific endocytosis during cytokinesis of zebrafish blastomeres. Exp Cell Res. 2002;279:14–20. doi: 10.1006/excr.2002.5579. [DOI] [PubMed] [Google Scholar]
- 21.Joo E, Tsang CW, Trimble WS. Septins: traffic control at the cytokinesis intersection. Traffic. 2005;6:626–634. doi: 10.1111/j.1600-0854.2005.00305.x. [DOI] [PubMed] [Google Scholar]
- 22.Mullins JM, Biesele JJ. Terminal phase of cytokinesis in D-98s cells. J Cell Biol. 1977;73:672–684. doi: 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poteryaev D, Squirrell JM, Campbell JM, White JG, Spang A. Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Mol Biol Cell. 2005;16:2139–2153. doi: 10.1091/mbc.E04-08-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otegui MS, Mastronarde DN, Kang BH, Bednarek SY, Staehelin LA. Three-dimensional analysis of syncytial-type cell plates during endosperm cellularization visualized by high resolution electron tomography. Plant Cell. 2001;13:2033–2051. doi: 10.1105/TPC.010150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otegui MS, Staehelin LA. Electron tomographic analysis of postmeiotic cytokinesis during pollen development in Arabidopsis thaliana. Planta. 2004;218:501–515. doi: 10.1007/s00425-003-1125-1. [DOI] [PubMed] [Google Scholar]
- 26.Samuels AL, Giddings TH, Staehelin LA. Cytokinesis in tobacco BY-2 and root tip cells: a new model of cell plate formation in higher plants. J Cell Biol. 1995;130:1345–1357. doi: 10.1083/jcb.130.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segui-Simarro JM, Austin JR, II, White EA, Staehelin LA. Electron tomographic analysis of somatic cell plate formation in meristematic cells of Arabidopsis preserved by high-pressure freezing. Plant Cell. 2004;16:836–856. doi: 10.1105/tpc.017749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verma DPS. Cytokinesis and building of the cell plate. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:571–584. doi: 10.1146/annurev.arplant.52.1.751. [DOI] [PubMed] [Google Scholar]
- 29.Lukowitz W, Mayer U, Jurgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;84:61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- 30.Assaad FF, Huet Y, Mayer U, Jurgens G. The cytokinesis gene KEULE encodes a Sec1 protein that binds the syntaxin KNOLLE. J Cell Biol. 2001;152:531–543. doi: 10.1083/jcb.152.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heese M, Gansel X, Sticher L, Wick P, Grebe M, Granier F, Jurgens G. Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33 and its role in plant cytokinesis. J Cell Biol. 2001;155:239–249. doi: 10.1083/jcb.200107126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng H, Bednarek SY, Sanderfoot AA, Alonso J, Ecker JR, Raikhel NV. NPSN11 is a cell plate-associated SNARE protein that interacts with the syntaxin KNOLLE. Plant Physiol. 2002;129:530–539. doi: 10.1104/pp.003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jurgens G. The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol. 1997;139:1485–1493. doi: 10.1083/jcb.139.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waizenegger I, Lukowitz W, Assaad F, Schwarz H, Jurgens G, Mayer U. The Arabidopsis KNOLLE and KEULE genes interact to promote vesicle fusion during cytokinesis. Curr Biol. 2000;10:1371–1374. doi: 10.1016/s0960-9822(00)00775-2. [DOI] [PubMed] [Google Scholar]
- 35.Valster AH, Pierson ES, Valenta R, Hepler PK, Emons A. Probing the plant actin cytoskeleton during Cytokinesis and interphase by profilin microinjection. Plant Cell. 1997;9:1815–1824. doi: 10.1105/tpc.9.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingouff M, Fitz Gerald JN, Guerin C, Robert H, Sorensen MB, Van Damme D, Geelen D, Blanchoin L, Berger F. Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat Cell Biol. 2005;7:374–380. doi: 10.1038/ncb1238. [DOI] [PubMed] [Google Scholar]
- 37.Molchan TM, Valster AH, Hepler PK. Actomyosin promotes cell plate alignment and late lateral expansion in Tradescantia stamen hair cells. Planta. 2002;214:683–693. doi: 10.1007/s004250100672. [DOI] [PubMed] [Google Scholar]
- 38.Shpetner HS, Vallee RB. Identification of dynamin, a novel mechanochemical enzyme that mediates interactions between microtubules. Cell. 1989;59:421–432. doi: 10.1016/0092-8674(89)90027-5. [DOI] [PubMed] [Google Scholar]
- 39.Praefcke GJ, McMahon HT. The dynamin superfamily. universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 40.Takei K, Yoshida Y, Yamada H. Regulatory mechanisms of dynamin-dependent endocytosis. J Biochem (Tokyo) 2005;137:243–247. doi: 10.1093/jb/mvi052. [DOI] [PubMed] [Google Scholar]
- 41.Muhlberg AB, Warnock DE, Schmid SL. Domain structure and intramolecular regulation of dynamin GTPase. EMBO J. 1997;16:6676–6683. doi: 10.1093/emboj/16.22.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang P, Hinshaw JE. Three-dimensional reconstruction of dynamin in the constricted state. Nat Cell Biol. 2001;3:922–926. doi: 10.1038/ncb1001-922. [DOI] [PubMed] [Google Scholar]
- 43.Hinshaw JE. Dynamin and its role in membrane fission. Annu Rev Cell Dev Biol. 2000;16:483–519. doi: 10.1146/annurev.cellbio.16.1.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong Z, Bednarek SY, Blumwald E, Hwang I, Jurgens G, Menzel D, Osteryoung KW, Raikhel NV, Shinozaki K, Tsutsumi N, Verma DP. A unified nomenclature for Arabidopsis dynamin-related large GTPases based on homology and possible functions. Plant Mol Biol. 2003;53:261–265. doi: 10.1023/b:plan.0000007000.29697.81. [DOI] [PubMed] [Google Scholar]
- 45.Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I. A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell. 2001;13:1511–1526. doi: 10.1105/TPC.000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang BH, Busse JS, Dickey C, Rancour DM, Bednarek SY. The arabidopsis cell plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development. Plant Physiol. 2001;126:47–68. doi: 10.1104/pp.126.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peters C, Baars TL, Buhler S, Mayer A. Mutual control of membrane fission and fusion proteins. Cell. 2004;119:667–678. doi: 10.1016/j.cell.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 48.Yu X, Cai M. The yeast dynamin-related GTPase Vps1p functions in the organization of the actin cytoskeleton via interaction with Sla1p. J Cell Sci. 2004;117:3839–3853. doi: 10.1242/jcs.01239. [DOI] [PubMed] [Google Scholar]
- 49.Damke H, Baba T, Warnock DE, Schmid SL. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herskovits JS, Burgess CC, Obar RA, Vallee RB. Effects of mutant rat dynamin on endocytosis. J Cell Biol. 1993;122:565–578. doi: 10.1083/jcb.122.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Bliek AM, Meyerowitz EM. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature. 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- 53.Grigliatti TA, Hall L, Rosenbluth R, Suzuki DT. Temperature-sensitive mutations in Drosophila melanogaster. XIV. A selection of immobile adults. Mol Gen Genet. 1973;120:107–114. doi: 10.1007/BF00267238. [DOI] [PubMed] [Google Scholar]
- 54.Kosaka T, Ikeda K. Reversible blockage of membrane retrieval and endocytosis in the garland cell of the temperature-sensitive mutant of Drosophila melanogaster, shibirets1. J Cell Biol. 1983;97:499–507. doi: 10.1083/jcb.97.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- 56.Marks B, Stowell MH, Vallis Y, Mills IG, Gibson A, Hopkins CR, McMahon HT. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature. 2001;410:231–235. doi: 10.1038/35065645. [DOI] [PubMed] [Google Scholar]
- 57.McNiven MA. Dynamin: a molecular motor with pinchase action. Cell. 1998;94:151–154. doi: 10.1016/s0092-8674(00)81414-2. [DOI] [PubMed] [Google Scholar]
- 58.Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 59.Zheng J, Cahill SM, Lemmon MA, Fushman D, Schlessinger J, Cowburn D. Identification of the binding site for acidic phospholipids on the pH domain of dynamin: implications for stimulation of GTPase activity. J Mol Biol. 1996;255:14–21. doi: 10.1006/jmbi.1996.0002. [DOI] [PubMed] [Google Scholar]
- 60.Tuma PL, Stachniak MC, Collins CA. Activation of dynamin GTPase by acidic phospholipids and endogenous rat brain vesicles. J Biol Chem. 1993;268:17240–17246. [PubMed] [Google Scholar]
- 61.Gold ES, Underhill DM, Morrissette NS, Guo J, McNiven MA, Aderem A. Dynamin 2 is required for phagocytosis in macrophages. J Exp Med. 1999;190:1849–1856. doi: 10.1084/jem.190.12.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao H, Thompson HM, Krueger EW, McNiven MA. Disruption of Golgi structure and function in mammalian cells expressing a mutant dynamin. J Cell Sci. 2000;113:1993–2002. doi: 10.1242/jcs.113.11.1993. [DOI] [PubMed] [Google Scholar]
- 63.Cao H, Weller S, Orth JD, Chen J, Huang B, Chen JL, Stamnes M, McNiven MA. Actin and Arf1-dependent recruitment of a cortactin-dynamin complex to the Golgi regulates post-Golgi transport. Nat Cell Biol. 2005;7:483–492. doi: 10.1038/ncb1246. [DOI] [PubMed] [Google Scholar]
- 64.Jones SM, Howell KE, Henley JR, Cao H, McNiven MA. Role of dynamin in the formation of transport vesicles from the trans-Golgi network. Science. 1998;279:573–577. doi: 10.1126/science.279.5350.573. [DOI] [PubMed] [Google Scholar]
- 65.Lauvrak SU, Torgersen ML, Sandvig K. Efficient endosome-to-Golgi transport of Shiga toxin is dependent on dynamin and clathrin. J Cell Sci. 2004;117:2321–2331. doi: 10.1242/jcs.01081. [DOI] [PubMed] [Google Scholar]
- 66.Meier O, Greber UF. Adenovirus endocytosis. J Gene Med. 2004;6(Suppl. 1):S152–S163. doi: 10.1002/jgm.553. [DOI] [PubMed] [Google Scholar]
- 67.Schafer DA. Regulating actin dynamics at membranes: a focus on dynamin. Traffic. 2004;5:463–469. doi: 10.1111/j.1600-0854.2004.00199.x. [DOI] [PubMed] [Google Scholar]
- 68.Krueger EW, Orth JD, Cao H, McNiven MA. A dynamin-cortactin-Arp2/3 complex mediates actin reorganization in growth factor-stimulated cells. Mol Biol Cell. 2003;14:1085–1096. doi: 10.1091/mbc.E02-08-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hussain NK, Jenna S, Glogauer M, Quinn CC, Wasiak S, Guipponi M, Antonarakis SE, Kay BK, Stossel TP, Lamarche-Vane N, McPherson PS. Endocytic protein intersectin-1 regulates actin assembly via Cdc42 and N-WASP. Nat Cell Biol. 2001;3:927–932. doi: 10.1038/ncb1001-927. [DOI] [PubMed] [Google Scholar]
- 70.Salazar MA, Kwiatkowski AV, Pellegrini L, Cestra G, Butler MH, Rossman KL, Serna DM, Sondek J, Gertler FB, De Camilli P. Tuba, a novel protein containing bin/amphiphysin/Rvs and Dbl homology domains, links dynamin to regulation of the actin cytoskeleton. J Biol Chem. 2003;278:49031–49043. doi: 10.1074/jbc.M308104200. [DOI] [PubMed] [Google Scholar]
- 71.Erickson JW, Cerione RA. Multiple roles for Cdc42 in cell regulation. Curr Opin Cell Biol. 2001;13:153–157. doi: 10.1016/s0955-0674(00)00192-7. [DOI] [PubMed] [Google Scholar]
- 72.Drechsel DN, Hyman AA, Hall A, Glotzer M. A requirement for Rho and Cdc42 during cytokinesis in Xenopus embryos. Curr Biol. 1997;7:12–23. doi: 10.1016/s0960-9822(06)00023-6. [DOI] [PubMed] [Google Scholar]
- 73.Watanabe N, Higashida C. Formins: processive cappers of growing actin filaments. Exp Cell Res. 2004;301:16–22. doi: 10.1016/j.yexcr.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 74.Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14:461–469. doi: 10.1016/j.tcb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 75.Witke W, Podtelejnikov AV, Di Nardo A, Sutherland JD, Gurniak CB, Dotti C, Mann M. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 1998;17:967–976. doi: 10.1093/emboj/17.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dean SO, Rogers SL, Stuurman N, Vale RD, Spudich JA. Distinct pathways control recruitment and maintenance of myosin II at the cleavage furrow during cytokinesis. Proc Natl Acad Sci USA. 2005;102:13473–13478. doi: 10.1073/pnas.0506810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Severson AF, Baillie DL, Bowerman B. A Formin Homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr Biol. 2002;12:2066–2075. doi: 10.1016/s0960-9822(02)01355-6. [DOI] [PubMed] [Google Scholar]
- 78.Noda Y, Nakata T, Hirokawa N. Localization of dynamin. Widespread distribution in mature neurons and association with membranous organelles. Neuroscience. 1993;55:113–127. doi: 10.1016/0306-4522(93)90459-s. [DOI] [PubMed] [Google Scholar]
- 79.Thompson HM, Cao H, Chen J, Euteneuer U, McNiven MA. Dynamin 2 binds gamma-tubulin and participates in centrosome cohesion. Nat Cell Biol. 2004;6:335–342. doi: 10.1038/ncb1112. [DOI] [PubMed] [Google Scholar]
- 80.Pelissier A, Chauvin JP, Lecuit T. Trafficking through Rab11 endosomes is required for cellularization during Drosophila embryogenesis. Curr Biol. 2003;13:1848–1857. doi: 10.1016/j.cub.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 81.Clark SG, Shurland DL, Meyerowitz EM, Bargmann CI, van der Bliek AM. A dynamin GTPase mutation causes a rapid and reversible temperature-inducible locomotion defect in C. elegans. Proc Natl Acad Sci USA. 1997;94:10438–10443. doi: 10.1073/pnas.94.19.10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gu X, Verma DP. Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. EMBO J. 1996;15:695–704. [PMC free article] [PubMed] [Google Scholar]
- 83.Gu X, Verma DP. Dynamics of phragmoplastin in living cells during cell plate formation and uncoupling of cell elongation from the plane of cell division. Plant Cell. 1997;9:157–169. doi: 10.1105/tpc.9.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hong Z, Geisler-Lee CJ, Zhang Z, Verma DP. Phragmoplastin dynamics: multiple forms, microtubule association and their roles in cell plate formation in plants. Plant Mol Biol. 2003;53:297–312. doi: 10.1023/b:plan.0000006936.50532.3a. [DOI] [PubMed] [Google Scholar]
- 85.Lam BC, Sage TL, Bianchi F, Blumwald E. Regulation of ADL6 activity by its associated molecular network. Plant J. 2002;31:565–576. doi: 10.1046/j.1365-313x.2002.01377.x. [DOI] [PubMed] [Google Scholar]
- 86.Sever S, Damke H, Schmid SL. Garrotes, springs, ratchets, and whips: putting dynamin models to the test. Traffic. 2000;1:385–392. doi: 10.1034/j.1600-0854.2000.010503.x. [DOI] [PubMed] [Google Scholar]
- 87.Bednarek SY, Falbel TG. Membrane trafficking during plant cytokinesis. Traffic. 2002;3:621–629. doi: 10.1034/j.1600-0854.2002.30904.x. [DOI] [PubMed] [Google Scholar]
- 88.Kang BH, Rancour DM, Bednarek SY. The dynamin-like protein ADL1C is essential for plasma membrane maintenance during pollen matur-ation. Plant J. 2003;35:1–15. doi: 10.1046/j.1365-313x.2003.01775.x. [DOI] [PubMed] [Google Scholar]
- 89.Hong Z, Delauney AJ, Verma DP. A cell plate-specific callose synthase and its interaction with phragmoplastin. Plant Cell. 2001;13:755–768. doi: 10.1105/tpc.13.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McNiven MA, Kim L, Krueger EW, Orth JD, Cao H, Wong TW. Regulated interactions between dynamin and the actin-binding protein cortactin modulate cell shape. J Cell Biol. 2000;151:187–198. doi: 10.1083/jcb.151.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]