Abstract
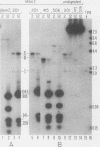
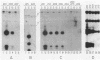
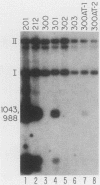
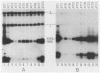
Herpes simplex virus (HSV) types 1 and 2 contain two classes of origins of DNA replication, oriS and oriL, which are closely related. A series of plasmids was constructed which contained specifically altered versions of the HSV type 2 oriS replication origin. Their ability to replicate in an in vivo replicon assay allowed a core origin of 75 base pairs (bp) to be defined. It included both arms of a 56-bp palindrome and from 13 to 20 bp of sequence leftward of the palindrome. The AT-rich sequence at the center of the palindrome was essential. Sequences on either side of the core origin enhanced replication. When additional copies of the -AT-dinucleotide were introduced progressively into the center of the palindrome, an oscillating effect on origin function was observed. These and other data implicate a linear rather than a cruciform conformation of the oriS palindrome in the initiation of HSV replication.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C. Transient gene expression control: effects of transfected DNA stability and trans-activation by viral early proteins. Mol Cell Biol. 1985 May;5(5):1034–1042. doi: 10.1128/mcb.5.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein J. A., Porter J. M., Challberg M. D. Template requirements for in vivo replication of adenovirus DNA. Mol Cell Biol. 1986 Jun;6(6):2115–2124. doi: 10.1128/mcb.6.6.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg M. D., Rawlins D. R. Template requirements for the initiation of adenovirus DNA replication. Proc Natl Acad Sci U S A. 1984 Jan;81(1):100–104. doi: 10.1073/pnas.81.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLucia A. L., Deb S., Partin K., Tegtmeyer P. Functional interactions of the simian virus 40 core origin of replication with flanking regulatory sequences. J Virol. 1986 Jan;57(1):138–144. doi: 10.1128/jvi.57.1.138-144.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb S., DeLucia A. L., Baur C. P., Koff A., Tegtmeyer P. Domain structure of the simian virus 40 core origin of replication. Mol Cell Biol. 1986 May;6(5):1663–1670. doi: 10.1128/mcb.6.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb S., DeLucia A. L., Koff A., Tsui S., Tegtmeyer P. The adenine-thymine domain of the simian virus 40 core origin directs DNA bending and coordinately regulates DNA replication. Mol Cell Biol. 1986 Dec;6(12):4578–4584. doi: 10.1128/mcb.6.12.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb S., Doelberg M. A 67-base-pair segment from the Ori-S region of herpes simplex virus type 1 encodes origin function. J Virol. 1988 Jul;62(7):2516–2519. doi: 10.1128/jvi.62.7.2516-2519.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias P., Lehman I. R. Interaction of origin binding protein with an origin of replication of herpes simplex virus 1. Proc Natl Acad Sci U S A. 1988 May;85(9):2959–2963. doi: 10.1073/pnas.85.9.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias P., O'Donnell M. E., Mocarski E. S., Lehman I. R. A DNA binding protein specific for an origin of replication of herpes simplex virus type 1. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6322–6326. doi: 10.1073/pnas.83.17.6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray C. P., Kaerner H. C. Sequence of the putative origin of replication in the UL region of herpes simplex virus type 1 ANG DNA. J Gen Virol. 1984 Dec;65(Pt 12):2109–2119. doi: 10.1099/0022-1317-65-12-2109. [DOI] [PubMed] [Google Scholar]
- Greaves D. R., Patient R. K., Lilley D. M. Facile cruciform formation by an (A-T)34 sequence from a Xenopus globin gene. J Mol Biol. 1985 Oct 5;185(3):461–478. doi: 10.1016/0022-2836(85)90064-6. [DOI] [PubMed] [Google Scholar]
- Guggenheimer R. A., Stillman B. W., Nagata K., Tamanoi F., Hurwitz J. DNA sequences required for the in vitro replication of adenovirus DNA. Proc Natl Acad Sci U S A. 1984 May;81(10):3069–3073. doi: 10.1073/pnas.81.10.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. Transition of a cloned d(AT)n-d(AT)n tract to a cruciform in vivo. Nucleic Acids Res. 1985 Jun 25;13(12):4343–4363. doi: 10.1093/nar/13.12.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Eghtedarzadeh M. K. Conserved arrangement of nested genes at the Drosophila Gart locus. Genetics. 1987 Dec;117(4):711–725. doi: 10.1093/genetics/117.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland L. E., Sandri-Goldin R. M., Goldin A. L., Glorioso J. C., Levine M. Transcriptional and genetic analyses of the herpes simplex virus type 1 genome: coordinates 0.29 to 0.45. J Virol. 1984 Mar;49(3):947–959. doi: 10.1128/jvi.49.3.947-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. A., Myers R. M., Tjian R. Mutational analysis of simian virus 40 large T antigen DNA binding sites. EMBO J. 1984 Dec 20;3(13):3247–3255. doi: 10.1002/j.1460-2075.1984.tb02286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinka M., Yaniv M. DNA replication origin of polyoma virus: early proximal boundary. J Virol. 1983 Jul;47(1):244–248. doi: 10.1128/jvi.47.1.244-248.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf C. W., Spies B., Kaerner H. C. The DNA replication origins of herpes simplex virus type 1 strain Angelotti. Nucleic Acids Res. 1986 Nov 11;14(21):8655–8667. doi: 10.1093/nar/14.21.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lally C., Dörper T., Gröger W., Antoine G., Winnacker E. L. A size analysis of the adenovirus replicon. EMBO J. 1984 Feb;3(2):333–337. doi: 10.1002/j.1460-2075.1984.tb01807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. J., Peden K. W., Dixon R. A., Kelly T. Functional organization of the simian virus 40 origin of DNA replication. Mol Cell Biol. 1986 Apr;6(4):1117–1128. doi: 10.1128/mcb.6.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshon D., Galloway D. A. Cloning and characterization of oriL2, a large palindromic DNA replication origin of herpes simplex virus type 2. J Virol. 1986 May;58(2):513–521. doi: 10.1128/jvi.58.2.513-521.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthman H., Nilsson M. G., Magnusson G. Non-contiguous segments of the polyoma genome required in cis for DNA replication. J Mol Biol. 1982 Nov 15;161(4):533–550. doi: 10.1016/0022-2836(82)90406-5. [DOI] [PubMed] [Google Scholar]
- Muller W. J., Mueller C. R., Mes A. M., Hassell J. A. Polyomavirus origin for DNA replication comprises multiple genetic elements. J Virol. 1983 Sep;47(3):586–599. doi: 10.1128/jvi.47.3.586-599.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie A. I., Lilley D. M. The mechanism of cruciform formation in supercoiled DNA: initial opening of central basepairs in salt-dependent extrusion. Nucleic Acids Res. 1987 Dec 10;15(23):9641–9654. doi: 10.1093/nar/15.23.9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie M. J., McGeoch D. J. DNA sequence analysis of an immediate-early gene region of the herpes simplex virus type 1 genome (map coordinates 0.950 to 0.978). J Gen Virol. 1982 Sep;62(Pt 1):1–15. doi: 10.1099/0022-1317-62-1-1. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Tjian R. Construction and analysis of simian virus 40 origins defective in tumor antigen binding and DNA replication. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6491–6495. doi: 10.1073/pnas.77.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polvino-Bodnar M., Orberg P. K., Schaffer P. A. Herpes simplex virus type 1 oriL is not required for virus replication or for the establishment and reactivation of latent infection in mice. J Virol. 1987 Nov;61(11):3528–3535. doi: 10.1128/jvi.61.11.3528-3535.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J. P., McGeoch D. J. DNA sequence of the region in the genome of herpes simplex virus type 1 containing the genes for DNA polymerase and the major DNA binding protein. Nucleic Acids Res. 1985 Nov 25;13(22):8143–8163. doi: 10.1093/nar/13.22.8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins D. R., Rosenfeld P. J., Wides R. J., Challberg M. D., Kelly T. J., Jr Structure and function of the adenovirus origin of replication. Cell. 1984 May;37(1):309–319. doi: 10.1016/0092-8674(84)90327-1. [DOI] [PubMed] [Google Scholar]
- Reisman D., Yates J., Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985 Aug;5(8):1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: analyses of cis-acting replication functions. Proc Natl Acad Sci U S A. 1985 Feb;82(3):694–698. doi: 10.1073/pnas.82.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B., Gerard R. D., Guggenheimer R. A., Gluzman Y. T antigen and template requirements for SV40 DNA replication in vitro. EMBO J. 1985 Nov;4(11):2933–2939. doi: 10.1002/j.1460-2075.1985.tb04026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D., Davison A. J. Identification of a varicella-zoster virus origin of DNA replication and its activation by herpes simplex virus type 1 gene products. J Gen Virol. 1986 Aug;67(Pt 8):1613–1623. doi: 10.1099/0022-1317-67-8-1613. [DOI] [PubMed] [Google Scholar]
- Stow N. D. Localization of an origin of DNA replication within the TRS/IRS repeated region of the herpes simplex virus type 1 genome. EMBO J. 1982;1(7):863–867. doi: 10.1002/j.1460-2075.1982.tb01261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D., McMonagle E. C. Characterization of the TRS/IRS origin of DNA replication of herpes simplex virus type 1. Virology. 1983 Oct 30;130(2):427–438. doi: 10.1016/0042-6822(83)90097-1. [DOI] [PubMed] [Google Scholar]
- Stow N. D. Mutagenesis of a herpes simplex virus origin of DNA replication and its effect on viral interference. J Gen Virol. 1985 Jan;66(Pt 1):31–42. doi: 10.1099/0022-1317-66-1-31. [DOI] [PubMed] [Google Scholar]
- Stow N. D., Wilkie N. M. An improved technique for obtaining enhanced infectivity with herpes simplex virus type 1 DNA. J Gen Virol. 1976 Dec;33(3):447–458. doi: 10.1099/0022-1317-33-3-447. [DOI] [PubMed] [Google Scholar]
- Strauss F., Gaillard C., Prunell A. Helical periodicity of DNA, Poly(dA) . poly(dT) and poly(dA-dT). poly(dA-dT) in solution. Eur J Biochem. 1981 Aug;118(2):215–222. doi: 10.1111/j.1432-1033.1981.tb06389.x. [DOI] [PubMed] [Google Scholar]
- Suggs J. W., Wagner R. W. Nuclease recognition of an alternating structure in a d(AT)14 plasmid insert. Nucleic Acids Res. 1986 May 12;14(9):3703–3716. doi: 10.1093/nar/14.9.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamanoi F., Stillman B. W. Initiation of adenovirus DNA replication in vitro requires a specific DNA sequence. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6446–6450. doi: 10.1073/pnas.80.21.6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triezenberg S. J., Folk W. R. Essential nucleotides in the polyomavirus origin region. J Virol. 1984 Aug;51(2):437–444. doi: 10.1128/jvi.51.2.437-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyndall C., La Mantia G., Thacker C. M., Favaloro J., Kamen R. A region of the polyoma virus genome between the replication origin and late protein coding sequences is required in cis for both early gene expression and viral DNA replication. Nucleic Acids Res. 1981 Dec 11;9(23):6231–6250. doi: 10.1093/nar/9.23.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlazny D. A., Frenkel N. Replication of herpes simplex virus DNA: localization of replication recognition signals within defective virus genomes. Proc Natl Acad Sci U S A. 1981 Feb;78(2):742–746. doi: 10.1073/pnas.78.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorlícková M., Kypr J. Conformational variability of poly(dA-dT).poly(dA-dT) and some other deoxyribonucleic acids includes a novel type of double helix. J Biomol Struct Dyn. 1985 Aug;3(1):67–83. doi: 10.1080/07391102.1985.10508399. [DOI] [PubMed] [Google Scholar]
- Wang K., Pearson G. D. Adenovirus sequences required for replication in vivo. Nucleic Acids Res. 1985 Jul 25;13(14):5173–5187. doi: 10.1093/nar/13.14.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson R. J., Vande Woude G. F. DNA sequence of an immediate-early gene (IEmRNA-5) of herpes simplex virus type I. Nucleic Acids Res. 1982 Feb 11;10(3):979–991. doi: 10.1093/nar/10.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller S. K., Spadaro A., Schaffer J. E., Murray A. W., Maxam A. M., Schaffer P. A. Cloning, sequencing, and functional analysis of oriL, a herpes simplex virus type 1 origin of DNA synthesis. Mol Cell Biol. 1985 May;5(5):930–942. doi: 10.1128/mcb.5.5.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitton J. L., Clements J. B. Replication origins and a sequence involved in coordinate induction of the immediate-early gene family are conserved in an intergenic region of herpes simplex virus. Nucleic Acids Res. 1984 Feb 24;12(4):2061–2079. doi: 10.1093/nar/12.4.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wides R. J., Challberg M. D., Rawlins D. R., Kelly T. J. Adenovirus origin of DNA replication: sequence requirements for replication in vitro. Mol Cell Biol. 1987 Feb;7(2):864–874. doi: 10.1128/mcb.7.2.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. A., Nelson N. J., McGeoch D. J., Challberg M. D. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J Virol. 1988 Feb;62(2):435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yates J., Warren N., Reisman D., Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers J., Schaffner W., Tyndall C., Lupton S., Kamen R. Polyoma virus DNA replication requires an enhancer. Nature. 1984 Nov 15;312(5991):242–246. doi: 10.1038/312242a0. [DOI] [PubMed] [Google Scholar]
- de Vries E., van Driel W., Tromp M., van Boom J., van der Vliet P. C. Adenovirus DNA replication in vitro: site-directed mutagenesis of the nuclear factor I binding site of the Ad2 origin. Nucleic Acids Res. 1985 Jul 11;13(13):4935–4952. doi: 10.1093/nar/13.13.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]