Previews
Virus replication requires lipid metabolism, but how lipids mediate virus infection remains obscure. In this issue, Amini-Bavil-Olyaee et al. (2013) reveal that IFITM proteins disturb cholesterol homeostasis to block virus entry. Previously in Cell, Morita and colleagues (2013) showed the antiviral potency of the lipid mediator protectin D1.
Viruses are obligate intracellular parasites and crucially depend on host cell machineries for efficient replication. Virus replication is a complex interplay between host and viral factors, in which cellular metabolic homeostasis can be disrupted to generate energy and macromolecular precursors for efficient virus production. For example, modulation of de novo fatty acid and lipid biosynthesis represents a common feature of virus replication (Chukkapalli et al., 2012). In particular, enveloped viruses consisting of a host derived lipid bilayer interfere with host cell lipid metabolism and hijack membrane lipid domains for biogenesis of their envelopes. Besides being mediators of virus replication and assembly/budding, host lipids in virus envelopes are also indispensable for structural integrity and functionality of virions. Enrichment of certain lipids in virus envelopes ensures maintenance of biophysical characteristics such as membrane fluidity (cholesterol) and membrane curvature (phopshatidylethanolamine; PE) which are important for membrane fusion during virus infection into host cells. Therefore, the enormous structural and functional diversities of lipids highlight their great potential as attractive targets for host-virus interactions at various life cycle stages. For example, tens of thousands of different lipid species are estimated to exist; aggregates of lipids can form plasma membrane microdomains and lipid droplets implicated in cell signaling, trafficking and storage, and single entities can act by way of inflammatory mediators such as platelet activating factor and eicosanoids) (Wenk, 2010).
In this issue, Amini-Bavil-Olyaee and colleagues (Amini-Bavil-Olyaee et al, 2013) unveil an intriguing antiviral mechanism of how perturbations in host cell cholesterol homeostasis block viral entry. They show that interferon-inducible transmembrane proteins (IFITMs) exert their antiviral activity by mediating the accumulation of cholesterol in late endosomal compartments. IFITMs are a group of interferon stimulated genes (ISGs) identified in many genome wide siRNA screens which block entry of a plethora of viruses along the endosomal trafficking pathway. A single-nucleotide polymorphism (SNP) of IFTIM3 correlated with increased morbidity in influenza-infected patients, demonstrating the biological significance of IFITM3 in restricting virus pathogenesis (Everitt et al., 2012; Zhang et al., 2013). Therefore, dissecting and understanding the fundamental mechanisms of how IFITMs mediate their antiviral activity is of substantial interest. Using a yeast two-hybrid assay, the authors first identified vesicle-associated membrane protein (VAMP)-associated protein A (VAPA) as cognate interaction partner for IFITM3. Although it is not obvious how the implicated transmembrane domain interactions between IFITM3 and VAPA could initially be detected by a yeast two-hybrid assay, the veracity of this IFITM3-VAPA interaction is borne out by a plethora of downstream specificity and functional experiments.
VAPA is implicated in endosomal cholesterol homeostasis through its interaction with oxysterol-binding protein (OSBP). The VAPA-OSBP complex mediates the transport of de novo synthesized cholesterol from the endoplasmic reticulum (ER) to other intracellular organelles. The authors further revealed that increasing IFITM3-VAPA interaction antagonized formation of VAPA-OSBP complexes. In combination with confocal microscopy, the authors concluded that IFITM3-mediated disruption of VAPA-OSBP interaction is the underlying cause of cholesterol accumulation in late endosmal compartments which consequently inhibits virus entry (Figure 1). Through a series of impressive electron microcopy experiments, the authors conclusively demonstrated the IFITM3 mediated antiviral activity of cholesterol accumulation on vesicular stomatitis virus (VSV) and influenza virus entry. Intriguingly, two independent studies recently reported that host cells evolved a sterol based antiviral strategy, whereby interferon induced production of the natural oxysterol 25-hydroxycholesterol (25HC) inhibits entry of enveloped viruses (Blanc et al., 2013; Liu et al., 2013). Considering evidence of 25HC mediated translocation of OSBP from the ER to the Golgi (Goto et al., 2012) (Figure 1), it will thus be interesting to decipher whether the IFITM3-VAPA mediated antiviral activity identified by Amini-Bavil-Olyaee and colleagues is part of a complex innate immune pathway interfering with cholesterol homeostasis to inhibit virus entry.
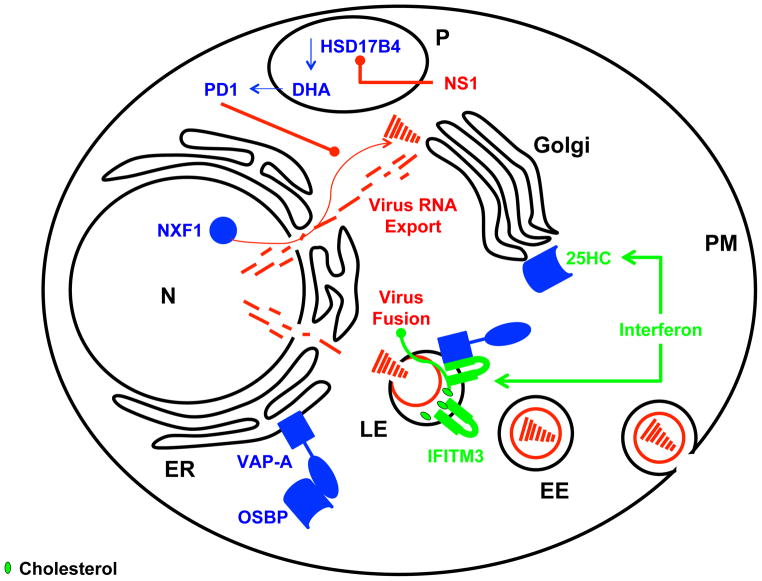
Figure 1.
The interferon response upregulates expression of IFITM proteins and increases the conversion of cholesterol to 25HC
IFITM3 expressed at late endosomal compartments (LE) interacts with vesicle-associated membrane protein (VAMP)-associated protein A (VAPA) disrupting the endoplasmic reticulum (ER) resident complex between oxysterol binding protein (OSBP) and VAPA. 25-hydroxycholesterol (25HC) mediates the translocation of OSBP to the Golgi and together, these intracellular changes lead to cholesterol accumulation in LE, blocking virus entry. The docosahexaenoic acid (DHA) derived lipid mediator protectin D1 (PD1) inhibits nuclear export of viral RNAs mediated by NXF1. Biosynthesis of DHA is crucially dependent on peroxisomal β-oxidation catalyzed by hydroxysteroid (17-beta) dehydrogenase 4 (HSD17B4). Influenza virus NS1 antagonizes metabolism of DHA derived lipid mediators by its inhibitory interaction with HSD17B4. Host factors, viral factors and interferon stimulated factors are colored in blue, red and green respectively; arrows indicate activation/stimulation or inhibition (rounded ends); plasma membrane (PM), early endosome (EE), late endosome (LE), endoplasmic reticulum (ER), nucleus (N) and peroxisome (P).
The findings by Amini-Bavil-Olyaee and colleagues were consistent with impaired intra-endosomal trafficking in cholesterol-laden late endosomal compartments and suggested that antiviral activity of IFITM3 is specific for viruses fusing and penetrating at late endosomal membranes. This observed specificity raises an interesting idea considering the heterogeneous distribution of lipids across intracellular membranes. For example, cholesterol is markedly enriched at the plasma membrane but its enrichment gradually decreases along the endosomal trafficking pathway. While Amini-Bavil-Olyaee and colleagues provide coherent evidence that virus fusion at late endosomal compartments is inhibited by cholesterol accumulation, previous studies implied that cholesterol stimulates entry of viruses such as human immunodeficiency virus (HIV) which fuse at the plasma membrane and/or at early endosomal compartments. Therefore, one could envision that different lipid requirements during virus entry correlate with the diverse entry pathways hijacked by several viruses.
A recent report in Cell (Morita et al., 2013) also underscores the biological significance of host cell lipid metabolites as modulators of virus infections. Harnessing a bioactive lipid screen, Morita and colleagues first observed a potent inhibition of influenza virus replication by the omega-3 polyunsaturated fatty acid (PUFA)-derived lipid mediator protectin D1 (PD1). They subsequently performed lipidomics analyses and quantified over 250 endogenous PUFA-derived lipid mediators in lungs of influenza virus infected mice, revealing a strong reduction of PD1 which correlated with virus pathogenicity. Remarkably, PD1 treatment protected against severe influenza in vivo, even 48 hours post infection when administrated in combination with peramivir. These findings collectively highlighted the therapeutic potential and antiviral activity of PD1 which is synthesized from docosahexaenoic acid (DHA) by the key enzyme arachidonate 15-lipoxygenase (ALOX15 or 12/15-LOX). Surprisingly, its antiviral activity on influenza virus replication was not due to its possible anti-inflammatory capacities. Instead, the authors elegantly discovered a novel PD1 mediated antiviral mechanism inhibiting nuclear export of virus RNAs. Their results showed that PD1 inhibited the direct interaction between viral RNAs and the mRNA exporter NXF1 which, in cooperation with the key docking protein Nup62, facilitates nuclear export of influenza virus RNAs (Figure 1). However, the precise inhibitory mechanism of PD1 and especially, how influenza virus interferes with PD1 production still remains unclear.
Considering the multifunctional nature of influenza virus non-structural protein 1 (NS1) in mediating virus replication, inhibiting host lipid metabolism and antagonizing the host immune response, it would be not be surprising if NS1 functionally intersects with DHA/PD1 metabolism. Peroxisomal β-oxidation catalyzed by hydroxysteroid (17-beta) dehydrogenase 4 (HSD17B4) is vital for the retroconversion step in DHA biosynthesis. Interestingly, a direct interaction between NS1 and HSD17B4 has been previously reported and overexpression of HSD17B4 inhibited influenza virus protein expression (Wolff et al., 1996), consistent with PD1 mediated impairment of nuclear export. These observations are further supported by recent siRNA screens identifying several enzymes involved in peroxisomal β-oxidation as antiviral. The possibility that influenza virus NS1 might interfere with the peroxisomal step of DHA/PD1 biosynthesis is reinforced by the identification of peroxisomes as initial sites for antiviral signaling (Dixit et al., 2010).
While Amini-Bavil-Olyaee and colleagues unveil a novel IFITM3-cholesterol based host defense mechanism against virus entry, Morita and colleagues discover PD1 as a potent inhibitor of influenza virus replication exhibiting great therapeutic potential. Together, these new studies highlight the increasing realization that sterol/lipid composition and metabolism play critical roles in host-virus interactions at different life-cycle stages, opening new and exciting future research avenues. Lipid involvement during viral infections currently is an emerging field which, with the recent advances in technology, holds great promise to unravel novel mechanisms and lipid factors involved in host-virus interactions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blanc M, Hsieh WY, Robertson KA, Kropp KA, Forster T, Shui G, Lacaze P, Watterson S, Griffiths SJ, Spann NJ, et al. The transcription factor STAT-1 couples macrophage synthesis of 25-hydroxycholesterol to the interferon antiviral response. Immunity. 2013;38:106–118. doi: 10.1016/j.immuni.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V, Heaton NS, Randall G. Lipids at the interface of virus-host interactions. Current opinion in microbiology. 2012;15:512–518. doi: 10.1016/j.mib.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto A, Liu X, Robinson CA, Ridgway ND. Multisite phosphorylation of oxysterol-binding protein regulates sterol binding and activation of sphingomyelin synthesis. Molecular biology of the cell. 2012;23:3624–3635. doi: 10.1091/mbc.E12-04-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SY, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK, Pernet O, Guo H, Nusbaum R, Zack JA, et al. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity. 2013;38:92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Kuba K, Ichikawa A, Nakayama M, Katahira J, Iwamoto R, Watanebe T, Sakabe S, Daidoji T, Nakamura S, et al. The Lipid Mediator Protectin D1 Inhibits Influenza Virus Replication and Improves Severe Influenza. Cell. 2013 doi: 10.1016/j.cell.2013.02.027. [DOI] [PubMed] [Google Scholar]
- Wenk MR. Lipidomics: new tools and applications. Cell. 2010;143:888–895. doi: 10.1016/j.cell.2010.11.033. [DOI] [PubMed] [Google Scholar]
- Wolff T, O’Neill RE, Palese P. Interaction cloning of NS1-I, a human protein that binds to the nonstructural NS1 proteins of influenza A and B viruses. Journal of virology. 1996;70:5363–5372. doi: 10.1128/jvi.70.8.5363-5372.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YH, Zhao Y, Li N, Peng YC, Giannoulatou E, Jin RH, Yan HP, Wu H, Liu JH, Liu N, et al. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nature communications. 2013;4:1418. doi: 10.1038/ncomms2433. [DOI] [PMC free article] [PubMed] [Google Scholar]